Fermentation, the microbial degradation of organic compounds without net oxidation, is an important process in the global carbon cycle and is also exploited worldwide for the production and preservation of food. It is one of the oldest food-processing technologies known, with some records dating back to 6,000 b.c. (50). The link between food and microbiology was laid by Pasteur, who found that yeasts were responsible for alcoholic fermentation (106). Since that discovery, scientific and industrial interests in food microbiology started to grow and continue to increase today. The number of food products that rely on fermentation in one or more steps of their production is tremendous. They form an important constituent of the daily diet and rank among the most innovative product categories in the food industry.
Most of the important microorganisms applied in the production of fermented foods have been studied for decades, yielding a wealth of information on their physiology and genetics in relation to product functionalities, such as the development of flavor, taste, and texture. The recent emergence of genomics has opened new avenues for the systematic analysis of microbial metabolism and the responses of microorganisms to their environment. Additionally, genomics has boosted research on important food microbes (22, 90, 93). Much of this research focuses on the performance of a single strain, including its interactions with the food matrix. However, food fermentations are typically carried out by mixed cultures consisting of multiple strains or species. Population dynamics play a crucial role in the performance of mixed-culture fermentations, and for many years, studies on mixed-culture food fermentations have focused on analyzing population dynamics using classical and molecular methods. Many of these studies are mainly descriptive, and relatively little is known about the mechanisms governing population dynamics in general and the molecular interactions that occur between the consortium members in particular. The availability of genome sequences for several species that are of industrial importance as well as technological advances in functional genomics enable new approaches to study food microbiology beyond the single species level and allow an integral analysis of the interactions and metabolic activity in mixed cultures.
Here we review the current knowledge on important food fermentation processes, focusing on the bacterial interactions. In addition, we illustrate how genomics approaches may contribute to the elucidation of the interaction networks between microbes, including interactions with the food environment. This information may find application in the industry through rational optimization and increased control over mixed-culture fermentations.
MIXED-CULTURE FOOD FERMENTATIONS—INDUSTRIAL PRACTICE AND CHALLENGES
Traditional fermentation processes relied on the transfer of knowledge and methodologies associated with manufacturing from generation to generation. The industrialization of food production together with the blossoming of microbiology in the middle of the 19th century led to the optimization and upscaling of many fermentation processes. Similarly, industrially produced starter cultures have emerged, leading to improved and reproducible product quality. Nowadays, the total economic value of fermented food products is huge, and the worldwide turnover of fermented fresh products in the dairy segment alone represents a total annual economic value of $54.2 billion (including yogurt [$34 billion], fermented dairy drinks [$4.3 billion], and fromage frais and quark [$7.4 billion]), whereas the cheese market is even larger ($74.4 billion) (111). In recent years, there has been massive product diversification, and many prebiotic and probiotic products with a high added value have emerged. Simultaneously, artisanal products have gained popularity due to their particular flavor and aroma characteristics (23).
At least two distinct product categories can be distinguished in which control of mixed-culture performance directly relates to the key challenges of innovators in the food industry. The first relates to the dairy market, which includes important products such as cheese and fermented milks. This market is characterized by the rapid growth of product varieties with distinct organoleptic properties. Examples include numerous applications in semihard cheeses, where adjunct cultures are added to introduce additional flavor notes (49, 114). Additionally, there is an increasing number of products appearing in response to current health trends, such as low-fat and low-salt product varieties (46). Here, it is important to develop such products while maintaining good organoleptic properties. In low-fat cheese, texture may be improved by the application of exopolysaccharide (EPS)-producing starter cultures (34). In fermented milks and yogurts containing probiotic microbes, off-flavor problems may appear due to undesired metabolic activities (99, 152). Furthermore, the success of the replacement or addition of the desired probiotic strain in mixed-culture fermentation may largely depend on the interaction of this strain with the other strains in the starter culture (64). In general, high numbers of viable probiotic bacteria are desired in these products at the moment of consumption. Typically between 5 and 8 logs CFU per gram of product is considered acceptable (99). Therefore, the growth, survival, and activity of the probiotic strain in the product environment are of key importance, and these traits are influenced both by specific environmental conditions (35, 152) and by interactions with the starter organisms (72, 99).
A second important product category is formed by food ingredients. Fermentation is widely applied to produce a broad range of ingredients, such as amino acids and organic acids. Some of these fermentations are carried out with mixed cultures (54, 155). Challenges in this area include improvement of productivity and stability and the elimination of unwanted by-products that interfere with downstream processing. Moreover, such processes may become economically more attractive if cheaper raw substrates can be used with new (combinations of) strains. An example here is the improved production of lactic acid from glucose by a mixed culture of Lactobacillus delbrueckii NRRL-B445 and Lactobacillus helveticus NRRL-B1937, of which the first is a good lactate producer and stimulated by the latter (86). Another example deals with a Saccharomyces cerevisiae strain that was engineered with l-arabinose utilization genes from Lactobacillus plantarum, allowing it to utilize the l-arabinose moiety of lignocellulosic fractions of plant-derived biomass (174).
Finally, we are seeing a rapid increase in the industrialization of non-Western fermented food products in Asia, Latin America, and Africa (2, 144). As a result of demographic changes in Europe, such products are also of increasing importance in the Western market, especially since some are believed to bring specific health benefits (48, 113). Examples include fermented products produced from dairy, cassava, cereals, beans, meat, and fish (7, 167). The challenges here relate to the stability, reproducibility, and productivity of fermentations.
MIXED CULTURES VERSUS PURE CULTURES— ECOLOGICAL CONSIDERATIONS
With few exceptions, food fermentations rely on mixed cultures of microorganisms. There are a number of important considerations that are at the basis of the ecological success of mixed cultures, and these will be discussed in this section. Microorganisms evolve to optimize their fitness, and this is often achieved by specialization, e.g., optimization of their metabolism. This is exemplified in a number of elegant experimental evolution studies with Escherichia coli in well-defined and homogeneous laboratory systems. In one study in a continuous culture sequentially fed with glucose and acetate, this organism differentiated into two ecotypes that displayed a large difference in lag phase when switching to growth on acetate after the depletion of glucose (142). Another example with E. coli is that a single strain cultured for a prolonged period with glucose limitation diverged into two or three clonal variants in which one variant ferments the glucose and the fermentation products, acetate and glycerol, serve as growth substrates for the other strains (65, 132).
Most substrates for food fermentations have a highly heterogeneous physicochemical composition which offers the possibility for the simultaneous occupation of multiple niches by “specialized” strains, for instance, through the utilization of different carbon sources. In these substrates, coexisting strains often interact through trophic or nutritional relations via multiple mechanisms, as will be discussed below.
Many food fermentations rely on spontaneous fermentation by the indigenous microbiota present in the food substrate. This implies that variations in the indigenous biota may affect the composition and activity of the fermenting community. This has a direct effect on product quality and the reproducibility of fermentations. A recent study showed large variations in the flavor and texture profiles of cheddar blocks produced at different factories (24). This is at least partially due to variations in proteolysis in the cheddar blocks. The application of starter cultures reduces the chance of unexpected population shifts and thereby ensures constant product characteristics and quality. Moreover, in combination with sterilization or pasteurization, it allows the food to be fermented by species or strains that would be outcompeted otherwise.
CLASSIFYING INTERACTIONS ON THE BASIS OF MUTUALLY BENEFICIAL AND DETRIMENTAL EFFECTS ON FITNESS
Microbial interactions in mixed cultures occur via multiple mechanisms. Such interactions may be direct, such as through physical contact, or via signaling molecules. Alternatively, indirect interactions may occur where changes in the physicochemical properties of the environment induced by one strain trigger a response in another strain (21, 51). The effects of such interactions on the fitness of the strains involved may either be positive, neutral, or negative. The mutual effects on fitness of interacting strains are an effective means of classifying interactions (68). These can be divided into five main classes—amensalism, competition, commensalism, parasitism, and mutualism—all of which will be discussed below and illustrated with relevant examples from food fermentations (Table 1).
TABLE 1.
Microbial (interspecies) interactions observed during food fermentations and in the fermented products
| Type of interaction and product or environment | Organisms involved | Reference(s) |
|---|---|---|
| Mutualism | ||
| Yogurt | S. thermophilus, L. delbrueckii subsp. bulgaricus | 1, 31-33, 43, 53, 66, 119, 140, 151, 154, 163, 179 |
| Dairy | LAB | 108, 153 |
| Cold milk | Lactococcus lactis subsp. cremoris, Pseudomonas fluorescens | 80 |
| Dairy, sourdough, laboratory medium | LAB, yeasts, fungi | 26, 108, 137 |
| Milk | LAB, yeasts | 52, 108 |
| Sourdough | Saccharomyces exiguous, Candida humilis, Lactobacillus sanfranciscensis | 39, 56, 57 |
| Surface-ripened cheese | Different species of molds, yeasts, and bacteria | 30 |
| Amensalism | ||
| Dairy, vegetable broth | LAB, Listeria monocytogenes, pseudomonads, Staphylococcus sp., Yersinia sp., Bacillus sp. | 9, 13, 14, 16, 71, 89, 94, 95, 102, 143 |
| Broth culture, laboratory medium, vegetable broth | LAB, Escherichia coli, Aspergillus spp., Enterobacter sp., Listeria monocytogenes, Vibrio sp., Salmonella sp. | 10, 11, 16, 25, 55, 63, 77, 92, 94, 95, 103, 115, 148, 159 |
| Wine | Lactobacillus hilgardii, Pediococcus pentosaceus | 94, 131 |
| Yogurt | S. thermophilus, L. delbrueckii subsp. bulgaricus | 71, 117, 127, 179 |
| Meat | LAB, Listeria monocytogenes | 13 |
| Wine | Malolactic bacteria, yeasts | 3 |
| Surface-ripened cheese | Lactobacillus plantarum, Listeria monocytogenes | 91 |
| Lettuce | LAB, Listeria monocytogenes | 4 |
| Commensalism | ||
| Dairy | LAB | 108, 153, 175 |
| Yogurt | S. thermophilus, L. delbrueckii subsp. bulgaricus, propionibacteria | 175 |
| Yogurt, dairy | LAB, propionibacteria | 28, 175 |
| Milk | LAB, yeasts | 52, 108 |
| Wine | Malolactic bacteria, yeasts | 3 |
| Surface-ripened cheese | LAB, Debaryomyces hansenii, Geotrichum candidum, Arthrobacter sp., Brevibacterium linens, Corynebacterium ammoniagenes, staphylococci | 107 |
| Laboratory medium | Yeasts, Bacterium linens | 124 |
| Fermented milks, yogurt, cheeses | Yeasts, bacteria | 165 |
| Competition | ||
| Yogurt | S. thermophilus, L. delbrueckii subsp. bulgaricus | 105, 179 |
| Dairy | LAB, yeasts | 52, 108 |
| Parasitism | ||
| Laboratory medium | Bacterium, phage | 29, 137 |
| Milk | Bacterium, phage | 20, 145 |
| Aquatic environments | Bacterium, phage | 169, 170 |
Amensalism is an interspecies interaction in which one organism adversely affects the other organism without being affected itself. It frequently occurs in food fermentations since the major end products of primary metabolism such as carboxylic acids and alcohols are effective growth inhibitors of indigenous microbiota and spoilage organisms (23, 89). In fact, metabolism by lactic acid bacteria (LAB) is optimized for fast acid production rather than efficient growth (158). Another example is the production of antimicrobial compounds, such as bacteriocins, that are produced by many food-fermenting LAB and that play an important role in mixed-culture population dynamics. Typically, bacteriocin-producing strains produce a dedicated immunity system that protects the host from detrimental effects. Lantibiotics, a special class of bacteriocins produced by LAB and other gram-positives, have drawn specific attention. Nisin is a well-known lantibiotic produced by Lactococcus lactis and broadly applied as a food preservative. Its activity is based on the permeabilization of the cytoplasmic membrane, leading to its depolarization (47, 70). Other potent bacteriocins include plantaricin and pediocin, which are widely distributed among L. plantarum and pediococci, respectively (41, 172). The broad activity spectrum of bacteriocins has been exploited for the inhibition of the outgrowth of spoilage microbes and pathogens (4, 91).
The second class of interactions is competition. Microorganisms compete for energy sources and nutrients during fermentation. Carbon sources are often present in high concentrations in food substrates, and competition therefore relates to the rapid uptake of nutrients and conversion into biomass. In dairy fermentations, nitrogen is limiting, and here organisms initially compete for the free amino acids and small peptides available in milk. In the later stages of fermentation, they compete for the peptides released by the actions of proteolytic enzymes. For this, they produce proteases, transport systems, and peptidases. Growth rate and population dynamics in mixed dairy fermentations are largely determined by the ability to utilize amino acids efficiently (73, 75). Micronutrients such as iron have also been reported to be limiting for strains in the biota of smear cheeses. Strains compete for iron pools through the use of specialized molecular systems for harvesting iron, including siderophores (109).
Commensalism is the third class of interactions. This is a situation in which one organism benefits from the interaction while the other strain is not affected. This also occurs in many food fermentations, for instance, through trophic interactions. In Swiss-type cheeses, propionic acid bacteria utilize the lactic acid produced by starter LAB (28). Similarly, in surface-ripened cheeses, lactic acid is consumed by yeasts, in particular Debaryomyces hansenii, and by the filamentous fungus Geotrichum candidum (107). This leads to the deacidification of the cheese surface, enabling the outgrowth of aerobic bacteria such as Arthrobacter species, Brevibacterium linens, Corynebacterium ammoniagenes, and staphylococci. One could argue that in this case, the aerobic bacteria benefit while D. hansenii and G. candidum are unaffected. However, it may be difficult to prove that there is no effect if it cannot be measured in terms of growth or survival. Another possible form of commensalism takes place in starters for Gouda cheese, where PrtP− L. lactis strains benefit from the peptides that are released from milk protein through the action of extracellular proteases (PrtP) produced by PrtP+ strains (69, 74) while the PrtP+ strains do not seem directly affected. In milk, PrtP+ strains produce more biomass than their isogenic PrtP− variants lacking plasmids containing the protease gene, but growth is slower due to the cost of expressing this protease (173). In pure cultures of PrtP+ strains grown in milk, PrtP− variants rapidly occur. The outcome of the long-term propagation of PrtP+ and PrtP− strains in a protein-containing medium like milk is that the strain that makes the least use of the resources in the medium, namely, the PrtP− strain, will become dominant. In this case, the immediate gain for the PrtP− strain is traded for the long-term community benefit. This particular example is also known as the “prisoner's dilemma” in evolutionary game theory (6, 120). The population dynamics of PrtP+ and PrtP− isolates are highly dependent on the growth conditions that influence the costs and benefits of the proteolytic phenotype (69).
The fourth class of interactions is known as parasitism. Parasitism is the interaction in which one species benefits at the expense of another. A well-known example of parasitism in the microbial world is represented by bacteriophages. It is well established that food fermentations, especially those repeatedly carried out with the same equipment, are highly vulnerable to phages. Phage attack may suddenly inactivate dominant strains in a fermenting culture, leading to failure and product losses in industrial fermentations (145). In recent years, our understanding of phage biology and the interactions of phages with their hosts has increased significantly. The biology of bacteriophages has been studied extensively for LAB such as L. lactis and Streptococcus thermophilus (20, 145). This work has benefited much from genome sequencing efforts, as, for instance, the genomes of at least seven phages specific for S. thermophilus have been sequenced (146). The diversity and fast evolution of phages typically result in the appearance of strains harboring different phage resistance phenotypes (170). Moreover, the recombination machinery of bacteriophages and their ability to transfer DNA from one bacterial cell to another may accelerate evolutionary processes in bacterial communities and contribute to the diversity in mixed-culture fermentation processes, especially when back slopping, the sequential transfer of cultures to fresh medium, is applied (169, 170). Recent studies showed a thus-far-unknown system present in archaea and bacteria, among which is S. thermophilus, that is involved in phage resistance (12, 38). In this clustered regularly interspaced short-palindromic-repeat system, bacteria acquire resistance to phages by incorporating phage-specific short transcribed nucleotide sequences into regions of clustered regularly interspaced short palindromic repeats. It was shown that these regions evolve very rapidly, probably driven by the rapid evolution of phages (161).
Finally, during mutualism, both participating microorganisms derive a benefit from the interaction. Many food fermentations rely on mutualistic interactions. Probably the best example is the yogurt consortium, consisting of the LAB S. thermophilus and Lactobacillus delbrueckii subsp. bulgaricus, which will be discussed in detail in the next section. The interactions among yogurt bacteria are also often referred to as synergism or protocooperation, interactions in which enhanced growth rate is the main mutual benefit. Cultures consisting of yeasts, LAB, and filamentous fungi are of key importance in a broad range of fermented foods in which mutualism is an important mode of interaction. For instance, in kefir granules S. cerevisiae raises the pH by utilizing the lactic acid produced by Lactobacillus kefiranofaciens as the carbon source enabling more growth of L. kefiranofaciens (26). In sourdough fermentation, there is a synergistic interaction between yeasts such as Saccharomyces exiguous or Candida humilis and LAB, especially Lactobacillus sanfranciscensis (56, 57). Yeast amylase releases maltose from starch which is fermented by L. sanfranciscensis. Part of the glucose derived from maltose is excreted by L. sanfranciscensis and is used as a carbon source by maltose-negative yeasts. In return, the yeasts stimulate the growth of L. sanfranciscensis by increasing the availability of amino acids and peptides, either through proteolysis or as a consequence of accelerated autolysis (39, 56). In wine, the interactions between yeasts and LAB also play a major role, and these have been reviewed recently by Alexandre et al. (3).
YOGURT CULTURES—THE MIXED-CULTURE PARADIGM IN FOOD FERMENTATION
Yogurt is the product of milk fermented by a defined mixed culture of two thermophilic LAB, Streptococcus thermophilus (15) and Lactobacillus delbrueckii subsp. bulgaricus (93, 163). There are also many fermented milk products that contain only one or neither of these strains, but in several countries, the name yogurt is only allowed for those products that are produced with cultures containing both (82, 151, 179). Nowadays, many yogurts and yogurt-based drinks are produced that contain probiotic strains, but these do not necessarily contribute to the fermentation. This mixed-culture fermentation is of huge economic importance. It represents an attractive model system for research on interactions due to its relatively small complexity. Some aspects of yogurt microbiology have already been reviewed elsewhere (23, 62, 151, 179). Here we present an updated review of a comparative analysis with other mixed-culture food fermentations.
Although S. thermophilus and L. delbrueckii subsp. bulgaricus are also able to ferment milk individually, both species were found to be stimulated in growth and acid production in mixed cultures compared to in single-strain cultures (119). Proteolysis plays an important role in yogurt as is illustrated by the growth of proteolytic S. thermophilus strains in milk (88). After inoculation, the cells start growing exponentially using the amino acids, dipeptides, tripeptides, and oligopeptides that are freely available. Subsequently, amino acids become limiting, and the culture enters into a nonexponential growth phase in which the synthesis of extracellular protease is initiated. Finally, in a second exponential phase, the proteolytic system is able to supply sufficient peptides for exponential growth, but here the growth rate is lower than in the first exponential phase, probably due to a limited capacity of the peptide uptake system (88).
Most commonly, yogurt cultures consist of proteolytic L. delbrueckii subsp. bulgaricus and nonproteolytic S. thermophilus (32, 119). During the first exponential phase of S. thermophilus, almost no growth of L. delbrueckii subsp. bulgaricus is observed. In the second phase, S. thermophilus growth decreases while L. delbrueckii subsp. bulgaricus starts to grow exponentially and protease expression is initiated. The cell-wall anchored protease PrtB mainly catalyzes the hydrolysis of the hydrophobic caseins into small peptides, which are subsequently taken up using various peptide transport systems (76, 118). In the cytoplasm, the peptides are further hydrolyzed into free amino acids by several endopeptidases and aminopeptidases (133, 135). The growth of L. delbrueckii subsp. bulgaricus continues in the third growth phase. At this stage, the peptides released from milk casein also serve as a source of amino acids for S. thermophilus, supporting a second exponential growth phase (31, 179).
Most S. thermophilus strains exhibit few amino acid auxotrophies, and the fact that they have fewer nutritional requirements than L. delbrueckii subsp. bulgaricus may explain their preferential growth in milk. Only histidine is required by most strains, and growth may be enhanced when methionine, proline, glutamic acid, and valine are added to growth media (67, 87). With few exceptions, S. thermophilus strains do not possess extracellular proteases (31, 67). As a result, the growth of most S. thermophilus strains, except some highly proteolytic strains, is strongly stimulated in cocultures with L. delbrueckii subsp. bulgaricus strains expressing prtB (31, 136).
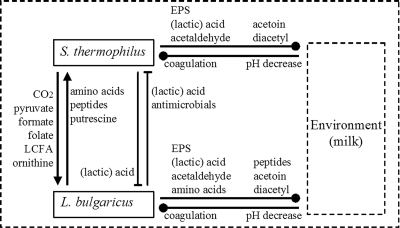
The mutualistic coexistence of S. thermophilus and L. delbrueckii subsp. bulgaricus is also based on other interactions, such as the exchange of several growth-stimulating factors (Fig. 1). S. thermophilus provides L. delbrueckii subsp. bulgaricus with formic acid, pyruvic acid, folic acid (33), and carbon dioxide (43). The positive effects of formic acid and folic acid on the growth of L. delbrueckii subsp. bulgaricus are related to the biosynthesis of purines (53). Formic acid is a precursor for purine synthesis, and L. delbrueckii subsp. bulgaricus lacks a pyruvate-formate lyase, which may explain why it relies on other sources for formate (32). Pyruvate-formate lyase is a highly abundant protein in S. thermophilus grown in milk, indicating that S. thermophilus may supply L. delbrueckii subsp. bulgaricus with formate during cocultivation (37). Folic acid is involved as a cofactor in purine and amino acid biosynthesis (149, 168) and was shown to be excreted by S. thermophilus and consumed by L. delbrueckii subsp. bulgaricus (33). Genome sequence analysis of L. delbrueckii subsp. bulgaricus strain ATCC 11842 has shown the absence of a biosynthetic pathway for para-aminobenzoic acid, and therefore, the biosynthetic pathway for folate in this strain is incomplete (163). S. thermophilus is capable of producing both para-aminobenzoic acid and folate, and hence, L. delbrueckii subsp. bulgaricus may benefit from elevated levels of either compound. A recent report on L. lactis shows the involvement of folate in the stimulation of a proteinase-positive strain by a proteinase-negative strain (121). Carbon dioxide is a precursor for the synthesis of aspartate (128, 166), glutamate (100), arginine, and nucleotides (17). In heat-treated milk, carbon dioxide levels may be too low for L. delbrueckii subsp. bulgaricus (43), and therefore, it profits from the carbon dioxide released by S. thermophilus from the urea that is present in milk. In addition, the urea catabolism plays a role in the synthesis of aspartate and glutamine, both essential amino acids (5, 104).
FIG. 1.
Schematic representation of the validated and hypothesized interactions that occur between Streptococcus thermophilus, Lactobacillus delbrueckii subsp. bulgaricus, their environment, and the compounds relevant for yogurt characteristics. ▾, positive interactions; ⊥, negative interactions; •, interactions that do not specifically promote or decrease the growth of the other species. LCFA, long-chain fatty acids. See text for references.
Yet other compounds may contribute to the mutualistic interaction between the yogurt LAB. For instance, Partanen et al. (116) reported that several long-chain fatty acids are stimulatory to L. delbrueckii subsp. bulgaricus. This is probably due to the fact that L. delbrueckii subsp. bulgaricus lacks part of the biosynthetic machinery required for de novo synthesis of long-chain unsaturated fatty acids, and one may speculate that S. thermophilus is also able to supply L. delbrueckii subsp. bulgaricus with long-chain fatty acids. In the recent paper describing the genome sequence of L. delbrueckii subsp. bulgaricus ATCC 11842, Van de Guchte et al. hypothesized that ornithine and putrescine may be produced by S. thermophilus and L. delbrueckii subsp. bulgaricus, respectively, and that the exchange of these metabolites mutually increases their resistance to oxidative stress (130, 163). Ornithine is involved in the metabolism of urea, and putrescine turns S-adenosyl methionine into spermine via the intermediate spermidine. Spermine and spermidine are involved in the stabilization of DNA and DNA replication, respectively.
Growth-detrimental interactions have also been reported. Reddy and Shahani (127) reported that some strains of L. delbrueckii subsp. bulgaricus produce the bacteriocin bulgarican that inhibits the growth of S. thermophilus (117). Moreover, some S. thermophilus strains were reported to produce peptide bacteriocins (71). Of the seven strains of L. delbrueckii subsp. bulgaricus tested, one was inhibited by this peptide.
A key sensory attribute that is introduced with fermentation during yogurt production is its texture. Due to acidification by LAB, proteins coagulate and thereby change the viscosity of the milk. Furthermore, the bacteria, and mainly S. thermophilus, produce EPS that form a matrix with the milk proteins, resulting in the final yogurt structure. In kefir, kefiran production by LAB is stimulated by the presence of yeasts induced via direct physical contact (26). Similarly, the interactions between the yogurt strains may influence EPS production, for instance, by increasing the availability of nitrogen sources or by interacting with non-EPS-producing S. thermophilus strains (40, 177). In general, the associations of yeasts and LAB are of key importance for a broad range of fermented foods. However, surprisingly, little information is available about molecular-interaction mechanisms (165).
QUORUM SENSING AND PHYSICAL INTERACTIONS
Besides the interactions mentioned so far, microorganisms may produce diffusible chemicals for the purpose of communication. This includes a process referred to as quorum sensing (QS) that is widely spread among gram-negatives and gram-positives and allows regulation at the population level of a wide range of traits, including competence, virulence, and stress responses (58, 78, 150). Two recent reviews link QS to motility, EPS production, biofilm formation, and toxin production, which are all important phenotypes in food fermentation (44, 59). As described previously, lantibiotics may play an important role in mixed-culture population dynamics, and their production is often regulated in a density-dependent way via QS (125, 129). Nisin production in L. lactis has been studied extensively, and it has been shown that it acts as an autoinducer. It regulates its own production at the transcriptional level with the involvement of a two-component regulatory system (for a review, see reference 83). More recently, the involvement of QS in the regulation of EPS production and monospecies biofilm formation by L. plantarum and Lactobacillus rhamnosus was reported (85, 147).
Whereas intraspecies communication is quite common, fewer examples exist of interspecies communication, and most examples are not related to food microbiology (78). An example of chemical communication has been observed in dental biofilms where Veillonella atypica and Streptococcus gordonii degrade complex carbohydrates in a way that benefits both species. V. atypica produces a yet-to-be-identified signal molecule that induces amylase production in S. gordonii and thereby increases the degradation rate (45). A QS system that is present in a major fraction of the bacterial population and allows interspecies signaling is the luxS system. The LuxS enzyme is responsible for the production of a precursor of autoinducer 2, a signaling molecule involved in the regulation of gene expression of, for instance, virulence factors, competence for genetic transformation, the production of antibiotics and secondary metabolites, and biofilm formation (36, 176). In cocultures of the hyperthermophiles Thermotoga maritima and Methanococcus jannaschii, growth and EPS production of the former was stimulated by the presence of the latter. Additionally, a QS signaling peptide in T. maritima was upregulated. It was shown that EPS expression was enhanced in the presence of this signaling peptide (72). Similar processes may also play an important role in biofilm formation in mixed-culture food fermentations.
An example of interspecies communication via QS among LAB is represented by L. plantarum NC8, in which not only plantaricin itself but also plantaricin-like peptides produced by other gram-positive bacteria were shown to induce the production of plantaricin by L. plantarum (96, 97).
Microbes may also interact with and influence each others' metabolism via physical contact, and a few examples of such interactions have been described for mixed-culture food fermentations. Cheirsilp et al. demonstrated that the production of the capsular EPS kefiran was enhanced by physical contact between Lactobacillus kefiranofaciens and S. cerevisiae (26). The molecular basis of this effect remains to be established. It was postulated that the bacteria and yeasts may benefit from the enhanced kefiran production through interactions that occur in the kefir granules where interspecies contact or the exchange of growth factors is facilitated through physical contact.
GENOMICS APPROACHES FOR MIXED- CULTURE RESEARCH
The genomic revolution has opened new avenues for research on mixed cultures, for an increasing number of relevant genome sequences for LAB are available (90, 93). In several genomes for LAB, among which L. plantarum (84), S. thermophilus (15), and Lactobacillus salivarius subsp. salivarius (27) are included, there is evidence for horizontal gene transfer, and interestingly, the acquired sequences appear in many cases to originate from species that frequently coexist and interact. In the genome of L. delbrueckii subsp. bulgaricus ATCC 11842, a region with a GC content of 38% and carrying an operon encoding an ABC transporter that could serve as an uptake transporter for putrescine and/or spermidine was found and proposed to be involved in the interactions between S. thermophilus and L. delbrueckii subsp. bulgaricus (163). This GC content is significantly lower than the average value of 49.7% observed for the entire genome. Interestingly, this locus has only been found in a few bacterial genomes, whereas it is not even present in the genome of another L. delbrueckii subsp. bulgaricus strain, ATCC BAA-365 (93), indicating that it may have been acquired recently via horizontal gene transfer (163).
Only a limited number of studies is available where genomics approaches are used to study the interactions in mixed-culture food fermentations. Such studies pose at least two technical challenges that need to be addressed. The first relates to the complexity of most food fermentation substrates. The physicochemical composition and especially the high protein or fat content may interfere with experimental procedures for RNA and protein isolation that work well with laboratory media. A number of studies have appeared that describe successful transcriptome or proteome analyses on samples from fermented substrates and even from highly complex materials such as fecal samples (81, 178). Most studies of dairy fermentations use skim milk, in which the precipitation of casein may be prevented by pretreatment with sodium citrate (126, 139). These studies have revealed several previously undescribed metabolic adaptations upon growth in milk, such as the induction of pyruvate-formate lyase in S. thermophilus that may serve as the supply of formate required for the biosynthesis of purine bases or other anabolic processes (37) (Fig. 1). A recent study deals with the transcriptome analysis of L. lactis grown in milk in coculture with S. cerevisiae (98). Although no difference in growth was observed between the coculture and a L. lactis mono culture, a number of genes were differentially expressed, in particular genes involved in pyrimidine metabolism. Several other regulatory responses could be assigned to the ethanol produced by the yeast.
The potential of using functional genomics approaches for analyzing interactions is well illustrated by recent studies describing the genome-wide analysis of interactions of commensal or pathogenic microbes with their hosts. With the availability of microarray platforms for several plant species and soil bacteria, it is possible to elucidate the responses of both the host and the microorganism upon interaction as exemplified by the induction of defense proteins in Arabidopsis thaliana by Agrobacterium tumefaciens (42). Host-microbe interactions in the gastrointestinal tract are of crucial importance for human health. The overwhelming complexity of this system with respect to composition and activity of the microbiota as well as its heterogeneity and poor accessibility to sampling requires inventive research approaches. The use of germfree animal models has greatly facilitated analysis of the genome-wide responses of microorganisms as well as hosts upon colonization (18, 134). Other studies report the in vivo time and spatial resolution of the expression of genes specifically expressed in the gastrointestinal tract in the model probiotic L. plantarum (19). Recently, Sonnenburg et al. have extended these approaches in a study where they cocolonized germfree mice with Bacteroides thetaiotaomicron, a prominent component of the adult gut microbiota, and Bifidobacterium longum, a frequently used probiotic microorganism. The results showed that cocolonization prompted B. thetaiotaomicron to increase the expression of genes involved in the acquisition and metabolism of polysaccharides (141).
Most of the studies described above relate to the performance of the interactions of microbes in simplified model systems or defined systems composed of a limited microbial complexity. Recent advances in the field of metagenomics provide a radically new approach for very complex ecosystems as well as for ecosystems dominated by a moderate number of species and strains (162, 164). Random sequencing of environmental samples supplies information on the (amount of) species present in an environment, including uncultured microorganisms, as well as information on known and previously unknown genes that occur in that environment. By comparing habitat-specific fingerprints of genes present in various known environments, it is possible to interpret other environments. Tringe et al. (160) clustered similar environmental samples together and found only a few genes specific for a certain environment. Based on relative abundance, it was clear that systems for the transport of ions and inorganic compounds, energy production, and (interspecies) communication were most discriminative between samples from the Sargasso Sea, deep-sea whale falls, and farm soil. Currently, various groups are sequencing the metagenomes of gut and oral microbiota (http://www.genomesonline.org/gold.cgi?want=Metagenomes), which will undoubtedly boost research on dynamics and interactions within these complex microbial populations and support the development of prebiotics and probiotics.
DESCRIPTIVE AND PREDICTIVE MODELING IN THE GENOMICS ERA
Modeling has played an important role in food microbiology, and an extensive review describing different modeling categories has been published recently (156). Historically, most of these studies aim at developing predictive models for the growth of desired or undesired microorganisms in the food matrix. As the demand for minimally processed foods increases, the risk of the outgrowth of spoilage or pathogenic microbes rises. Accurate empirical models are of great value as they assist in the definition of processing conditions minimizing the risk of growth of these bacteria such as Bacillus cereus, lactobacilli, or E. coli (95, 101, 122). Similarly, Sodini et al. used black-box modeling for predicting the acidification of mixed cultures of S. thermophilus and L. delbrueckii subsp. bulgaricus and quantifying the interactions between the two species (140). In another example, growth dynamics in a mixed yeast culture of killer and sensitive strains was reported (123). Here, the lethal action of the killer strain showed a lag phase probably due to the necessary accumulation of the toxin before it reached a lethal dose. The existing model was adapted for this effect.
Despite the value of such models in process optimization, their predictive value is often limited to specific substrates and conditions, and they do not provide additional mechanistic insights such as interaction effects. Other modeling strategies aim at predicting the performances of microbes in fermentations on the basis of their metabolic pathways and networks. Such “white-box” or mechanistic models have been successfully applied for the optimization of industrial fermentations, including food ingredients such as lactic acid and amino acids (8, 138). To our knowledge, there are no examples of the integration of interaction effects in such models to the level where they can be used to predict the performance of mixed-culture fermentations. However, Gregory et al. developed a computing system that allows the modeling of interactions and evolution in bacterial communities (60, 61). This model includes several aspects, such as growth stimulatory interactions, antibiotic sensitivity, the occurrence of antibiotic-resistant mutants, and growth on nutrients derived from killed cells, in one model.
With the emergence of genomics, a radically different modeling approach has drawn increasing attention (reference 156 and references therein). Here, genome-scale metabolic models are constructed that allow a systematic exploration of metabolic capacities, and a number of such models have appeared in recent years for important microbes in mixed-culture food fermentations like L. plantarum and L. lactis (112, 157). These may serve as references for future metabolic models, thereby accelerating the process of model construction (110). A genome-scale metabolic model and associated constraint-based modeling techniques were used to analyze the physiology of the growth of L. plantarum in a complex medium revealing the importance of amino acid catabolic pathways previously not associated with free-energy metabolism (158). With respect to mixed-culture fermentations, it will be interesting to see whether it is possible to connect genome-scale metabolic models of the individual components of mixed cultures through a limited number of interactions. Such multigenome scale models should be effective tools for the optimization of mixed-culture performance with respect to growth and metabolite production.
CONCLUSIONS AND FUTURE PROSPECTS
Mixed-culture food fermentations are of primary economic importance. The performance of such cultures, consisting of LAB, yeasts, and/or filamentous fungi, is not the simple result of “adding up” the individual single-strain functionalities but is largely determined by interactions at the level of substrates, the exchange of metabolites and growth factors or inhibiting compounds.
Technological breakthroughs in the postgenomic era open up new avenues to study microbial communities and interaction networks beyond simple descriptive models. These are now mainly applied to ecological studies of highly complex systems, such as the gastrointestinal tract or complex environmental ecosystems. On the other hand, studies aiming at understanding the more fundamental ecological principles underlying the success of evolutionary strategies typically make use of more artificial laboratory strains and ecosystems (79, 120). Food fermentations may provide a valuable alternative model with a high practical relevance. They typically have moderate microbial complexity and offer excellent possibilities for process control. Moreover, the availability of advanced genomics and genetic tools will allow the integration of mechanistic and evolutionary approaches (171).
Footnotes
Published ahead of print on 20 June 2008.
REFERENCES
- 1.Accolas, J. P., M. Veaux, and J. Auclaur. 1971. Etude des interactions entre diverses bactéries lactiques thermophiles et mésophiles, en relation avec la fabrication des fromages à pâté cuite. Lait 57:1-23. [Google Scholar]
- 2.Aidoo, K. E., M. J. Nout, and P. K. Sarkar. 2006. Occurrence and function of yeasts in Asian indigenous fermented foods. FEMS Yeast Res. 6:30-39. [DOI] [PubMed] [Google Scholar]
- 3.Alexandre, H., P. J. Costello, F. Remize, J. Guzzo, and M. Guilloux-Benatier. 2004. Saccharomyces cerevisiae-Oenococcus oeni interactions in wine: current knowledge and perspectives. Int. J. Food Microbiol. 93:141-154. [DOI] [PubMed] [Google Scholar]
- 4.Allende, A., B. Martinez, V. Selma, M. I. Gil, J. E. Suarez, and A. Rodriguez. 2007. Growth and bacteriocin production by lactic acid bacteria in vegetable broth and their effectiveness at reducing Listeria monocytogenes in vitro and in fresh-cut lettuce. Food Microbiol. 24:759-766. [DOI] [PubMed] [Google Scholar]
- 5.Arioli, S., C. Monnet, S. Guglielmetti, C. Parini, I. De Noni, J. Hogenboom, P. M. Halami, and D. Mora. 2007. Aspartate biosynthesis is essential for the growth of Streptococcus thermophilus in milk, and aspartate availability modulates the level of urease activity. Appl. Environ. Microbiol. 73:5789-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axelrod, R., and W. D. Hamilton. 1981. The evolution of cooperation. Science 211:1390-1396. [DOI] [PubMed] [Google Scholar]
- 7.Azokpota, P., D. J. Hounhouigan, and M. C. Nago. 2006. Microbiological and chemical changes during the fermentation of African locust bean (Parkia biglobosa) to produce afitin, iru and sonru, three traditional condiments produced in Benin. Int. J. Food Microbiol. 107:304-309. [DOI] [PubMed] [Google Scholar]
- 8.Bai, D. M., X. M. Zhao, X. G. Li, and S. M. Xu. 2004. Strain improvement and metabolic flux analysis in the wild-type and a mutant Lactobacillus lactis strain for l(+)-lactic acid production. Biotechnol. Bioeng. 88:681-689. [DOI] [PubMed] [Google Scholar]
- 9.Balasubramanyam, B. V., and M. C. Varadaraj. 1998. Cultural conditions for the production of bacteriocin by a native isolate of Lactobacillus delbruecki ssp. bulgaricus CFR 2028 in milk medium. J. Appl. Microbiol. 84:97-102. [DOI] [PubMed] [Google Scholar]
- 10.Barefoot, S. F., and T. R. Klaenhammer. 1983. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl. Environ. Microbiol. 45:1808-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barefoot, S. F., and T. R. Klaenhammer. 1984. Purification and characterization of the Lactobacillus acidophilus bacteriocin lactacin B. Antimicrob. Agents Chemother. 26:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 13.Benkerroum, N., A. Daoudi, T. Hamraoui, H. Ghalfi, C. Thiry, M. Duroy, P. Evrart, D. Roblain, and P. Thonart. 2005. Lyophilized preparations of bacteriocinogenic Lactobacillus curvatus and Lactococcus lactis subsp. lactis as potential protective adjuncts to control Listeria monocytogenes in dry-fermented sausages. J. Appl. Microbiol. 98:56-63. [DOI] [PubMed] [Google Scholar]
- 14.Benkerroum, N., H. Oubel, and L. B. Mimoun. 2002. Behavior of Listeria monocytogenes and Staphylococcus aureus in yogurt fermented with a bacteriocin-producing thermophilic starter. J. Food Prot. 65:799-805. [DOI] [PubMed] [Google Scholar]
- 15.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breidt, F., and H. P. Fleming. 1998. Modeling of the competitive growth of Listeria monocytogenes and Lactococcus lactis in vegetable broth. Appl. Environ. Microbiol. 64:3159-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bringel, F., and J.-C. Hubert. 2003. Extent of genetic lesions of the arginine and pyrimidine biosynthetic pathways in Lactobacillus plantarum, L. paraplantarum, L. pentosus, and L. casei: prevalence of CO2-dependent auxotrophs and characterization of deficient arg genes in L. plantarum. Appl. Environ. Microbiol. 69:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brosch, R., A. S. Pym, S. V. Gordon, and S. T. Cole. 2001. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 9:452-458. [DOI] [PubMed] [Google Scholar]
- 20.Brussow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 21.Bull, A. T., and J. H. Slater. 1982. Microbial interactions and communities. Academic Press, London, United Kingdom.
- 22.Canchaya, C., M. J. Claesson, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology 152:3185-3196. [DOI] [PubMed] [Google Scholar]
- 23.Caplice, E., and G. F. Fitzgerald. 1999. Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol. 50:131-149. [DOI] [PubMed] [Google Scholar]
- 24.Carunchia Whetstine, M. E., P. J. Luck, M. A. Drake, E. A. Foegeding, P. D. Gerard, and D. M. Barbano. 2007. Characterization of flavor and texture development within large (291 kg) blocks of cheddar cheese. J. Dairy Sci. 90:3091-3109. [DOI] [PubMed] [Google Scholar]
- 25.Chapuis, C., and J. P. Flandrois. 1994. Mathematical model of the interactions between Micrococcus spp. and Pseudomonas aeruginosa on agar surface. J. Appl. Bacteriol. 77:727-732. [DOI] [PubMed] [Google Scholar]
- 26.Cheirsilp, B., H. Shoji, H. Shimizu, and S. Shioya. 2003. Interactions between Lactobacillus kefiranofaciens and Saccharomyces cerevisiae in mixed culture for kefiran production. J. Biosci. Bioeng. 96:279-284. [DOI] [PubMed] [Google Scholar]
- 27.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeno-Tarraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 103:6718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Codon, S., T. M. Cogan, P. Piveteau, J. O'Callaghan, and B. Lyons. 2001. Stimulation of propionic acid bacteria by lactic acid bacteria in cheese manufacture. Irish Agriculture and Food Development Authority, Cork, Ireland.
- 29.Contois, D. E., and L. D. Yango. 1964. Studies of steady-state, mixed microbial populations, abstr. Q17. Abstr. 148th Meet. Am. Chem. Soc. American Chemical Society, Washington, DC.
- 30.Corsetti, A., J. Rossi, and M. Gobbetti. 2001. Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int. J. Food Microbiol. 69:1-10. [DOI] [PubMed] [Google Scholar]
- 31.Courtin, P., V. Monnet, and F. Rul. 2002. Cell-wall proteinases PrtS and PrtB have a different role in Streptococcus thermophilus/Lactobacillus bulgaricus mixed cultures in milk. Microbiology 148:3413-3421. [DOI] [PubMed] [Google Scholar]
- 32.Courtin, P., and F. Rul. 2004. Interactions between microorganisms in a simple ecosystem: yogurt bacteria as a study model. Lait 84:125-134. [Google Scholar]
- 33.Crittenden, R. G., N. R. Martinez, and M. J. Playne. 2003. Synthesis and utilisation of folate by yoghurt starter cultures and probiotic bacteria. Int. J. Food Microbiol. 80:217-222. [DOI] [PubMed] [Google Scholar]
- 34.Dabour, N., E. Kheadr, N. Benhamou, I. Fliss, and G. LaPointe. 2006. Improvement of texture and structure of reduced-fat cheddar cheese by exopolysaccharide-producing lactococci. J. Dairy Sci. 89:95-110. [DOI] [PubMed] [Google Scholar]
- 35.Dave, R. I., and N. P. Shah. 1998. Ingredient supplementation effects on viability of probiotic bacteria in yogurt. J. Dairy Sci. 81:2804-2816. [DOI] [PubMed] [Google Scholar]
- 36.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 37.Derzelle, S., A. Bolotin, M. Y. Mistou, and F. Rul. 2005. Proteome analysis of Streptococcus thermophilus grown in milk reveals pyruvate formate-lyase as the major upregulated protein. Appl. Environ. Microbiol. 71:8597-8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deveau, H., R. Barrangou, J. E. Garneau, J. Labonté, C. Fremaux, P. Boyaval, D. A. Romero, P. Horvath, and S. Moineau. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Vuyst, L., and P. Neysens. 2005. The sourdough microflora: biodiversity and metabolic interactions. Trends Food Sci. Technol. 16:43-56. [Google Scholar]
- 40.De Vuyst, L., F. Vanderveken, S. Van de Ven, and B. Degeest. 1998. Production by and isolation of exopolysaccharides from Streptococcus thermophilus grown in a milk medium and evidence for their growth-associated biosynthesis. J. Appl. Microbiol. 84:1059-1068. [DOI] [PubMed] [Google Scholar]
- 41.Diep, D. B., L. Godager, D. Brede, and I. F. Nes. 2006. Data mining and characterization of a novel pediocin-like bacteriocin system from the genome of Pediococcus pentosaceus ATCC 25745. Microbiology 152:1649-1659. [DOI] [PubMed] [Google Scholar]
- 42.Ditt, R. F., K. F. Kerr, P. de Figueiredo, J. Delrow, L. Comai, and E. W. Nester. 2006. The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant-Microbe Interact. 19:665-681. [DOI] [PubMed] [Google Scholar]
- 43.Driessen, F. M., F. Kingma, and J. Stadhouders. 1982. Evidence that Lactobacillus bulgaricus in yogurt is stimulated by carbon dioxide produced by Streptococcus thermophilus. Neth. Milk Dairy J. 36:135-144. [Google Scholar]
- 44.Dunn, A. K., and E. V. Stabb. 2007. Beyond quorum sensing: the complexities of prokaryotic parliamentary procedures. Anal. Bioanal. Chem. 387:391-398. [DOI] [PubMed] [Google Scholar]
- 45.Egland, P. G., R. J. Palmer, Jr., and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 101:16917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eiben, G., C. S. Andersson, E. Rothenberg, V. Sundh, B. Steen, and L. Lissner. 2004. Secular trends in diet among elderly Swedes—cohort comparisons over three decades. Public Health Nutr. 7:637-644. [DOI] [PubMed] [Google Scholar]
- 47.Entian, K. D., and W. M. de Vos. 1996. Genetics of subtilin and nisin biosyntheses: biosynthesis of lantibiotics. Antonie van Leeuwenhoek 69:109-117. [DOI] [PubMed] [Google Scholar]
- 48.Farnworth, E. R., I. Mainville, M. P. Desjardins, N. Gardner, I. Fliss, and C. Champagne. 2007. Growth of probiotic bacteria and bifidobacteria in a soy yogurt formulation. Int. J. Food Microbiol. 116:174-181. [DOI] [PubMed] [Google Scholar]
- 49.Ferreira, A. D., and B. C. Viljoen. 2003. Yeasts as adjunct starters in matured cheddar cheese. Int. J. Food Microbiol. 86:131-140. [DOI] [PubMed] [Google Scholar]
- 50.Fox, P. F. 1993. Cheese: chemistry, physics and microbiology, p. 1-36. In P. F. Fox (ed.), Cheese: an overview, 2nd ed., vol. 1. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 51.Fredrickson, A. G. 1977. Behavior of mixed cultures of microorganisms. Annu. Rev. Microbiol. 31:63-87. [DOI] [PubMed] [Google Scholar]
- 52.Gadaga, T. H., A. N. Mutukumira, and J. A. Narvhus. 2001. The growth and interaction of yeasts and lactic acid bacteria isolated from Zimbabwean naturally fermented milk in UHT milk. Int. J. Food Microbiol. 68:21-32. [DOI] [PubMed] [Google Scholar]
- 53.Galesloot, T. E., F. Hassing, and H. A. Veringa. 1968. Symbiosis in yogurt. (1) Stimulation of Lactobacillus bulgaricus by a factor produced by Streptococcus thermophilus. Neth. Milk Dairy J. 22:50-63. [Google Scholar]
- 54.Garde, A., G. Jonsson, A. S. Schmidt, and B. K. Ahring. 2002. Lactic acid production from wheat straw hemicellulose hydrolysate by Lactobacillus pentosus and Lactobacillus brevis. Bioresour. Technol. 81:217-223. [DOI] [PubMed] [Google Scholar]
- 55.Garrod, L. P., H. P. Lambert, F. O'Grady, and P. M. Waterworth. 1973. Antibiotic and chemotherapy, 4th ed. Churchill Livingstone, Edinburgh, United Kingdom.
- 56.Gobbetti, M., and A. Corsetti. 1997. Lactobacillus sanfrancisco a key sourdough lactic acid bacterium: a review. Food Microbiol. 14:175-187. [Google Scholar]
- 57.Gobbetti, M., A. Corsetti, and J. Rossi. 1994. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: metabolism of carbohydrates. Appl. Microbiol. Biotechnol. 41:456-460. [DOI] [PubMed] [Google Scholar]
- 58.Gobbetti, M., M. De Angelis, R. Di Cagno, F. Minervini, and A. Limitone. 2007. Cell-cell communication in food related bacteria. Int. J. Food Microbiol. 120:34-45. [DOI] [PubMed] [Google Scholar]
- 59.Gonzalez, J. E., and N. D. Keshavan. 2006. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 70:859-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gregory, R., R. Paton, J. Saunders, and Q. H. Wu. 2004. Parallelising a model of bacterial interaction and evolution. BioSystems 76:121-131. [DOI] [PubMed] [Google Scholar]
- 61.Gregory, R., V. A. Saunders, and J. R. Saunders. 2008. Rule-based computing system for microbial interactions and communications: evolution in virtual bacterial populations. BioSystems 91:216-230. [DOI] [PubMed] [Google Scholar]
- 62.Guarner, F., G. Perdigon, G. Corthier, S. Salminen, B. Koletzko, and L. Morelli. 2005. Should yoghurt cultures be considered probiotic? Br. J. Nutr. 93:783-786. [DOI] [PubMed] [Google Scholar]
- 63.Harris, L. J., H. P. Fleming, and T. R. Klaenhammer. 1991. Sensitivity and resistance of Listeria monocytogenes ATCC 19115, Scott A, and UAL500 to nisin. J. Food Prot. 54:836-840. [DOI] [PubMed] [Google Scholar]
- 64.Heller, K. J. 2001. Probiotic bacteria in fermented foods: product characteristics and starter organisms. Am. J. Clin. Nutr. 73:374S-379S. [DOI] [PubMed] [Google Scholar]
- 65.Helling, R. B., C. N. Vargas, and J. Adams. 1987. Evolution of Escherichia coli during growth in a constant environment. Genetics 116:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Higashio, K., T. Kikuchi, and E. Furuichi. 1978. Symbiose entre Lactobacillus bulgaricus et Streptococcus thermophilus dans le yoghourt, p. 522-523. 20th International Dairy Congress France 1978. Congrilait, Paris, France.
- 67.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, S. Dusko Ehrlich, E. Guedon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29:435-463. [DOI] [PubMed] [Google Scholar]
- 68.Hugenholtz, J. 1986. Population dynamics of mixed starter cultures. Neth. Milk Dairy J. 40:129-140. [Google Scholar]
- 69.Hugenholtz, J., R. Splint, W. N. Konings, and H. Veldkamp. 1987. Selection of protease-positive and protease-negative variants of Streptococcus cremoris. Appl. Environ. Microbiol. 53:309-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hyde, A. J., J. Parisot, A. McNichol, and B. B. Bonev. 2006. Nisin-induced changes in Bacillus morphology suggest a paradigm of antibiotic action. Proc. Natl. Acad. Sci. USA 103:19896-19901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ivanova, I., V. Miteva, T. Stefanova, A. Pantev, I. Budakov, S. Danova, P. Moncheva, I. Nikolova, X. Dousset, and P. Boyaval. 1998. Characterization of a bacteriocin produced by Streptococcus thermophilus 81. Int. J. Food Microbiol. 42:147-158. [DOI] [PubMed] [Google Scholar]
- 72.Johnson, M. R., C. I. Montero, S. B. Conners, K. R. Shockley, S. L. Bridger, and R. M. Kelly. 2005. Population density-dependent regulation of exopolysaccharide formation in the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 55:664-674. [DOI] [PubMed] [Google Scholar]
- 73.Juillard, V., C. Foucaud, M. Desmazeaud, and J. Richard. 1996. Utilization of nitrogen sources during growth of Lactococcus lactis in milk. Lait 76:13-24. [Google Scholar]
- 74.Juillard, V., S. Furlan, C. Foucaud, and J. Richard. 1996. Mixed cultures of proteinase-positive and proteinase-negative strains of Lactococcus lactis in milk. J. Dairy Sci. 79:964-970. [Google Scholar]
- 75.Juillard, V., D. Le Bars, E. R. Kunji, W. N. Konings, J. C. Gripon, and J. Richard. 1995. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Appl. Environ. Microbiol. 61:3024-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Juille, O., D. Le Bars, and V. Juillard. 2005. The specificity of oligopeptide transport by Streptococcus thermophilus resembles that of Lactococcus lactis and not that of pathogenic streptococci. Microbiology 151:1987-1994. [DOI] [PubMed] [Google Scholar]
- 77.Kabuki, T., T. Saito, Y. Kawai, J. Uemura, and T. Itoh. 1997. Production, purification and characterization of reutericin 6, a bacteriocin with lytic activity produced by Lactobacillus reuteri LA6. Int. J. Food Microbiol. 34:145-156. [DOI] [PubMed] [Google Scholar]
- 78.Keller, L., and M. G. Surette. 2006. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4:249-258. [DOI] [PubMed] [Google Scholar]
- 79.Kerr, B., M. A. Riley, M. W. Feldman, and B. J. Bohannan. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171-174. [DOI] [PubMed] [Google Scholar]
- 80.Kives, J., D. Guadarrama, B. Orgaz, A. Rivera-Sen, J. Vazquez, and C. SanJose. 2005. Interactions in biofilms of Lactococcus lactis ssp. cremoris and Pseudomonas fluorescens cultured in cold UHT milk. J. Dairy Sci. 88:4165-4171. [DOI] [PubMed] [Google Scholar]
- 81.Klaassens, E. S., W. M. de Vos, and E. E. Vaughan. 2007. Metaproteomics approach to study the functionality of the microbiota in the human infant gastrointestinal tract. Appl. Environ. Microbiol. 73:1388-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klaver, F. A. M., and F. Kingma. 1989. De bereiding van yoghurt door membraandialysefermentatie. Overdruk van Voedingsmiddelentechnologie 22:23-26. [Google Scholar]
- 83.Kleerebezem, M. 2004. Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25:1405-1414. [DOI] [PubMed] [Google Scholar]
- 84.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lebeer, S., S. C. De Keersmaecker, T. L. Verhoeven, A. A. Fadda, K. Marchal, and J. Vanderleyden. 2007. Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J. Bacteriol. 189:860-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee, K., J. Lee, Y. H. Kim, S. H. Moon, and Y. H. Park. 2001. Unique properties of four lactobacilli in amino acid production and symbiotic mixed culture for lactic acid biosynthesis. Curr. Microbiol. 43:383-390. [DOI] [PubMed] [Google Scholar]
- 87.Letort, C. 2001. Relation entre croissance et nutrition azotée de deux bactéries lactiques thermophiles: Streptococcus thermophilus et Lactobacillus delbrueckii subsp. bulgaricus. Ph.D. thesis. Université de Poitiers, Poitiers, France.
- 88.Letort, C., M. Nardi, P. Garault, V. Monnet, and V. Juillard. 2002. Casein utilization by Streptococcus thermophilus results in a diauxic growth in milk. Appl. Environ. Microbiol. 68:3162-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lindgren, S. E., and W. J. Dobrogosz. 1990. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol. Rev. 7:149-163. [DOI] [PubMed] [Google Scholar]
- 90.Liu, M., F. H. van Enckevort, and R. J. Siezen. 2005. Genome update: lactic acid bacteria genome sequencing is booming. Microbiology 151:3811-3814. [DOI] [PubMed] [Google Scholar]
- 91.Loessner, M., S. Guenther, S. Steffan, and S. Scherer. 2003. A pediocin-producing Lactobacillus plantarum strain inhibits Listeria monocytogenes in a multispecies cheese surface microbial ripening consortium. Appl. Environ. Microbiol. 69:1854-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Luchese, R. H., and W. F. Harrigan. 1990. Growth of and aflatoxin production by Aspergillus parasiticus when in the presence of either Lactococcus lactis or lactic acid and at different initial pH values. J. Appl. Bacteriol. 69:512-519. [DOI] [PubMed] [Google Scholar]
- 93.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Malakar, P. K., G. C. Barker, M. H. Zwietering, and K. van't Riet. 2003. Relevance of microbial interactions to predictive microbiology. Int. J. Food Microbiol. 84:263-272. [DOI] [PubMed] [Google Scholar]
- 95.Malakar, P. K., D. E. Martens, M. H. Zwietering, C. Beal, and K. van 't Riet. 1999. Modelling the interactions between Lactobacillus curvatus and Enterobacter cloacae. II. Mixed cultures and shelf life predictions. Int. J. Food Microbiol. 51:67-79. [DOI] [PubMed] [Google Scholar]
- 96.Maldonado, A., R. Jimenez-Diaz, and J. L. Ruiz-Barba. 2004. Induction of plantaricin production in Lactobacillus plantarum NC8 after coculture with specific gram-positive bacteria is mediated by an autoinduction mechanism. J. Bacteriol. 186:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maldonado, A., J. L. Ruiz-Barba, and R. Jimenez-Diaz. 2004. Production of plantaricin NC8 by Lactobacillus plantarum NC8 is induced in the presence of different types of gram-positive bacteria. Arch. Microbiol. 181:8-16. [DOI] [PubMed] [Google Scholar]
- 98.Maligoy, M., M. Mercade, M. Cocaign-Bousquet, and P. Loubiere. 2008. Transcriptome analysis of Lactococcus lactis in coculture with Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maragkoudakis, P. A., C. Miaris, P. Rojez, N. Manalis, F. Magkanari, G. Kalantzopoulos, and E. Tsakalidou. 2006. Production of traditional Greek yoghurt using Lactobacillus strains with probiotic potential as starter adjuncts. Int. Dairy J. 16:52-60. [Google Scholar]
- 100.McFadden, B. A. 1973. Autotrophic CO2 assimilation and the evolution of ribulose diphosphate carboxylase. Bacteriol. Rev. 37:289-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mellefont, L. A., T. A. McMeekin, and T. Ross. 2003. Performance evaluation of a model describing the effects of temperature, water activity, pH and lactic acid concentration on the growth of Escherichia coli. Int. J. Food Microbiol. 82:45-58. [DOI] [PubMed] [Google Scholar]
- 102.Miteva, V., I. Ivanova, I. Budakov, A. Pantev, T. Stefanova, S. Danova, P. Moncheva, V. Mitev, X. Dousset, and P. Boyaval. 1998. Detection and characterization of a novel antibacterial substance produced by a Lactobacillus delbrueckii strain 1043. J. Appl. Microbiol. 85:603-614. [DOI] [PubMed] [Google Scholar]
- 103.Miteva, V., T. Stefanova, I. Budakov, I. Ivanova, V. Mitev, A. Gancheva, and M. Ljubenov. 1998. Characterization of bacteriocins produced by strains from traditional Bulgarian dairy products. Syst. Appl. Microbiol. 21:151-161. [DOI] [PubMed] [Google Scholar]
- 104.Monnet, C., D. Mora, and G. Corrieu. 2005. Glutamine synthesis is essential for growth of Streptococcus thermophilus in milk and is linked to urea catabolism. Appl. Environ. Microbiol. 71:3376-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moon, N. J., and G. W. Reinbold. 1976. Commensalism and competition in mixed cultures of Lactobacillus bulgaricus and Streptococcus thermophilus. J. Milk Food Technol. 39:337-341. [Google Scholar]
- 106.Mortimer, R. K. 2000. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 10:403-409. [DOI] [PubMed] [Google Scholar]
- 107.Mounier, J., R. Gelsomino, S. Goerges, M. Vancanneyt, K. Vandemeulebroecke, B. Hoste, S. Scherer, J. Swings, G. F. Fitzgerald, and T. M. Cogan. 2005. Surface microflora of four smear-ripened cheeses. Appl. Environ. Microbiol. 71:6489-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Narvhus, J. A., and T. H. Gadaga. 2003. The role of interaction between yeasts and lactic acid bacteria in African fermented milks: a review. Int. J. Food Microbiol. 86:51-60. [DOI] [PubMed] [Google Scholar]
- 109.Noordman, W. H., R. Reissbrodt, R. S. Bongers, J. L. Rademaker, W. Bockelmann, and G. Smit. 2006. Growth stimulation of Brevibacterium sp. by siderophores. J. Appl. Microbiol. 101:637-646. [DOI] [PubMed] [Google Scholar]
- 110.Notebaart, R. A., F. H. van Enckevort, C. Francke, R. J. Siezen, and B. Teusink. 2006. Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics 7:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.O'Brien, J. W. 2004. Global dairy demand—where do we go? Eur. Dairy Mag. 16:22-25. [Google Scholar]
- 112.Oliveira, A. P., J. Nielsen, and J. Forster. 2005. Modeling Lactococcus lactis using a genome-scale flux model. BMC Microbiol. 5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Otieno, D. O., J. F. Ashton, and N. P. Shah. 2007. Isoflavone phytoestrogen degradation in fermented soymilk with selected beta-glucosidase producing L. acidophilus strains during storage at different temperatures. Int. J. Food Microbiol. 115:79-88. [DOI] [PubMed] [Google Scholar]
- 114.Oumer, B. A., P. Gaya, E. Fernandez-Garcia, R. Marciaca, S. Garde, M. Medina, and M. Nunez. 2001. Proteolysis and formation of volatile compounds in cheese manufactured with a bacteriocin-producing adjunct culture. J. Dairy Res. 68:117-129. [DOI] [PubMed] [Google Scholar]
- 115.Park, J. H., S. H. Seok, S. A. Cho, M. W. Baek, H. Y. Lee, D. J. Kim, M. J. Chung, S. D. Kim, U. P. Hong, and J. H. Park. 2005. Antimicrobial effect of lactic acid producing bacteria culture condensate mixture (LCCM) against Salmonella enteritidis. Int. J. Food Microbiol. 101:111-117. [DOI] [PubMed] [Google Scholar]
- 116.Partanen, L., N. Marttinen, and T. Alatossava. 2001. Fats and fatty acids as growth factors for Lactobacillus delbrueckii. Syst. Appl. Microbiol. 24:500-506. [DOI] [PubMed] [Google Scholar]
- 117.Peirera Martins, J. F., and R. H. Luchese. 1988. The assessment of growth compatibility between strains of Lactobacillus bulgaricus and Streptococcus thermophilus. Rev. Inst. Lactic Cândido Tostes (Brasil) 43:11-13. [Google Scholar]
- 118.Peltoniemi, K., E. Vesanto, and A. Palva. 2002. Genetic characterization of an oligopeptide transport system from Lactobacillus delbrueckii subsp. bulgaricus. Arch. Microbiol. 177:457-467. [DOI] [PubMed] [Google Scholar]
- 119.Pette, J. W., and H. Lolkema. 1950. Yoghurt. I. Symbiosis and antibiosis in mixed cultures of Lb bulgaricus and Se thermophilus. Neth. Milk Dairy J. 4:197-208. [Google Scholar]
- 120.Pfeiffer, T., and S. Schuster. 2005. Game-theoretical approaches to studying the evolution of biochemical systems. Trends Biochem. Sci. 30:20-25. [DOI] [PubMed] [Google Scholar]
- 121.Picon, A., and M. Nunez. 2007. Growth stimulation of a proteinase positive Lactococcus lactis strain by a proteinase negative Lactococcus lactis strain. Int. J. Food Microbiol. 119:308-313. [DOI] [PubMed] [Google Scholar]
- 122.Pinon, A., M. Zwietering, L. Perrier, J. M. Membre, B. Leporq, E. Mettler, D. Thuault, L. Coroller, V. Stahl, and M. Vialette. 2004. Development and validation of experimental protocols for use of cardinal models for prediction of microorganism growth in food products. Appl. Environ. Microbiol. 70:1081-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pommier, S., P. Strehaiano, and M. L. Delia. 2005. Modelling the growth dynamics of interacting mixed cultures: a case of amensalism. Int. J. Food Microbiol. 100:131-139. [DOI] [PubMed] [Google Scholar]
- 124.Purko, M., W. O. Nelson, and W. A. Wood. 1951. The associative action between certain yeasts and Bacterium linens. J. Dairy Sci. 23:699-701. [Google Scholar]
- 125.Quadri, L. E. 2002. Regulation of antimicrobial peptide production by autoinducer-mediated quorum sensing in lactic acid bacteria. Antonie van Leeuwenhoek 82:133-145. [PubMed] [Google Scholar]
- 126.Raynaud, S., R. Perrin, M. Cocaign-Bousquet, and P. Loubiere. 2005. Metabolic and transcriptomic adaptation of Lactococcus lactis subsp. lactis biovar diacetylactis in response to autoacidification and temperature downshift in skim milk. Appl. Environ. Microbiol. 71:8016-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reddy, G. V., and K. M. Shahani. 1971. Isolation of an antibiotic from Lactobacillus bulgaricus. J. Dairy Sci. 54:748. [Google Scholar]
- 128.Reiter, B., and J. D. Oram. 1962. Nutritional studies on cheese starters. I. Vitamin and amino acid requirements of single strain starters. J. Dairy Res. 29:63-67. [Google Scholar]
- 129.Risoen, P. A., M. B. Brurberg, V. G. Eijsink, and I. F. Nes. 2000. Functional analysis of promoters involved in quorum sensing-based regulation of bacteriocin production in Lactobacillus. Mol. Microbiol. 37:619-628. [DOI] [PubMed] [Google Scholar]
- 130.Rochat, T., J.-J. Gratadoux, A. Gruss, G. Corthier, E. Maguin, P. Langella, and M. van de Guchte. 2006. Production of a heterologous nonheme catalase by Lactobacillus casei: an efficient tool for removal of H2O2 and protection of Lactobacillus bulgaricus from oxidative stress in milk. Appl. Environ. Microbiol. 72:5143-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rodriguez, A. V., and C. M. Manca de Nadra. 1995. Effect of pH and hydrogen peroxide produced by Lactobacillus hilgardii on Pediococcus pentosaceus growth. FEMS Microbiol. Lett. 128:59-62. [Google Scholar]
- 132.Rosenzweig, R. F., R. R. Sharp, D. S. Treves, and J. Adams. 1994. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137:903-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rul, F., and V. Monnet. 1997. Presence of additional peptidases in Streptococcus thermophilus CNRZ 302 compared to Lactococcus lactis. J. Appl. Microbiol. 82:695-704. [DOI] [PubMed] [Google Scholar]
- 134.Samuel, B. S., E. E. Hansen, J. K. Manchester, P. M. Coutinho, B. Henrissat, R. Fulton, P. Latreille, K. Kim, R. K. Wilson, and J. I. Gordon. 2007. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl. Acad. Sci. USA 104:10643-10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sasaki, M., B. W. Bosman, and P. S. Tan. 1995. Comparison of proteolytic activities in various lactobacilli. J. Dairy Res. 62:601-610. [DOI] [PubMed] [Google Scholar]
- 136.Savijoki, K., H. Ingmer, and P. Varmanen. 2006. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 71:394-406. [DOI] [PubMed] [Google Scholar]
- 137.Shindala, A., H. R. Bungay III, N. R. Krieg, and K. Culbert. 1965. Mixed-culture interactions. I. Commensalism of Proteus vulgaris with Saccharomyces cerevisiae in continuous culture. J. Bacteriol. 89:693-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shirai, T., A. Nakato, N. Izutani, K. Nagahisa, S. Shioya, E. Kimura, Y. Kawarabayasi, A. Yamagishi, T. Gojobori, and H. Shimizu. 2005. Comparative study of flux redistribution of metabolic pathway in glutamate production by two coryneform bacteria. Metab. Eng. 7:59-69. [DOI] [PubMed] [Google Scholar]
- 139.Smeianov, V. V., P. Wechter, J. R. Broadbent, J. E. Hughes, B. T. Rodriguez, T. K. Christensen, Y. Ardo, and J. L. Steele. 2007. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 73:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sodini, I., E. Latrille, and G. Corrieu. 2000. Identification of interacting mixed cultures of lactic acid bacteria by their exclusion from a model predicting the acidifying activity of non-interacting mixed cultures. Appl. Microbiol. Biotechnol. 54:715-718. [DOI] [PubMed] [Google Scholar]
- 141.Sonnenburg, J. L., C. T. Chen, and J. I. Gordon. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4:e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Spencer, C. C., M. Bertrand, M. Travisano, and M. Doebeli. 2007. Adaptive diversification in genes that regulate resource use in Escherichia coli. PLoS Genet. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stecchini, M. L., I. Sarais, and M. de Bertoldi. 1991. The influence of Lactobacillus plantarum culture inoculation on the fate of Staphylococcus aureus and Salmonella typhimurium in Montasio cheese. Int. J. Food Microbiol. 14:99-109. [DOI] [PubMed] [Google Scholar]
- 144.Steinkraus, K. H. 1989. Industrialization of indigenous fermented foods. Marcel Dekker, New York, NY.
- 145.Sturino, J. M., and T. R. Klaenhammer. 2004. Bacteriophage defense systems and strategies for lactic acid bacteria. Adv. Appl. Microbiol. 56:331-378. [DOI] [PubMed] [Google Scholar]
- 146.Sturino, J. M., and T. R. Klaenhammer. 2006. Engineered bacteriophage-defence systems in bioprocessing. Nat. Rev. Microbiol. 4:395-404. [DOI] [PubMed] [Google Scholar]
- 147.Sturme, M. H., J. Nakayama, D. Molenaar, Y. Murakami, R. Kunugi, T. Fujii, E. E. Vaughan, M. Kleerebezem, and W. M. de Vos. 2005. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 187:5224-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Suma, K., M. C. Misra, and M. C. Varadaraj. 1998. Plantaricin LP84, a broad spectrum heat-stable bacteriocin of Lactobacillus plantarum NCIM 2084 produced in a simple glucose broth medium. Int. J. Food Microbiol. 40:17-25. [DOI] [PubMed] [Google Scholar]
- 149.Sybesma, W., M. Starrenburg, L. Tijsseling, M. H. Hoefnagel, and J. Hugenholtz. 2003. Effects of cultivation conditions on folate production by lactic acid bacteria. Appl. Environ. Microbiol. 69:4542-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Taga, M. E., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tamime, A. Y. 2002. Fermented milks: a historical food with modern applications—a review. Eur. J. Clin. Nutr. 56(Suppl. 4):S2-S15. [DOI] [PubMed] [Google Scholar]
- 152.Tamime, A. Y., V. M. Marshall, and R. K. Robinson. 1995. Microbiological and technological aspects of milks fermented by bifidobacteria. J. Dairy Res. 62:151-187. [DOI] [PubMed] [Google Scholar]
- 153.Tamime, A. Y., and V. M. E. Marshall. 1997. Microbiology and technology of fermented milks, p. 57-152. In B. A. Law (ed.), Microbiology and biochemistry of cheese and fermented milk. Blackie Academic & Professional, London, United Kingdom.
- 154.Tamime, A. Y., and R. K. Robinson. 1999. Yoghurt science and technology, 2nd ed. Woodhead Publishing, Cambridge, United Kingdom.
- 155.Taniguchi, M., T. Tokunaga, K. Horiuchi, K. Hoshino, K. Sakai, and T. Tanaka. 2004. Production of l-lactic acid from a mixture of xylose and glucose by co-cultivation of lactic acid bacteria. Appl. Microbiol. Biotechnol. 66:160-165. [DOI] [PubMed] [Google Scholar]
- 156.Teusink, B., and E. J. Smid. 2006. Modelling strategies for the industrial exploitation of lactic acid bacteria. Nat. Rev. Microbiol. 4:46-56. [DOI] [PubMed] [Google Scholar]
- 157.Teusink, B., F. H. van Enckevort, C. Francke, A. Wiersma, A. Wegkamp, E. J. Smid, and R. J. Siezen. 2005. In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: comparing predictions of nutrient requirements with those from growth experiments. Appl. Environ. Microbiol. 71:7253-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Teusink, B., A. Wiersma, D. Molenaar, C. Francke, W. M. de Vos, R. J. Siezen, and E. J. Smid. 2006. Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J. Biol. Chem. 281:40041-40048. [DOI] [PubMed] [Google Scholar]
- 159.Thomas, L. V., J. W. Wimpenny, and G. C. Barker. 1997. Spatial interactions between subsurface bacterial colonies in a model system: a territory model describing the inhibition of Listeria monocytogenes by a nisin-producing lactic acid bacterium. Microbiology 143:2575-2582. [DOI] [PubMed] [Google Scholar]
- 160.Tringe, S. G., C. von Mering, A. Kobayashi, A. A. Salamov, K. Chen, H. W. Chang, M. Podar, J. M. Short, E. J. Mathur, J. C. Detter, P. Bork, P. Hugenholtz, and E. M. Rubin. 2005. Comparative metagenomics of microbial communities. Science 308:554-557. [DOI] [PubMed] [Google Scholar]
- 161.Tyson, G. W., and J. F. Banfield. 2007. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10:200-207. [DOI] [PubMed] [Google Scholar]
- 162.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 163.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 165.Viljoen, B. C. 2001. The interaction between yeasts and bacteria in dairy environments. Int. J. Food Microbiol. 69:37-44. [DOI] [PubMed] [Google Scholar]
- 166.Wang, H., W. Yu, T. Coolbear, D. O'Sullivan, and L. L. McKay. 1998. A deficiency in aspartate biosynthesis in Lactococcus lactis subsp. lactis C2 causes slow milk coagulation. Appl. Environ. Microbiol. 64:1673-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang, J., and D. Y. Fung. 1996. Alkaline-fermented foods: a review with emphasis on pidan fermentation. Crit. Rev. Microbiol. 22:101-138. [DOI] [PubMed] [Google Scholar]
- 168.Wegkamp, A., M. Starrenburg, W. M. de Vos, J. Hugenholtz, and W. Sybesma. 2004. Transformation of folate-consuming Lactobacillus gasseri into a folate producer. Appl. Environ. Microbiol. 70:3146-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 170.Weinbauer, M. G., and F. Rassoulzadegan. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 171.West, S. A., A. S. Griffin, A. Gardner, and S. P. Diggle. 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4:597-607. [DOI] [PubMed] [Google Scholar]
- 172.Wiedemann, I., T. Bottiger, R. R. Bonelli, T. Schneider, H. G. Sahl, and B. Martinez. 2006. Lipid II-based antimicrobial activity of the lantibiotic plantaricin C. Appl. Environ. Microbiol. 72:2809-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Winkel, S. A., and G. H. Richardson. 1984. Cell mass and acid production of proteinase-positive and proteinase-negative lactic cultures in buffered nonfat milk. J. Dairy Sci. 67:2856-2859. [Google Scholar]
- 174.Wisselink, H. W., M. J. Toirkens, M. del Rosario Franco Berriel, A. A. Winkler, J. P. van Dijken, J. T. Pronk, and A. J. van Maris. 2007. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl. Environ. Microbiol. 73:4881-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xu, S., T. D. Boylston, and B. A. Glatz. 2005. Conjugated linoleic acid content and organoleptic attributes of fermented milk products produced with probiotic bacteria. J. Agric. Food Chem. 53:9064-9072. [DOI] [PubMed] [Google Scholar]
- 176.Yoshida, A., T. Ansai, T. Takehara, and H. K. Kuramitsu. 2005. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 71:2372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zisu, B., and N. P. Shah. 2003. Effects of pH, temperature, supplementation with whey protein concentrate, and adjunct cultures on the production of exopolysaccharides by Streptococcus thermophilus 1275. J. Dairy Sci. 86:3405-3415. [DOI] [PubMed] [Google Scholar]
- 178.Zoetendal, E. G., C. C. Booijink, E. S. Klaassens, H. G. Heilig, M. Kleerebezem, H. Smidt, and W. M. de Vos. 2006. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 1:954-959. [DOI] [PubMed] [Google Scholar]
- 179.Zourari, A., J. P. Accolas, and M. J. Desmazeaud. 1992. Metabolism and biochemical characteristics of yogurt bacteria—a review. Lait 72:1-34. [Google Scholar]