Abstract
Ceriporiopsis sp. strain MD-1, isolated from forest soil, produced several extracellular enzymes that decolorized human hair melanin. Among them, three enzymes (E1, E2-1, and E2-2) were purified to homogeneity and characterized. The enzymes required hydrogen peroxide in their enzyme reactions and, typical of other fungal peroxidases, oxidized various phenol compounds such as guaiacol, but not 3,4-dimethoxybenzyl alcohol. The spectra of the three enzymes showed an absorption maximum at 406 nm, indicating that they were heme proteins. However, the A406/A280 values of the enzymes were below 0.4, which was lower than those of other peroxidases. E2-1 and E2-2 were similar to each other in their molecular and catalytic properties, and they possibly represent products of posttranslational modifications and/or allelic variants of the same gene, mdcA. The corresponding cDNA was cloned and sequenced; the deduced amino acid sequence showed high identities to the manganese peroxidases from other microorganisms. The specific activities and Km values of E2-1 and E2-2 for synthetic and human hair melanins were much higher than those of Phanerochaete chrysosporium manganese peroxidase and lignin peroxidase.
Melanin is a black or brown polymer widely distributed in animals, plants, and microorganisms. Fungal melanin was reported to improve the resistance of its cells to dryness, high temperatures, UV rays, sterilizers, lytic enzymes, etc. (2), and is also involved in the fungal pathogenesis of plants (3). Melanin in plants is an important strengthening element of the cell walls (29).
In the human body, melanin is mainly contained in skin and hair, where it is believed that melanin absorbs excess light and protects them from damage caused by harmful light such as UV rays (11). The typical mammalian melanin, eumelanin, is biosynthesized from l-tyrosine through l-dopaquinone, dopachrome, etc. (10, 11). Although the overall structure of eumelanin is unclear, it is thought to be a heterogeneous polymer composed of 5,6-dihydroxyindole, indole-5,6-quinone, 5,6-dihydroxyindole-2-carboxylic acid, and indole-5,6-quinone-2-carboxylic acid units (27).
Because melanin in human skin is a black or brown pigment, the decolorization of melanin by enzymes has become of interest in the field of cosmetics in recent years. Woo et al. (41) observed that the lignin peroxidase from Phanerochaete chrysosporium decolorizes synthetic melanin. They suggest that the enzymatic decolorization of melanin is applicable to the development of new cosmetic whitening agents. Mohorcic et al. (24) isolated a Sporotrichum pruinosum strain that produces a melanin-bleaching enzyme on a screening medium containing synthetic melanin. They found that this enzyme can degrade human skin melanin. However, the enzyme was not purified to homogeneity, and thus its molecular and catalytic characteristics, including its gene structure, were not identified.
Human hair consists of protein, fat, melanin, and water (9). Over 90% of its dry weight is made up of a protein called keratin that contains many cysteine residues. Human hair contains around 10% water, which modifies its mechanical properties, and about 3% melanin. Structurally, hair consists of the medulla (the central space of the hair), cortex (the middle and largest layer), and cuticle (the outer layer of colorless cells). Melanin contained in the cortex gives hair its natural color. When human hair is bleached and colored using conventional oxidative hair dyes, hydrogen peroxide is used under alkaline conditions to decompose and decolorize melanin. This treatment causes damage to the hair due to the denaturing and decomposition of the keratin proteins and to the skin around the hair. An enzyme that easily decolorizes hair melanin might be useful for safely bleaching and coloring hair. Herein, we describe the purification, characterization, and gene cloning of enzymes from Ceriporiopsis sp. strain MD-1 that decolorize human hair melanin.
MATERIALS AND METHODS
Chemicals.
The chemicals used in this study and their sources are as follows: polypepton from Nihonseiyaku, Tokyo, Japan; 2,6-dimethoxyphenol, syringaldazine, ABTS [2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid)], resorcinol, catechol, pyrogallol, hydroquinone, 4-methoxyphenol, 4-aminophenol, l-3,4-dihydroxyphenylalanine (l-DOPA), and l-tyrosine from Wako Pure Chemicals, Osaka, Japan; 3,4-dimethoxybenzyl alcohol and 4-methoxyphenol from Nacalai Tesque, Kyoto, Japan; guaiacol from Kanto Chemical, Tokyo, Japan; vanillic acid from Tokyo Chemical Industry, Tokyo, Japan; synthetic melanin (prepared from l-tyrosine), manganese peroxidase (from P. chrysosporium; 50.0 U/mg), lignin peroxidase (from P. chrysosporium; 0.52 U/mg), and peroxidase (from horseradish; 259 U purpurogallin/mg) from Sigma-Aldrich, St. Louis, MO; DE52 cellulose from Whatman Chemical Separation, Clifton, NJ; DEAE-Toyopearl 650 S and phenyl-Toyopearl 650 S from Tosoh Corporation, Tokyo, Japan; and restriction endonucleases from Takara Bio Incorporation, Otsu, Japan. The purities of the manganese peroxidase, lignin peroxidase, and horseradish peroxidase were over 90% as proteins, judging from their electrophoretic patterns in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Screening of microorganisms.
For the screening medium, solutions A and B were separately prepared. Solution A contained 2 g malt extract and d-glucose, 0.11 g polypepton, and agar in 90 ml of water (pH 6.0). Solution B contained 0.05 g human hair melanin prepared as described below in 10 ml water (pH 6.0). Solution A was autoclaved, and solution B was sterilized by filtration. The two solutions were mixed at 45 to 50°C under sterile conditions, and the mixed solution was layered on a petri dish. Five hundred twenty-five separate soil samples collected in forests were suspended in a saline solution. The suspensions were stirred vigorously for 1 min and then allowed to stand for 2 min. One drop of each suspension was spread on the screening medium in a petri dish. The plate was incubated at 30°C for 1 to 2 weeks. Organisms growing on the medium and decolorizing human hair melanin around their colonies were isolated. Strain MD-1 efficiently decolorized the melanin and was subsequently identified as a Ceriporiopsis species on the basis of its morphological features and the sequence of the large ribosomal subunit RNA gene.
Microorganisms, bacteriophages, and plasmids.
The above-mentioned Ceriporiopsis sp. strain MD-1 was used throughout. A lambda Uni-Zap XR vector (Stratagene, La Jolla, CA) was used for the construction of the cDNA library. The pGEM-Teasy vector (Promega, Madison, WI) was used for cloning of the PCR products and subcloning of the DNA fragments.
Culture conditions.
Ceriporiopsis sp. strain MD-1 was cultured in medium (pH 6.0) containing 2 g malt extract and d-glucose in 100 ml of deionized water at 30°C with shaking. The cultural fluid and its cells were used for the enzyme purification and preparation of the total DNA and RNA, respectively.
Preparation of melanin solution.
The human hair melanin was prepared according to the method of Ito and Fujita (13). The synthetic and human hair melanins were suspended in water and dissolved with 1 N NaOH, and then the pH of the solution was adjusted to a designated value.
Enzyme assays.
The activity of the melanin-decolorizing enzyme was measured by monitoring a decrease in absorbance at 540 nm due to the oxidation of melanin. The reaction mixture (1,274 μl) consisted of 100 μl of a 0.05% (wt/vol) melanin solution (synthetic or human hair melanin), 900 μl of 50 mM lactate-succinate buffer (pH 4.5), 10 μl of 130 mM Mn2+, 14 μl of 9 mM H2O2, and 250 μl of the enzyme solution. The reaction was started at 30°C by adding H2O2. The decrease in absorbance at 540 nm was read at appropriate intervals. The activities of commercially obtained peroxidases toward melanins were assayed under the standard assay conditions. One unit of the enzyme activity was defined as the amount of enzyme that decreased the absorbance at 540 nm by 0.001 for 1 h. The specific activity was defined as units per mg protein. Protein concentrations were measured by the method of Lowry et al. (20).
The activities of the enzyme for 2,6-dimethoxyphenol (39), guaiacol (7), syringaldazine (19), ABTS (12), 3,4-dimethoxybenzyl alcohol (8), resorcinol (35), catechol (33), pyrogallol (16), vanillic acid (38), hydroquinone (25), 4-methoxyphenol (25), 4-aminophenol (25), l-DOPA (18), and l-tyrosine (30) were spectrophotometrically estimated.
Purification of enzyme.
All enzyme purification operations were done at 0 to 4°C, and all centrifugations were carried out at 27,000 × g for 10 min. The enzyme activity was measured using synthetic melanin as a substrate.
(i) Step 1: ammonium fractionation.
The culture (4,500 ml) of Ceriporiopsis sp. strain MD-1, incubated for 4 days, was filtered to remove any fungal pellets, and then the filtrate was centrifuged. Proteins in the supernatant were fractionated with a 55 to 85% saturation of ammonium sulfate. The fractionated precipitate, dissolved in 10 mM sodium-potassium phosphate buffer (pH 6.0) (buffer A), was dialyzed against buffer A (fraction I).
(ii) Step 2: chromatography on DE52 cellulose.
Fraction I was applied to a column (2.4 by 15 cm) of DE52 cellulose equilibrated with buffer A. Proteins were eluted with a linear gradient of NaCl in buffer A, and then the protein concentrations and enzyme activities were assayed. Three peaks (the E1, E2, and E3 enzymes) of enzyme activity appeared on the chromatogram. The active fractions of E1, E2, and E3 were separately pooled and dialyzed against buffer A (fractions II-E1, -E2, and -E3).
(iii) Step 3: chromatography on DEAE-Toyopearl 650 S.
Proteins in fractions II-E1, -E2, and -E3 were separately chromatographed on DEAE-Toyopearl 650 S essentially by the same methods as mentioned above. The active fractions were pooled and dialyzed against buffer A (fractions III-E1, -E2, and -E3).
(iv) Step 4: chromatography on phenyl-Toyopearl 650 S.
Fractions III-E1 and -E3 supplemented with ammonium sulfate were separately applied to a column (1.7 by 7 cm) of phenyl-Toyopearl 650 S equilibrated with buffer A containing 1 M ammonium sulfate. Proteins were eluted with the same buffer. The purity of the enzyme in each fraction was tested by PAGE (6). The fractions of E1 showing a one-protein band on the gel were pooled (fraction IV-E1). However, there were no fractions of E3 showing a one-protein band, and thus, E3 was not further purified. Proteins in fraction III-E2 were chromatographed on phenyl-Toyopearl 650 S essentially by the same methods as mentioned above. Two peaks (the E2-1 and E2-2 enzymes) of enzyme activity appeared on the chromatogram. Fractions passing the purity test by PAGE were pooled separately (fractions IV-E2-1 and -E2-2).
Effects of temperature and pH on the activity and stability of the purified enzymes.
The effect of temperature on the melanin-decolorizing activity was determined at 20 to 75°C in 5°C increments. The optimum pH of the enzymes was determined at 30°C using the following 35 mM buffers: lactic acid-disodium succinate (pH 2.0 to 5.0), sodium acetate (pH 5.0 to 6.0), sodium-potassium phosphate (pH 6.0 to 7.5), and Tris-HCl (pH 7.5 to 9.0). The effect of temperature on the stability of enzymes was determined by incubating the enzymes at 20 to 75°C for 10 min. The remaining activity was then measured at 30°C. The effect of pH on the stability of enzymes was determined by dialyzing the enzymes against buffers with various pHs at 4°C overnight and measuring the remaining activity at pH 4.5.
Determination of molecular masses.
Molecular masses of the native enzymes were measured by gel filtration. Those of the denatured enzymes were measured by SDS-PAGE (40). Size markers used for the gel filtration were those provided in the calibration proteins gel chromatography kit from Boehringer Mannheim (Mannheim, Germany). The LMW electrophoresis calibration kit (Amersham Pharmacia Biotech, Uppsala, Sweden) was used for SDS-PAGE.
Determination of NH2-terminal and inner amino acid sequences.
The E1 enzyme was treated with cyanogen bromide. SDS-PAGE was employed for the digested E1 (34) and the denatured E1, E2-1, and E2-2 enzymes. The peptides and proteins on the gel were electroblotted according to the method of Matsudaira (23) and then sequenced.
Cloning and characterization of mdcA.
Total RNA of Ceriporiopsis sp. strain MD-1 was prepared according to the protocol described by Kingston et al. (17). mRNA was purified from the total RNA by using the PolyATtract mRNA isolation systems (Promega). cDNA was prepared according to the protocols of the cDNA synthesis kit (Stratagene) and the ZAP-cDNA synthesis kit (Stratagene).
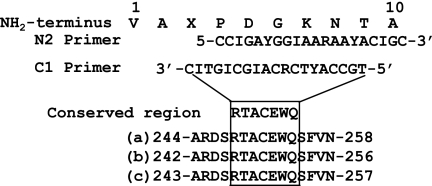
PCR primer design was based on NH2-terminal amino acid sequences of E2-1 and the conserved region of the peroxidases (see Fig. 4). The cDNA, used as a template, was synthesized with a RevertAid Moloney murine leukemia Virus reverse transcriptase (Fermentas, Burlington, Canada) using the C1 primer and mRNA of Ceriporiopsis sp. strain MD-1. For amplifying the fragments containing an E2-1 enzyme gene (mdcA gene) by PCR, Takara Ex Taq (Takara Bio Inc.) was added with the N2 primer. The cycling of PCR was performed at 95°C (30 s), 49°C (30 s), and 72°C (2 min) for 35 cycles.
FIG. 4.
NH2-terminal amino acid sequence of E2-1 and its corresponding N2 and C1 primer sequences. The C1 primer was synthesized on the basis of the conserved regions of manganese peroxidase from T. versicolor (a) (14), lignin peroxidase from P. chrysosporium (b) (28), and peroxidase from a Bjerkandera strain (c) (26).
The cDNA amplicons were ligated into a vector (Promega) and partially sequenced to ensure that the ligated fragments contained the mdcA gene. After sequencing, the recombinant plasmid containing the mdcA gene was digested with MboII. Then, the inserted DNA fragments were also digested with this enzyme. A 327-bp fragment was separated from the vector DNA and the other DNA fragments by using 2.5% (wt/vol) agarose gel electrophoresis, recovered from gel slices by the QIAEX II gel extraction kit (Qiagen, Hilden, Germany), and radiolabeled with [α-32P]dCTP (Amersham, Buckinghamshire, United Kingdom) and the random primer DNA labeling kit, version 2 (Takara Bio Inc.).
Plaque and Southern blot hybridizations for cDNA were performed by standard methods (32). Genomic DNA corresponding to mdcA was PCR amplified using total genomic DNA as a template. The forward primer was 5′-CCACTCAGGTAGCTCCAG-3′ and the reverse primer 5′-GGACTAGCAACGCAAATTCT-3′. PCR amplification was performed using a Pfu DNA polymerase. PCR cycling was performed at 95°C (30 s), 53°C (30 s), and 72°C (2.5 min) for 30 cycles.
Nucleotide accession numbers.
Nucleotide sequences reported in this paper have been submitted to the DDBJ/GenBank/EMBL databases and assigned the accession numbers AB362388 and AB362389 for the large ribosomal subunit RNA gene of Ceriporiopsis sp. strain MD-1 and the mdcA gene, respectively.
RESULTS
Purification of enzymes and their molecular properties.
Table 1 shows a summary of typical enzyme purification. The three melanin-decolorizing enzymes, E1, E2-1, and E2-2, were purified to homogeneity by the purification procedure. However, E3 was not purified to homogeneity. Specific activities of the final preparations of E1, E2-1, and E2-2 for synthetic melanin were 17 × 104, 21 × 104, and 26 × 104 units mg−1, respectively. The overall recovery of the three enzymes totaled 12%. One hundred seventy-, 210-, and 270-fold increases in the specific activities of E1, E2-1, and E2-2, respectively, were observed in the final step of the purification procedure. The final enzyme preparations of E1, E2-1, and E2-2 each showed a one-protein band on an SDS-polyacrylamide gel with molecular masses of 72, 43, and 41 kDa, respectively. The native molecular masses of E1, E2-1, and E2-2 were 70, 45, and 42 kDa, respectively, by gel filtration. These results indicate that the enzymes were monomers.
TABLE 1.
Summary of purification
| Fraction | Total activity (units)a | Total protein (mg) | Sp act (units mg−1) | Recovery (%) |
|---|---|---|---|---|
| Supernatant | 45 × 105 | 4,600 | 0.098 × 104 | 100 |
| Ammonium sulfate | 27 × 105 | 200 | 1.4 × 104 | 60 |
| E1 chromatographed on: | ||||
| DE52 | 4.7 × 105 | 12 | 3.9 × 104 | 10 |
| DEAE-Toyopearl | 3.7 × 105 | 3.1 | 12 × 104 | 8.2 |
| Phenyl-Toyopearl | 2.9 × 105 | 1.7 | 17 × 104 | 6.4 |
| E2 chromatographed on: | ||||
| DE52 | 6.3 × 105 | 7.0 | 9.0 × 104 | 14 |
| DEAE-Toyopearl | 5.2 × 105 | 3.1 | 17 × 104 | 12 |
| E2-1 chromatographed on phenyl-Toyopearl | 0.74 × 105 | 0.35 | 21 × 104 | 1.6 |
| E2-2 chromatographed on phenyl-Toyopearl | 1.6 × 105 | 0.62 | 26 × 104 | 3.6 |
| E3 chromatographed on DE52 | 17 × 105 | 24 | 7.1 × 104 | 38 |
The enzyme activity was measured using synthetic melanin as a substrate.
General properties of the purified enzymes.
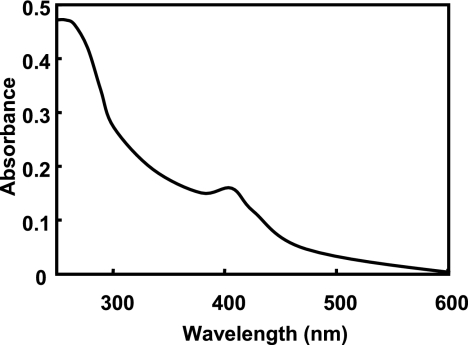
The optimum pHs, effects of temperature on enzyme activity, pH stabilities, and thermostabilities were similar for E1, E2-1, and E2-2 (Table 2). The absorption spectrum of the E2-1 enzyme had a small shoulder at 260 nm and a peak at 406 nm (Fig. 1). The E1 and E2-2 enzymes showed almost the same spectra as that of E2-1. The A406/A280 (RZ) values of E1, E2-1, and E2-2 were 0.22, 0.40, and 0.38, respectively.
TABLE 2.
Characteristics of the E1, E2-1, and E2-2 enzymes
| Parametera | Results for:
|
||
|---|---|---|---|
| E1 | E2-1 | E2-2 | |
| Molecular mass measured by SDS-PAGE (kDa) | 72 | 43 | 41 |
| Optimum pH | 3.5-4.0 | 4.0 | 4.0 |
| Temp with highest activity (°C) | 50 | 50 | 50 |
| pH stability (>50%b) | 2.5-8.0 | 3.5-8.0 | 3.5-8.0 |
| Thermostability (>60%b) (°C) | 55 | 60 | 60 |
| NH2-terminal amino acid sequence | SAG | VAXPDGKNTA | VAXPDG |
The enzyme activity was measured using synthetic melanin as a substrate.
Remaining activity. See the text for details.
FIG. 1.
Absorption spectrum of E2-1. The enzyme (1.1 mg) was measured in 2 ml of buffer A.
Decolorization of melanins.
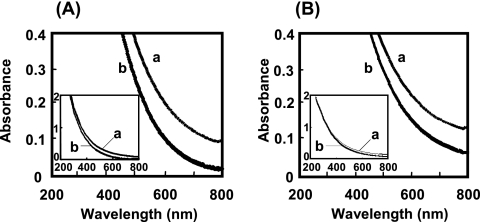
When the synthetic and human hair melanins were incubated with the E1 enzyme, decreases in absorbance arising from the melanin oxidation were observed at between 200 and 800 nm (Fig. 2). As the absorbance decreased, the dark-brown color of the incubation mixture became slightly pale. E2-1 and E2-2 also showed the same absorbance pattern as E1. The synthetic melanin was decolorized more rapidly than the human hair melanin by the three enzymes. The greatest differences in absorbance were observed at 540 nm for both types of melanins.
FIG. 2.
Decolorization of synthetic (A) and human hair (B) melanins by the E1 enzyme. The reaction was performed under standard conditions using 6 μg of E1. The absorption spectra of the reaction mixture were measured at 0 min (a) and 20 min (b) after the beginning of the reaction.
Effects of hydrogen peroxide and Mn2+ on enzyme activity.
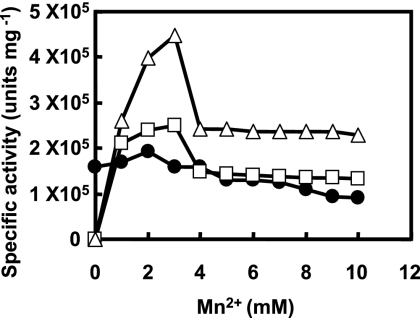
Purified E1, E2-1, and E2-2 did not decolorize the synthetic or human hair melanins in the absence of hydrogen peroxide. When hydrogen peroxide was added to the incubation mixture in the presence of Mn2+, the enzyme reaction steadily proceeded. The optimum concentrations of hydrogen peroxide for E1 and for E2-1 and E2-2 were 0.1 and 0.2 mM, respectively. However, E1 could also decolorize the synthetic and human hair melanins without Mn2+; the enzyme showed 80% of the activity for the synthetic melanin obtained in the presence of 2 mM Mn2+ (Fig. 3). On the other hand, E2-1 and E2-2 required Mn2+ for decolorizing the melanins. The partially purified E3 needed both Mn2+ and hydrogen peroxide for the enzyme reaction, as did E2-1 and E2-2.
FIG. 3.
Effects of concentrations of Mn2+ on the enzyme activities of E1 (filled circles), E2-1 (open squares), and E2-2 (open triangles). One microgram each of E1, E2-1, and E2-2 was incubated in the standard reaction mixture containing synthetic melanin and Mn2+ at concentrations of 0 to 10 mM.
Inhibition.
The effects of metal salts, sulfhydryl, and chelating agents on the enzyme activities were tested using synthetic melanin as a substrate (Table 3). Among the metal ions tested, E1, E2-1, and E2-2 were strongly inhibited by Fe2+, Ag+, and Cu2+, while Zn2+, Mg2+, Hg2+, and Co2+ did not inhibit the enzymes very much and Fe3+ increased the activity of E1. Sulfhydryl and chelating agents moderately inhibited the three enzymes. Sodium azide strongly repressed the enzyme activities.
TABLE 3.
Effects of various compounds on the enzyme activitya
| Compoundb | Concn (mM) | Remaining activity (%) of the indicated enzyme
|
||
|---|---|---|---|---|
| E1 | E2-1 | E2-2 | ||
| None | 100 | 100 | 100 | |
| Zn2+ | 1 | 79 | 87 | 83 |
| Fe2+ | 1 | 28 | 0 | 0 |
| Fe3+ | 1 | 145 | 57 | 57 |
| Ag+ | 1 | 0 | 0 | 0 |
| Mg2+ | 1 | 63 | 89 | 93 |
| Hg2+ | 1 | 68 | 98 | 98 |
| Cu2+ | 1 | 25 | 35 | 30 |
| Mn2+ | 1 | 51 | 70 | 70 |
| Ni2+ | 1 | 58 | 63 | 63 |
| Co2+ | 1 | 84 | 70 | 74 |
| CH2ICOOH | 1 | 62 | 70 | 70 |
| PCMB | 1 | 67 | 70 | 74 |
| DTNB | 1 | 65 | 89 | 87 |
| α,α′-Dipyridyl | 1 | 76 | 63 | 59 |
| N-Ethylmaleimide | 1 | 67 | 89 | 83 |
| Tiron | 1 | 43 | 46 | 46 |
| EDTA | 1 | 65 | 57 | 59 |
| o-Phenanthroline | 1 | 65 | 35 | 30 |
| NaN3 | 1 | 15 | 21 | 21 |
One microgram each of E1, E2-1, and E2-2 was incubated in 110 μl of buffer A containing a 1 mM compound at 30°C for 10 min. An aliquot (50 μl) of the incubation mixture was subjected to enzyme assay in the presence of 1 mM Mn2+ using synthetic melanin as a substrate.
PCMB, p-chloromercuribenzoic acid; DTNB, 5,5′-dithiobis(2-nitrobenzoic acid).
Substrate specificity.
Preferences for 14 substrates were examined (Table 4). The three enzymes oxidized 12 substrates among them, but not 3,4-dimethoxybenzyl alcohol or l-tyrosine. Of the 12 substrates oxidized, the enzymes showed the highest specific activity for hydroquinone and slight activities for resorcinol and l-DOPA. The E1 enzyme oxidized 2,6-dimethoxyphenol, guaiacol, syringaldazine, and ABTS without Mn2+. The Km values of the enzymes were determined for 2,6-dimethoxyphenol, guaiacol, syringaldazine, and ABTS (Table 5). The three enzymes showed the highest affinity for guaiacol of these four substrates examined.
TABLE 4.
Substrate specificity of the enzymes
| Substrate | Presence of 1 mM Mn2+a | Sp act (IU mg−1) of indicated enzyme
|
||
|---|---|---|---|---|
| E1 | E2-1 | E2-2 | ||
| 2,6-Dimethoxyphenol | + | 37 | 73 | 75 |
| 2,6-Dimethoxyphenol | − | 40 | 0 | 0 |
| Guaiacol | + | 60 | 93 | 88 |
| Guaiacol | − | 64 | 0 | 0 |
| Syringaldazine | + | 9.0 | 70 | 68 |
| Syringaldazine | − | 6.4 | 0 | 0 |
| ABTS | + | 97 | 63 | 65 |
| ABTS | − | 88 | 0 | 0 |
| 3,4-Dimethoxybenzyl alcohol | + | 0 | 0 | 0 |
| Resorcinol | + | 0.73 | 1.6 | 1.4 |
| Catechol | + | 15 | 29 | 29 |
| Pyrogallol | + | 25 | 42 | 37 |
| Vanillic acid | + | 41 | 36 | 34 |
| Hydroquinone | + | 130 | 230 | 230 |
| 4-Methoxyphenol | + | 53 | 80 | 73 |
| 4-Aminophenol | + | 57 | 150 | 150 |
| l-DOPA | + | 3.2 | 6.3 | 7.0 |
| l-Tyrosine | + | 0 | 0 | 0 |
+, assayed in the presence of 1mM Mn2+; −, assayed in the absence of Mn2+.
TABLE 5.
Km values of the enzymes
| Substrate |
Km value (μM) of indicated enzymea
|
||
|---|---|---|---|
| E1 | E2-1 | E2-2 | |
| 2,6-Dimethoxyphenol | 6.4 | 5.4 | 5.2 |
| Guaiacol | 1.4 | 1.6 | 2.8 |
| Syringaldazine | 67 | 57 | 58 |
| ABTS | 17 | 8.5 | 11 |
The enzyme activities were assayed in the presence of 1 mM Mn2+.
Kinetic properties of peroxidases toward melanins.
Using synthetic and human hair melanins as substrates, specific activities and Km values were determined for E1, E2-1, E2-2, P. chrysosporium manganese peroxidase, P. chrysosporium lignin peroxidase, and horseradish peroxidase (Table 6). E1, E2-1, and E2-2 exhibited specific activities for synthetic melanin 5- to 22-fold higher than did the peroxidases from P. chrysosporium and horseradish. For human hair melanin, specific activities of the three Ceriporiopsis enzymes were 5- to 12-fold higher. In addition, the enzymes from the Ceriporiopsis strain had higher affinity for the synthetic and human hair melanins than the peroxidases from P. chrysosporium and horseradish did.
TABLE 6.
Specific activities and Km values
| Enzymea | Sp act (units mg−1) for indicated type of melanin
|
Km value (% [wt/vol]) for indicated type of melanin
|
||
|---|---|---|---|---|
| Synthetic | Human hair | Synthetic | Human hair | |
| E1 | 17 × 104 | 7.1 × 104 | 6.5 × 10−4 | 15 × 10−4 |
| E2-1 | 21 × 104 | 8.4 × 104 | 4.3 × 10−4 | 10 × 10−4 |
| E2-2 | 26 × 104 | 11 × 104 | 4.0 × 10−4 | 10 × 10−4 |
| Manganese peroxidase | 1.2 × 104 | 0.9 × 104 | 11 × 10−4 | 16 × 10−4 |
| Lignin peroxidase | 3.3 × 104 | 1.5 × 104 | 40 × 10−4 | 34 × 10−4 |
| Peroxidase | 1.2 × 104 | 1.2 × 104 | 13 × 10−4 | 19 × 10−4 |
Manganese peroxidase, lignin peroxidase, and peroxidase were purchased from Sigma-Aldrich. Each of these enzyme proteins was assayed by the method of Lowry et al. (20) and used for the calculation of specific activity.
Cloning of the mdcA gene.
On the basis of NH2-terminal amino acid sequences of E2-1 (Table 2) and well-established conserved regions of peroxidases (Fig. 4), a 728-bp cDNA fragment was PCR amplified. A 327-bp MboII fragment was then used to probe a lambda Uni-Zap library, yielding 22 positive clones. One of these phages was selected for subcloning and sequencing of the mdcA gene. The NH2-terminal amino acid sequence of E1 was limited to 3 residues (Table 2), although a cyanogen bromide cleavage product yielded a longer internal peptide (TAWANNANNA). No E1 clone(s) was obtained.
Analysis of the mdcA gene.
The mdcA gene encodes 359 amino acid residues, of which 333 composed the mature MdcA. The deduced amino acid sequences from positions 27 to 28 (VA) and 30 to 36 (PDGKNTA) completely agreed with the NH2-terminal amino acid sequence of the purified E2-1. Those from positions 27 to 28 (VA) and 30 to 32 (PDG) agreed with the NH2-terminal amino acid sequence of the purified E2-2. The NH2-terminal and internal sequences of E1 did not match those of MdcA. We observed putative glycosylation residues in the deduced amino acid sequence of MdcA at positions 122 to 124 (NIT) and 183 to 185 (NFS). The predicted molecular mass of the mature MdcA was 35,493 Da.
DNA sequences of the genomic gene of melanin-decolorizing enzymes.
A 1,818-bp fragment was amplified by the PCR approach on the basis of the sequences of the mdcA gene (cDNA) using total genomic DNA as a template. The coding region of mdcA was interrupted by 10 relatively small introns (size range, 51 to 58 bp), including one intron that interrupted the signal peptide-coding region.
DISCUSSION
In this study, we purified and characterized three peroxidases, E1, E2-1, and E2-2, that decolorized human hair melanin from the culture of Ceriporiopsis sp. strain MD-1. Enzymes decolorizing synthetic melanin (41) and human skin melanin (24) were previously reported, but the enzymes and corresponding genes were not fully characterized.
The gene encoding E2-1 and E2-2 was cloned and sequenced. Although the NH2-terminal amino acid sequences of E2-1 and E2-2 were identical to each other, the molecular mass of E2-2 was slightly lower than that of E2-1. Since the two enzymes were definitely separated by chromatography on phenyl-Toyopearl, it is likely that E2-2 lacked the COOH-terminal region of E2-1 or was differently glycosylated from E2-1. However, there is another possibility, i.e., that the two enzymes are encoded by closely related genes or by allelic variants of the same gene (14).
The purified E1, E2-1, and E2-2 enzymes had optimum pHs for the synthetic melanin on the acidic side (Table 2). The three enzymes also acted on the human hair melanin with optimum pHs on the acidic side (data not shown). This is a convenient characteristic for the enzymes when they are used as bleaching agents of human hair, because acidic conditions cause much less damage to the hair and the skin around the hair than alkaline conditions.
The enzymatic removal of phenolic compounds from wastewater has been studied using peroxidases (1, 5). During the treatment with peroxidase, phenols are oxidized in the presence of hydrogen peroxide to form the corresponding radicals, which react to form insoluble polymers within several minutes (37). The insoluble polymers can then be separated from the wastewater as a precipitate. The synthetic and human hair melanins used in the present study are composed of phenolic components, such as 5,6-dihydroxyindole, 5,6-dihydroxyindole-2-carboxylic acid, and benzothiazinylalanine (27). E1, E2-1, and E2-2 probably oxidized the hydroxyl groups of these phenolic components to form radicals with hydrogen peroxide in the presence or absence of Mn2+. The radicals would nonenzymatically and quickly form insoluble polymers, which results in a decreased absorbance in the UV- and visible-light ranges. In this study, we observed a small quantity of precipitates after centrifugation of the enzyme reaction mixture containing human hair and synthetic melanins (Fig. 2). The experiments using O-isotope-labeled H2O2 may be available to further verify whether the melanins were transformed into the insoluble polymers.
Besides the synthetic and human hair melanins, the E1, E2-1, and E2-2 enzymes oxidized common peroxidase and laccase substrates, including 2,6-dimethoxyphenol, guaiacol, syringaldazine, and ABTS. In contrast, the Ceriporiopsis enzymes were unable to oxidize the lignin peroxidase substrate, 3,4-dimethoxybenzyl alcohol. Substrate preferences and the Mn2+ requirement suggest that E2-1 and E2-2 are classified as manganese peroxidases.
In contrast, E1 oxidized the synthetic and human hair melanins, 2,6-dimethoxyphenol, guaiacol, syringaldazine, and ABTS in the absence of Mn2+. The manganese peroxidases from Pleurotus eryngii (4) and Ceriporiopsis subvermispora (36) were also reported to be Mn independent in the presence of certain phenols. When guaiacol is used as a substrate, however, these peroxidases require Mn2+. On the other hand, the E1 enzyme oxidized guaiacol without Mn2+. In addition, E1 did not oxidize 3,4-dimethoxybenzyl alcohol, as mentioned above, indicating that this enzyme is not a lignin peroxidase. In short, the characteristics of E1 are unlike previously reported peroxidases.
The deduced amino acid sequence of MdcA was 77.4 and 70.0% identical to manganese peroxidases from Trametes versicolor (MPGI) (14) and P. eryngii (MnPLI) (31), respectively. As expected of a manganese peroxidase, manganese binding residues E-63, E-67, and D-202 were identified by multiple alignments (21). The deduced molecular mass of the mature MdcA was 35,493 Da, somewhat lower than the estimates based on SDS-PAGE (41 to 43 kDa). As in the cases of other extracellular peroxidases, this difference is likely attributable to glycosylation.
The E2-1 and E2-2 enzymes from Ceriporiopsis sp. strain MD-1 were similar to peroxidases from other molds in molecular characteristics, including molecular mass and amino acid sequence. However, catalytic properties such as substrate specificity, specific activity, and Km value were very different. The RZ values of most peroxidases were reported to be greater than 3.0 (15, 22), and the lower RZ values of E1, E2-1, and E2-2 suggest that the Ceriporiopsis sp. strain MD-1 enzymes contain an unusual modification or prosthetic group, besides the typical heme. Detailed analysis of the prosthetic groups, including the heme reconstitution and Fe stoichiometry, is planned.
Future efforts will also focus on E1, which decolorizes human hair melanin in the absence of Mn2+. Because an excess of heavy metal ions may be harmful, E1 could be especially useful for hair bleaching.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.al-Kassim, L., K. E. Taylor, J. A. Nicell, J. K. Bewtra, and N. Biswas. 1994. Enzymatic removal of selected aromatic contaminants from wastewater by a fungal peroxidase from Coprinus macrorhizus in batch reactors. J. Chem. Technol. Biotechnol. 61:179-182. [DOI] [PubMed] [Google Scholar]
- 2.Butler, M. J., and A. W. Day. 1998. Destruction of fungal melanins by ligninases of Phanerochaete chrysosporium and other white rot fungi. Int. J. Plant Sci. 159:989-995. [Google Scholar]
- 3.Butler, M. J., and A. W. Day. 1998. Fungal melanins: a review. Can. J. Microbiol. 44:1115-1136. [Google Scholar]
- 4.Camarero, S., S. Sarkar, F. J. Ruiz-Dueñas, M. J. Martínez, and A. T. Martínez. 1999. Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. J. Biol. Chem. 274:10324-10330. [DOI] [PubMed] [Google Scholar]
- 5.Caza, N., J. K. Bewtra, N. Biswas, and K. E. Taylor. 1999. Removal of phenolic compounds from synthetic wastewater using soybean peroxidase. Water Res. 33:3012-3018. [Google Scholar]
- 6.Davis, B. J. 1964. Disc electrophoresis. II. Method and application to human serum proteins. Ann. N. Y. Acad. Sci. 121:404-427. [DOI] [PubMed] [Google Scholar]
- 7.Doerge, D. R., R. L. Divi, and M. I. Churchwell. 1997. Identification of the colored guaiacol oxidation product produced by peroxidases. Anal. Biochem. 250:10-17. [DOI] [PubMed] [Google Scholar]
- 8.Faison, B. D., and T. K. Kirk. 1985. Factors involved in the regulation of a ligninase activity in Phanerochaete chrysosporium. Appl. Environ. Microbiol. 49:299-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franbourg, A., and F. Leroy. 2005. Hair structure, function, and physicochemical properties, p. 1-65. In C. Bouillon and J. Wilkinson (ed.), The science of hair care, 2nd ed. CRC Press, Boca Raton, FL.
- 10.García-Molina, F., L. G. Fenoll, J. C. Morote, P. A. García-Ruiz, J. N. Rodríguez-López, F. García-Cánovas, and J. Tudela. 2005. Opposite effects of peroxidase in the initial stages of tyrosinase-catalysed melanin biosynthesis. Int. J. Biochem. Cell Biol. 37:1179-1196. [DOI] [PubMed] [Google Scholar]
- 11.Henson, J. M. 2001. Melanin, p. 229-246. In M. Hofrichter and A. Steinbuechel (ed.), Biopolymers, vol. 1. Wiley-VCH Verlag GmbH, Weinheim, Germany. [Google Scholar]
- 12.Hong, F., N. Q. Meinander, and L. J. Jönsson. 2002. Fermentation strategies for improved heterologous expression of laccase in Pichia pastoris. Biotechnol. Bioeng. 79:438-449. [DOI] [PubMed] [Google Scholar]
- 13.Ito, S., and K. Fujita. 1985. Microanalysis of eumelanin and pheomelanin in hair and melanomas by chemical degradation and liquid chromatography. Anal. Biochem. 144:527-536. [DOI] [PubMed] [Google Scholar]
- 14.Johansson, T., and P. O. Nyman. 1996. A cluster of genes encoding major isozymes of lignin peroxidase and manganese peroxidase from the white-rot fungus Trametes versicolor. Gene 170:31-38. [DOI] [PubMed] [Google Scholar]
- 15.Johansson, T., and P. O. Nyman. 1993. Isozymes of lignin peroxidase and manganese(II) peroxidase from the white-rot basidiomycete Trametes versicolor. Arch. Biochem. Biophys. 300:57-62. [DOI] [PubMed] [Google Scholar]
- 16.Jung, H., F. Xu, and K. Li. 2002. Purification and characterization of laccase from wood-degrading fungus Trichophyton rubrum LKY-7. Enzyme Microb. Technol. 30:161-168. [Google Scholar]
- 17.Kingston, R. E., P. Chomczynski, and N. Sacchi. 2002. Guanidinium methods for total RNA preparation, p. 4.2-4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology, 5th ed., vol. 1. John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- 18.Laufer, Z., R. P. Beckett, and F. V. Minibayeva. 2006. Co-occurrence of the multicopper oxidases tyrosinase and laccase in lichens in sub-order Peltigerineae. Ann. Bot. 98:1035-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonowicz, A., and K. Grzywnowicz. 1981. Quantitative estimation of laccase forms in some white-rot fungi using syringaldazine as a substrate. Enzyme Microb. Technol. 3:55-58. [Google Scholar]
- 20.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 21.Martínez, A. T. 2002. Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme Microb. Technol. 30:425-444. [Google Scholar]
- 22.Martínez, M. J., F. J. Ruiz-Dueñas, F. Guillén, and A. T. Martínez. 1996. Purification and catalytic properties of two manganese peroxidase isozymes from Pleurotus eryngii. Eur. J. Biochem. 237:424-432. [DOI] [PubMed] [Google Scholar]
- 23.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 24.Mohorcic, M., J. Friedrich, I. Renimel, P. Andre, D. Mandin, and J.-P. Chaumont. 2007. Production of melanin bleaching enzyme of fungal origin and its application in cosmetics. Biotechnol. Bioprocess Eng. 12:200-206. [Google Scholar]
- 25.Moreira, P. R., F. Bouillenne, E. Almeida-Vara, F. X. Malcata, J. M. Frère, and J. C. Duarte. 2006. Purification, kinetics and spectral characterisation of a new versatile peroxidase from a Bjerkandera sp. isolate. Enzyme Microb. Technol. 38:28-33. [Google Scholar]
- 26.Moreira, P. R., C. Duez, D. Dehareng, A. Antunes, E. Almeida-Vara, J. M. Frère, F. X. Malcata, and J. C. Duarte. 2005. Molecular characterisation of a versatile peroxidase from a Bjerkandera strain. J. Biotechnol. 118:339-352. [DOI] [PubMed] [Google Scholar]
- 27.Prota, G. 2000. Melanins, melanogenesis and melanocytes: looking at their functional significance from the chemist's viewpoint. Pigment Cell Res. 13:283-293. [DOI] [PubMed] [Google Scholar]
- 28.Reiser, J., I. S. Walther, C. Fraefel, and A. Fiechter. 1993. Methods to investigate the expression of lignin peroxidase genes by the white rot fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 59:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riley, P. A. 1997. Melanin. Int. J. Biochem. Cell Biol. 29:1235-1239. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez-López, J. N., J. Tudela, R. Varón, F. García-Carmona, and F. García-Cánovas. 1992. Analysis of a kinetic model for melanin biosynthesis pathway. J. Biol. Chem. 267:3801-3810. [PubMed] [Google Scholar]
- 31.Ruiz-Dueñas, F. J., M. J. Martínez, and A. T. Martínez. 1999. Molecular characterization of a novel peroxidase isolated from the ligninolytic fungus Pleurotus eryngii. Mol. Microbiol. 31:223-235. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 33.Shleev, S., O. Nikitina, A. Christenson, C. T. Reimann, A. I. Yaropolov, T. Ruzgas, and L. Gorton. 2007. Characterization of two new multiforms of Trametes pubescens laccase. Bioorg. Chem. 35:35-49. [DOI] [PubMed] [Google Scholar]
- 34.Swank, R. T., and K. D. Munkres. 1971. Molecular weight analysis of oligopeptide by electrophoresis in polyacrylamide gel with sodium dodecyl sulfate. Anal. Biochem. 39:462-477. [DOI] [PubMed] [Google Scholar]
- 35.Tsukamoto, K., H. Itakura, K. Sato, K. Fukuyama, S. Miura, S. Takahasi, H. Ikezawa, and T. Hosoya. 1999. Binding of salicylhydroxamic acid and several aromatic donor molecules to Arthromyces ramosus peroxidase, investigated by X-ray crystallography, optical difference spectroscopy, NMR relaxation, molecular dynamics, and kinetics. Biochemistry 38:12558-12568. [DOI] [PubMed] [Google Scholar]
- 36.Urzúa, U., F. Larrondo, S. Lobos, J. Larrain, and R. Vicuña. 1995. Oxidation reactions catalyzed by manganese peroxidase isoenzymes from Ceriporiopsis subvermispora. FEBS Lett. 371:132-136. [DOI] [PubMed] [Google Scholar]
- 37.Vasudevan, P. T., and L. O. Li. 1996. Peroxidase catalyzed polymerization of phenol. Appl. Biochem. Biotechnol. 60:73-82. [Google Scholar]
- 38.Ward, G., Y. Hardar, I. Bilkis, and C. G. Dosoretz. 2003. Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes. J. Biol. Chem. 278:39726-39734. [DOI] [PubMed] [Google Scholar]
- 39.Wariishi, H., K. Valli, and M. H. Gold. 1992. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. J. Biol. Chem. 267:23688-23695. [PubMed] [Google Scholar]
- 40.Weber, K., and M. Osborn. 1969. The reliability of molecular weight determination by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
- 41.Woo, S. H., J. S. Cho, B. S. Lee, and E. K. Kim. 2004. Decolorization of melanin by lignin peroxidase from Phanerochaete chrysosporium. Biotechnol. Bioprocess Eng. 9:256-260. [Google Scholar]