Abstract
The solvent-tolerant bacterium Pseudomonas putida S12 was engineered to utilize xylose as a substrate by expressing xylose isomerase (XylA) and xylulokinase (XylB) from Escherichia coli. The initial yield on xylose was low (9% [g CDW g substrate−1], where CDW is cell dry weight), and the growth rate was poor (0.01 h−1). The main cause of the low yield was the oxidation of xylose into the dead-end product xylonate by endogenous glucose dehydrogenase (Gcd). Subjecting the XylAB-expressing P. putida S12 to laboratory evolution yielded a strain that efficiently utilized xylose (yield, 52% [g CDW g xylose−1]) at a considerably improved growth rate (0.35 h−1). The high yield could be attributed in part to Gcd inactivity, whereas the improved growth rate may be connected to alterations in the primary metabolism. Surprisingly, without any further engineering, the evolved d-xylose-utilizing strain metabolized l-arabinose as efficiently as d-xylose. Furthermore, despite the loss of Gcd activity, the ability to utilize glucose was not affected. Thus, a P. putida S12-derived strain was obtained that efficiently utilizes the three main sugars present in lignocellulosic hydrolysate: glucose, xylose, and arabinose. This strain will form the basis for a platform host for the efficient production of biochemicals from renewable feedstock.
The increasing price of oil and imminent shortage of fossil fuels raise the necessity for the development of alternative technologies for the production of petrochemicals. The use of lignocellulosic biomass as feedstock for the chemical industry is a promising alternative that is being studied widely and extensively. Ethanol and other biochemicals are currently produced from glucose by organisms such as Zymomonas mobilis and Saccharomyces cerevisiae. Although glucose is the primary sugar in lignocellulosic biomass, a considerable fraction consists of xylose and arabinose, which can make up to 25% of the total sugar amount (14). Therefore, expanding the substrate range of whole-cell biocatalysts with these pentose sugars will greatly contribute to the economic feasibility of biochemical production from renewable feedstock.
Several approaches have been used to achieve the utilization of pentose sugars by whole-cell biocatalysts. Expressing xylose isomerase and/or xylulokinase, encoded by, respectively, xylA and xylB, has proven to be a successful strategy to enable phosphorylative growth on xylose (11, 12, 31). Also, genes encoding xylose reductase and xylitol dehydrogenase have been employed, especially for engineering yeast cells (6). Still, problems like redox imbalance or an incomplete pentose phosphate (PP) pathway have been encountered, hampering efficient xylose utilization (6, 31). In addition to xylose utilization, microorganisms have been engineered to utilize arabinose, e.g., by expressing the AraBAD pathway from Escherichia coli (10) or Lactobacillus plantarum (30). Ultimately, microorganisms should be engineered to efficiently and concomitantly utilize glucose, xylose, and arabinose to attain cost-effective production of biochemicals.
Our laboratory is developing Pseudomonas putida S12 as a platform for the production of chemicals from renewable feedstock via central metabolites as the precursor (17, 18, 26, 29). P. putida S12 is exceptionally tolerant to organic solvents (1), which makes this strain an excellent host for the production of chemicals that are generally toxic to other bacterial cells, such as substituted aromatic compounds. For these compounds, the use of mixtures of hexoses and pentoses as substrate may offer a specific advantage, as they are derived from the aromatic amino acids l-phenylalanine and l-tyrosine (17, 18, 26, 29). The key precursors for the aromatic amino acids are phospho-enol pyruvate and erythrose-4-phosphate, which are respectively derived efficiently from hexoses (via the Entner-Doudoroff pathway) and pentoses (via the PP pathway). The aim of this study was to construct a P. putida S12 strain that is capable of utilizing glucose, xylose, and arabinose to serve as an optimized host strain for efficient, green production of chemicals from renewable lignocellulose-derived feedstock.
MATERIALS AND METHODS
Culture conditions.
The strains and plasmids used in this study are presented in Table 1. The media used were Luria broth (22) and a phosphate-buffered mineral salts medium described previously (8). In the mineral salts medium, 10 mM glucose (MMG), 12 mM xylose (MMX), or 12 mM arabinose (MMA) was used as a sole carbon source unless stated otherwise. Antibiotics were added as required, in the following concentrations: ampicillin, 100 μg ml−1 (E. coli); gentamicin, 30 μg ml−1 for Luria broth or 10 μg ml−1 for mineral salts medium; and kanamycin 50 μg ml−1. Shake-flask experiments were performed in Boston bottles containing 20 ml MMG, MMX, or MMA in a horizontally shaking incubator at 30°C for P. putida S12 or 37°C for E. coli. For E. coli, 26.5 mg liter−1 thiamine was added to the mineral salts medium.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Wild type | Invitrogen |
| P. putida S12 | Wild type | Hartmans et al. (9) |
| P. putida S12xylAB | P. putida S12 containing plasmid pJTxylAB | This study |
| P. putida S12xylAB_FGH | P. putida S12 containing plasmid pJTxylAB_FGH | This study |
| P. putida S12xylAB2 | P. putida S12 containing plasmid pJTxylAB, evolved to efficient pentose utilizer | This study |
| P. putida S12xylAB2c | P. putida S12xylAB2 cured from pJTxylAB | This study |
| P. putida S12Δgcd | P. putida S12 glucose dehydrogenase knockout | This study |
| P. putida S12Δgcd_xylAB | P. putida S12 glucose dehydrogenase knockout containing plasmid pJTxylAB | This study |
| P. putida S12araFGH | P. putida S12 containing plasmid pBTaraFGH | This study |
| P. putida S12xylAB_araFGH | P. putida S12 containing plasmids pJTxylAB and pBTaraFGH | This study |
| Plasmids | ||
| pJNTmcs(t) | Apr Gmr; basic expression vector derived from plasmid pJWB1 (28) containing the salicylate-inducible promoter nagR-nagAa | Unpublished data |
| pJTTmcs | Apr Gmr; expression vector containing the constitutive tac promoter, derived from pJWB1 | Unpublished data |
| pJTmcs | Apr Gmr; expression vector containing the constitutive tac promoter without the tac RBS | This study |
| pBTmcs | Cmr; expression vector containing the constitutive tac promoter without the tac RBS | Unpublished data |
| pJTxylAB | pJTmcs containing the xylAB genes from E. coli DH5α | This study |
| pJTxylAB_FGH | pJTmcs containing the xylAB and xylFGH genes from E. coli DH5α | This study |
| pBTaraFGH | pBTmcs containing the araFGH genes from E. coli DH5α | This study |
| pJQ200SK | P15A ori sacB RP4 Gmr(pBluescriptSK); suicide vector | Quandt et al. (20) |
| pJQgcd::Kana | pJQ200SK containing a Kmr-disrupted copy of the gcd gene | This study |
Apr, ampicillin resistance; Gmr, gentamicin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; ori, origin of replication; sacB, gene producing levansucrase.
DNA techniques.
Genomic DNA was isolated by using a DNeasy tissue kit (Qiagen). Plasmid DNA was isolated with a QIAprep spin miniprep kit (Qiagen). DNA concentrations were measured with an ND-1000 spectrophotometer (Nanodrop). Agarose-trapped DNA fragments were isolated with a QIAEXII gel extraction kit (Qiagen). PCRs were performed with Accuprime Pfx polymerase (Invitrogen) according to the manufacturer's instructions. Plasmid DNA was introduced into electrocompetent cells by using a Gene Pulser electroporation device. DNA sequencing reactions were performed by MWG Biotech AG.
qPCR.
The mRNA levels of the glucose dehydrogenase (Gcd) gene gcd were analyzed by quantitative PCR (qPCR). Total RNA extractions were performed with an RNeasy kit (Qiagen). qPCR was performed with oligonucleotide primers 9 and 10 (Table 2) using mRNA of mid-log-phase samples of batch cultures with a spectrofluorimetric thermal cycler (iCycler thermal cycler equipped with optical module; Bio-Rad) using IQ Sybr green supermix (Bio-Rad) according to the manufacturer's protocols.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Target | Sequence (5′ → 3′)a | Characteristic |
|---|---|---|---|
| Primer 1 | xylA from E. coli | GCGGCGGGTACCATGCAAGCCTATTTTGACC | KpnI cohesive end |
| Primer 2 | xylB from E. coli | GCGGCGGCGGCCGCTTACGCCATTAATGGCAG | NotI cohesive end |
| Primer 3 | 5′ End of gcd | GCGGCGGCCGCTTACTCAGCTAATTTGTAAGCGAT | NotI cohesive end |
| Primer 4 | Positions 1154-1134 in gcd | GCGTCTAGACCAACATGTGGTCGATCGCCA | XbaI cohesive end |
| Primer 5 | Positions 1261-1281 in gcd | GCGTCTAGAGATCACCCCGGACGGCTCATT | XbaI cohesive end |
| Primer 6 | 3′ End of gcd | GCGGGATCCATGAGCACTGAAGGTGCGAACC | BamHI cohesive end |
| Primer 7 | 5′ End of loxP-Kmr-loxP | GCTCTAGAATAACTTCGTATAATGTATGCTATAC | XbaI cohesive end |
| Primer 8 | 3′ End of loxP-Kmr-loxP | CGCGCAATTAACCCTCACT | |
| Primer 9 | Positions 2088-2106 in gcd | ACGGTAGCAGCAGTACCAC | qPCR primer |
| Primer 10 | Positions 2266-2285 in gcd | TACTACCTGATCGCCGGTAT | qPCR primer |
| Primer 11 | Positions 588-608 in gcd | AATGCGCCAGGCCTCTTCCAG | Sequencing primer |
| Primer 12 | Positions 397-376 upstream of gcd | CGCCACCGTGCATGACAAGAAG | Sequencing primer |
The restriction sites used for cloning are underlined.
Construction of the expression vector pJTmcs.
Expression vector pJTmcs was constructed by using pJTTmcs, formerly named pTac (18) (Table 1), as the backbone. PCR on pJTTmcs was carried out to amplify the plasmid fragment containing the tac promoter site, omitting the tac ribosomal binding site (RBS). The PCR product was cloned into vector pJTTmcs using restriction sites KpnI and ScaI. The resulting expression vector, pJTmcs, has the same characteristics as pJTTmcs but contains no tac RBS.
Construction of recombinant plasmids.
XylAB was amplified by PCR using genomic DNA from E. coli DH5α as the template and oligonucleotide primers 1 and 2 (Table 2). The resulting 2.8-kb DNA fragment was ligated into vector pJTmcs using the restriction sites KpnI and NotI. The resulting plasmid was designated pJTxylAB.
The suicide vector pJQ200SK (20) was used to construct a gene replacement plasmid for the gcd gene as described below. Primers 3 to 6 (Table 2) were used to amplify 1,158 bp of the 5′ end (gcd1) and 951 bp of the 3′ end (gcd2) of the gcd gene. The kanamycin resistance gene, flanked by loxP recombination sites, was amplified by using primers 7 and 8 on pSK-kanalox (unpublished data) as the template. pJQ200SK was digested by using restriction enzymes NotI and BamHI, and gcd1 and gcd2 were digested with NotI/XbaI and XbaI/BamHI, respectively. The three resulting fragments were ligated in vector pJQ200SK to yield vector pJQgcd. pJQgcd was linearized with XbaI, and the cohesive ends were dephosphorylated. The loxP-kanaR-loxP fragment, digested with XbaI, was cloned into the linearized pJQgcd, yielding pJQgcd::kana. This vector was introduced into wild-type P. putida S12, and transformants were selected for kanamycin resistance. Double-crossover mutants were selected for kanamycin resistance (Kmr) and gentamicin sensitivity (Gms). The replacement of the native gcd by the KanaR-disrupted gcd was confirmed by colony PCR using primers 3 and 6 and sequence analysis of the disrupted gene. Plasmid pJTNcre (unpublished data) was introduced to cure P. putida S12Δgcd from KanaR. This plasmid encodes the Cre recombinase that catalyzes the site-specific recombination at the loxP target sites by which the DNA fragment enclosed by the two loxP sites is removed, in this case KanaR (23, 25). Vector pJTNcre was removed by overnight culturing in nonselective Luria broth.
Analytical methods.
Optical densities were measured at 600 nm using a Biowave cell density meter (WPA Ltd.). An optical density of 1.0 corresponds with a cell dry weight (CDW) of 465 mg liter−1. Sugars and organic acids were analyzed by ion chromatography (Dionex ICS3000 system) using a CarboPac PA20 column with 10 mM NaOH as the eluent for sugars or an IonPac ICE AS6 column with 0.4 mM heptafluorobutyric acid as the eluent for organic acids. d-Xylulose-5-phosphate (Xu5P) was assessed by using a transketolase activity assay.
Enzyme activity assays.
Cell extracts for enzyme assays were prepared by sonication of 5 ml of concentrated cell suspensions (0.9 g liter−1 CDW in 50 mM Tris-HCl buffer, pH 7.5) from overnight cultures. After centrifugation, supernatants were desalted by using PD-10 desalting columns (GE Healthcare). The resulting cell extracts were used for enzyme assays.
The activity of xylose isomerase (XylA) was determined as described by Gao et al. (5). In the assay, xylose isomerase activity is coupled to NADH consumption via sorbitol dehydrogenase. The assay was performed at 30°C in a total volume of 1 ml. The assay mixture contained 50 mM Tris-HCl buffer (pH 7.5), 10 mM MgSO4, 1 mM triethanolamine, 0.5 U sorbitol dehydrogenase, 0.2 mM NADH, and cell extract. The reaction was started by adding xylose to a final concentration of 50 mM.
The xylulokinase activity was determined as described by Eliasson et al. (3). Xylulokinase activity is coupled to the consumption of NADH in the reduction of pyruvate to lactate by lactate dehydrogenase. The assay was performed at 30°C, in a total volume of 1 ml. The reaction mixture contained 50 mM Tris-HCl buffer (pH 7.5), 2.0 mM MgCl2, 2.0 mM ATP, 0.2 mM phosphoenolpyruvate, 10 U pyruvate kinase, 10 U lactate dehydrogenase, 0.2 mM NADH, and cell extract. The reaction was started by adding xylulose to a final concentration of 10 mM.
The transketolase activity was measured to demonstrate d-xylulose-5-phosphate formation from l-arabinose or d-xylose. Transketolase couples d-xylulose-5-phosphate to d-ribulose-5-phosphate to form glyceraldehyde-3-phosphate and sedoheptulose-7-phosphate. The transketolase reaction is coupled to NADH consumption via glyceraldehyde-phosphate dehydrogenase. The assay was performed at 30°C, in a total volume of 1 ml. The reaction mixture contained 216 mM glycylglycine buffer (pH 7.7), 1.7 mM d-ribose-5-phosphate, 0.002% (wt/vol) cocarboxylase, 0.14 mM NADH, 15 mM MgCl2, 2.0 mM ATP, 20 U α-glyceraldehyde-phosphate dehydrogenase-triosephosphate isomerase (based on triosephosphate isomerase units), 0.05 U transketolase, and cell extract. The reaction was started by adding xylose or arabinose to a final concentration of 50 mM.
The pyrroloquinoline quinone (PQQ)-dependent Gcd activity was determined as described by Liu et al. (16). The activity of Gcd was determined spectrophotometrically by measuring the decrease in the absorbance of 2,6-dichlorophenolindophenol (DCPIP) at 600 nm. The assay was performed at 30°C, in a total volume of 1 ml. The reaction mixture contained 50 mM Tris-HCl buffer (pH 7.5), 15 mM NH4Cl, 80 μM DCPIP, 1 μM KCN, 0.33 mM phenazine methosulfate, and cell extract. The reaction was started by adding glucose to a final concentration of 1 mM.
For calculations of enzyme activities, the following molar extinction coefficients were used: 6.22 mM−1 cm−1 for NADH and 19 mM−1 cm−1 for reduced DCPIP. One unit is defined as the amount of enzyme that oxidizes 1 μmol of substrate per minute in the coupled assays described above.
RESULTS
Cloning and functional expression of xylAB in P. putida S12.
Genes xylA and xylB, part of the xyl operon of E. coli, were cloned into expression vector pJTmcs under the transcriptional control of the constitutive tac promoter. The resulting vector, pJTxylAB, was introduced into P. putida S12, yielding P. putida S12xylAB. The results of enzyme assays confirmed that xylose isomerase and xylulokinase were expressed as functional enzymes. The specific activities were 34 U g−1 protein for xylose isomerase and 134 U g−1 protein for xylulokinase. The overall activity of XylAB was 22 U g−1.
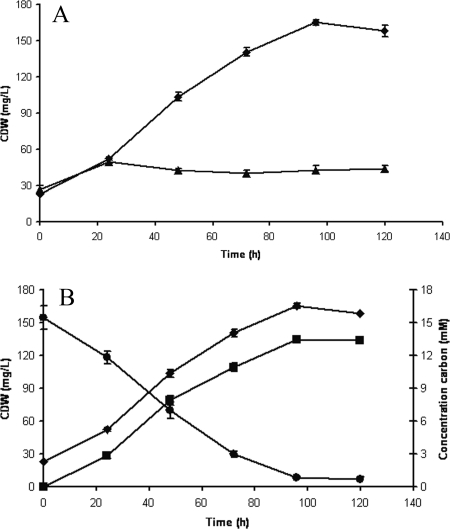
After demonstrating that XylAB were functionally expressed, P. putida S12xylAB was tested for its ability to utilize xylose as a carbon source. When strain S12xylAB was inoculated into xylose-containing medium, growth was observed. However, the biomass yield was low (9% [g CDW g substrate−1]) (Fig. 1A) compared to the biomass yield on glucose (typically 55%). The growth rate was also much lower on xylose than on glucose (0.01 h−1 versus 0.5 h−1). The same characteristics were found for strain S12xylAB_FGH, indicating that xylose transport was not limiting xylose utilization (Table 3). It was observed previously that P. putida S12 oxidizes xylose to xylonate, a reaction shown to be catalyzed in other P. putida strains by PQQ-dependent Gcd (7). Also in xylose-grown P. putida S12xylAB cultures, 81% of the initial amount of xylose was oxidized to xylonate (Fig. 1B), rendering a large part of the xylose unavailable for growth. When the biomass formation in such cultures was related to the amount of xylose that was not oxidized, an apparent yield of 47% (g CDW g substrate−1) was calculated.
FIG. 1.
(A) Growth of wild-type P. putida S12 (triangles) and P. putida S12xylAB (diamonds) cells in mineral salts medium with xylose as the sole carbon source. Data points are the averages of the results of duplicate measurements. Error bars represent the maximum deviations from the averages. (B) Xylonate (squares) and xylose (circles) concentrations in P. putida S12xylAB culture on MMX. P. putida S12xylAB cells were grown in mineral salts medium with xylose as the sole carbon source. CDW concentrations are presented by diamonds. Data points are the averages of the results of duplicate measurements. Error bars represent the maximum deviations of the averages. L, liter.
TABLE 3.
Overview of growth characteristics of pentose-utilizing P. putida S12-derived strainsa
| Strain | Xylose
|
Arabinose
|
||
|---|---|---|---|---|
| Biomass yield (%; g CDW g substrate−1) | Maximum growth rate (h−1) | Biomass yield (%; g CDW g substrate−1) | Maximum growth rate (h−1) | |
| S12xylAB | 9 | 0.01 | NG | NG |
| S12xylAB2 | 52 | 0.35 | 52 | 0.35 |
| S12xylAB2c | NG | NG | NG | NG |
| S12Δgcd_xylAB | 44 | 0.01 | ND | ND |
| S12xylAB_FGH | 10 | 0.01 | NG | NG |
| S12xylAB_araFGH | 14 | 0.01 | 13 | 0.01 |
| S12araFGH | ND | ND | NG | NG |
NG, no growth; ND, not determined.
Optimizing xylose utilization by laboratory evolution.
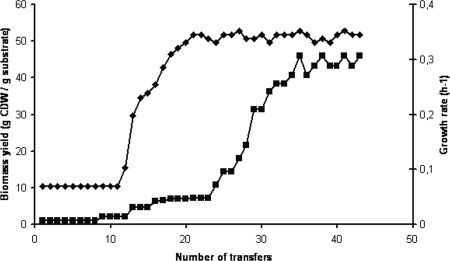
To optimize the biomass yield and growth rate on xylose, a laboratory evolution approach similar to that described by Kuyper et al. was applied (13). Two parallel cultures were maintained and repeatedly transferred to fresh minimal medium with xylose. Throughout, the best performing of the two parallel cultures was used as inoculum for the next transfer while the other was discarded. Three stages could be discriminated in the evolutionary process (Fig. 2). In the first stage, transformant S12xylAB strains were selected for increased biomass yield. When the culture entered the stationary phase, the culture with the highest biomass yield was selected for further evolution. After approximately 20 transfers, the biomass yield stabilized at 52%, comparable to the yield on glucose (55%). Although the growth rate increased with increasing biomass yield, it stabilized when the maximum yield had been achieved, at only 0.05 h−1. Since this was an order of magnitude lower than the growth rate on glucose, the evolutionary approach was continued to select for a strain with a higher growth rate. Instead of transferring the culture with the highest biomass yield, the faster-growing cultures were selected and reinoculated into fresh medium. After 10 transfers, a strain was obtained that exhibited a significantly higher growth rate of 0.35 h−1. At this stage of the evolutionary procedure, the cultures were found to lyse upon entering the stationary phase. Evolution was therefore prolonged to select for a strain that was less prone to lysis. With every transfer, the susceptibility to lysis decreased, ultimately leading to a nonlysing and efficiently xylose-utilizing P. putida S12-derived strain that was designated S12xylAB2.
FIG. 2.
Laboratory evolution of P. putida S12xylAB. Transformant cells were repeatedly transferred into fresh xylose-containing medium in order to optimize biomass yield (diamonds) and growth rate (squares) on MMX.
Characterization of P. putida S12xylAB2.
The increased biomass yield on xylose after laboratory evolution provided a strong indication that xylose oxidation was affected in P. putida S12xylAB2. Indeed, no xylonate formation was observed during growth on xylose, and also, glucose was no longer oxidized to its corresponding aldonic acids. This suggested that Gcd had become inactive during the evolutionary procedure, which was confirmed by the results of Gcd assays. Sequence analysis of the gcd gene showed no mutations, indicating that the absence of active Gcd was caused at a different level. qPCR was used to determine the gcd transcript levels. The mRNA concentration in P. putida S12 cells was 7.00 ± 1.1 ng μl−1 (average ± standard deviation of the results from three independent exponentially growing cultures); that in P. putida S12Δgcd cells was 10.62 ± 2.0 ng μl−1; and that in P. putida S12xylAB2 cells was 9.10 ± 0.8 ng μl−1. Surprisingly, the gcd mRNA level in S12xylAB2 was increased in comparison to that of the wild-type S12 strain, eliminating impaired transcription of gcd as an explanation for the absence of Gcd activity. Also, inefficient translation of gcd mRNA is unlikely to have caused the abolishment of Gcd activity as no mutations were found in the gcd RBS. Therefore, it is proposed that the inactivation of Gcd results from a yet-unidentified posttranslational event.
In order to investigate whether the improved growth characteristics of the evolved strain on xylose could be attributed to the absence of Gcd activity, the gcd gene was disrupted in wild-type P. putida S12. The resulting strain, P. putida S12Δgcd, was transformed with plasmid pJTxylAB to enable xylose utilization. Like P. putida S12xylAB2, strain S12Δgcd_xylAB utilized both glucose and xylose as the sole carbon source without formation of the associated aldonates. Also in strain S12Δgcd_xylAB, in which 107 bp were removed from gcd, the gcd mRNA level was increased with respect to the level in the wild-type S12 strain (see above). Compared to the yield of the nonevolved strain S12xylAB, the yield of strain S12Δgcd_xylAB was considerably improved (44% [g CDW g xylose−1]), but the growth rate was equally low (0.01 h−1). Thus, the absence of Gcd activity explains only part of the improved xylose utilization. Mutations in the xylAB genes of strain S12xylAB2 resulting in a higher xylose conversion rate could be excluded by sequence analysis, which was confirmed by the results of XylAB activity measurements (not shown).
Utilization of arabinose and mixtures of glucose and pentoses by P. putida S12xylAB2.
The evolved strain S12xylAB2 was able to efficiently utilize xylose, as well as glucose, despite the apparent loss of a key enzyme activity for direct oxidative glucose metabolism. For optimal utilization of lignocellulose-derived feedstock, l-arabinose should also be metabolized in addition to glucose and d-xylose. Although strain S12xylAB2 was not specifically engineered for arabinose utilization, the introduced xylose metabolic enzymes may show nonspecific activity toward l-arabinose, a C4 epimer of d-xylose (19, 21). The ability of strain S12xylAB2 to utilize l-arabinose was therefore assessed by growth in mineral salts medium containing 12 mM arabinose. Surprisingly, l-arabinose was utilized as a carbon source at an efficiency identical to that of growth on xylose, with a biomass yield of 52% (g CDW g arabinose−1) and a maximum growth rate of 0.35 h−1.
Strain S12xylAB2 lost the ability to utilize arabinose when cured from pJTxylAB (Table 3), demonstrating that the xylAB genes introduced for xylose consumption were also essential for arabinose consumption. Also, the evolutionary procedure apparently made a key contribution to efficient arabinose consumption since the nonevolved strain S12xylAB did not utilize arabinose (Table 3). The results of previous work demonstrated that P. putida S12 expressing the AraBAD pathway that converts l-arabinose into d-xylulose-5-phosphate did not grow on arabinose unless the high-efficiency arabinose transporter AraFGH was coexpressed (unpublished data). The coexpression of AraFGH and XylAB in wild-type P. putida S12 resulted in an arabinose-utilizing strain, whereas the introduction of araFGH alone did not establish growth on arabinose (Table 3). These results suggest that the evolutionary procedure improved arabinose transport efficiency. The effect of Gcd having become inactive probably plays a minor role, as wild-type P. putida S12 oxidizes arabinose only to a very limited extent (not shown).
The involvement of XylA and XylB in l-arabinose metabolism was further confirmed by the results of enzyme measurements (Table 4). NADH consumption was observed when l-arabinose was added as the substrate instead of d-xylose when assaying xylose isomerase and xylulokinase in the cell extract of strain S12xylAB2. In addition, d-xylulose-5-phosphate from l-arabinose could be detected with the transketolase assay (Table 4). Since the product of l-arabinose formed by XylAB is expected to be l-ribulose-5-phosphate, the formation of d-xylulose-5-phosphate suggests the presence of a C4 epimerase activity that remains to be identified. The observation that the nonevolved strain S12xylAB_araFGH also utilizes l-arabinose suggests that the C4 epimerase activity is endogenous to P. putida S12 and not the result of the evolutionary procedure.
TABLE 4.
Key enzyme activities for pentose utilization in cell extracts of P. putida S12xylAB2 with xylose or arabinose as substratea
| Enzyme | Activity (U g−1) on:
|
|
|---|---|---|
| Xylose | Arabinose | |
| XylA (pentose isomerase) | 34 | 1.5b |
| XylB (“pentulose” kinase) | 134 | NDc |
| XylAB (combined isomerase/kinase) | 22 | 16 |
| Putative C4 epimerased | ND | 194 |
ND, not determined.
The activity assay of XylA with l-arabinose was compromised by the low affinity of sorbitol dehydrogenase for l-ribulose.
The activity of XylB was not determined separately as l-ribulose is not commercially available.
The formation of d-xylulose-5-phosphate from l-arabinose was quantified by measuring transketolase activity.
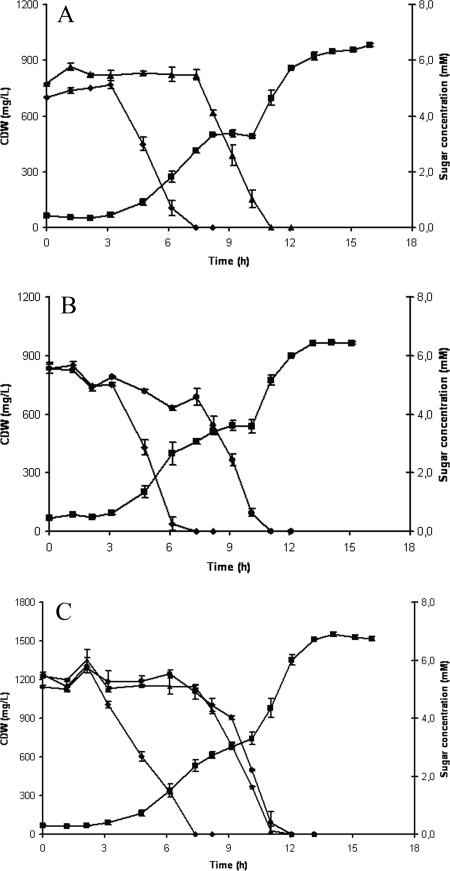
Finally, the growth of strain S12xylAB2 on mixtures of sugars was investigated. Cells were inoculated into mineral salts medium containing glucose and xylose (MMGX); glucose and arabinose (MMGA); or glucose, xylose, and arabinose (MMGXA). Timed samples were drawn and analyzed for CDW and sugar content. The results show that all sugars in the tested combinations are consumed (Fig. 3). A diauxic shift was observed in all cultures: only after glucose was depleted were the pentoses consumed. When both xylose and arabinose were present in addition to glucose, the pentose sugars were utilized simultaneously when cells were deprived of glucose.
FIG. 3.
Growth of P. putida S12xylAB2 (CDW is represented by squares) and consumption of glucose (diamonds), xylose (triangles), and arabinose (circles). P. putida S12xylAB2 was inoculated into mineral salts medium containing glucose and xylose (A); glucose and arabinose (B); or glucose, xylose, and arabinose (C). Data points are the averages of the results of duplicate measurements. Error bars represent the maximum deviations of the averages. L, liter.
DISCUSSION
A P. putida S12 strain was constructed that efficiently utilizes d-xylose, as well as l-arabinose. The expression of xylose isomerase and xylulokinase is essential for the utilization of both pentoses, but the subsequent laboratory evolution is key to the efficiency with which these pentoses are metabolized. The improved yield on xylose attained by the evolved strain could largely be attributed to Gcd having become inactive in the evolved strain, preventing xylose “loss” as a result of oxidation to xylonate. Although targeted disruption of the gcd gene in wild-type P. putida S12 resulted in an improved biomass yield on xylose, this strategy did not result in an improved growth rate, indicating that other changes occurred in the evolved strain. It may be speculated that mutations occurred that affected the metabolic fluxes through the PP pathway, which is the expected route by which xylose is metabolized. In P. putida, a complete PP pathway is present, but metabolic flux analyses on Pseudomonas fluorescens have shown that this pathway mainly serves to replenish biosynthetic intermediates (4). Similarly, modest flux distributions have been demonstrated for P. putida S12 (unpublished data). Therefore, the function of the PP pathway may have changed from anabolic to catabolic in the evolved P. putida S12xylAB2 strain.
The observation that d-xylose and l-arabinose are consumed with equal efficiency, both in terms of biomass yield and specific growth rate, suggests that both pentoses are consumed via the PP pathway. In addition, the evolved strain appears to have acquired an efficient l-arabinose uptake system, as wild-type P. putida S12 required the expression of both XylAB and the AraFGH transporter for arabinose utilization. Although coexpressing a high-affinity xylose transporter (XylFGH from E. coli) did not have a significant effect on xylose metabolism in the nonevolved strain (Table 3), the possibility that improved xylose uptake has contributed to more-efficient xylose utilization in strain S12xylAB2 cannot be excluded. Since pentose transporters have been shown to be promiscuous (24), it may be hypothesized that efficient l-arabinose uptake has coevolved with improved d-xylose uptake in strain S12xylAB2.
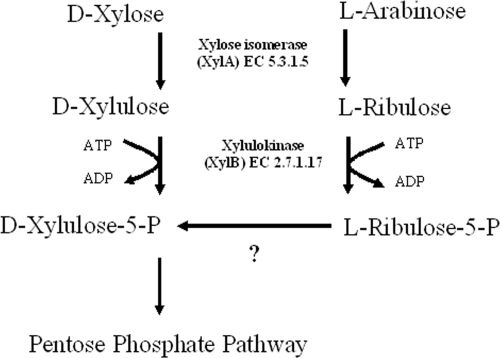
At this point it is unclear how arabinose is converted into a PP pathway intermediate (Fig. 4). The results of enzyme assays showed that l-arabinose is a substrate for XylAB, and growth on arabinose did not occur without the expression of XylAB. The expected product, l-ribulose-5-phosphate, is not a central pathway intermediate, and a C4 epimerization would be required to form d-xylulose-5-phosphate. Indeed, the formation of d-xylulose-5-phosphate from l-arabinose was suggested by the results of the transketolase assay, but no indications that this strain contains an AraD homologue were found in the P. putida S12 genome sequence (unpublished data). Therefore, it is proposed that the endogenous ribulose-5-phosphate-3-epimerase shows nonspecific epimerization activity on l-ribulose-5-phosphate, converting the molecule into d-xylulose-5-phosphate. Further research is ongoing to confirm this hypothesis.
FIG. 4.
Utilization of d-xylose and l-arabinose may proceed via (partly) shared pathways. Indications were found that l-arabinose can be converted into d-xylulose-5-phosphate by the combined action of XylAB and a yet-unidentified C4 epimerase (indicated by the question mark).
Despite the loss of Gcd activity, glucose was still efficiently used as the sole carbon source by the evolved P. putida S12xylAB2. The glucose catabolism in P. putida operates through the action of three simultaneous pathways that converge at 6-phosphogluconate. Glucose is preferentially oxidized in the periplasm by Gcd to (2-keto-)gluconate and subsequently phosphorylated in the cytoplasm to yield 6-phosphogluconate (2, 15). Alternatively, glucose is imported by an ABC-transport system, phosphorylated by glucokinase, and oxidized to 6-phosphogluconate (2, 15). Apparently, the evolved xylose-utilizing strain can readily switch to this alternative pathway for glucose oxidation without affecting the yield or growth rate.
The absence of active Gcd may itself provide an explanation for the increased gcd transcription levels observed in both the evolved strain and the gcd knockout strain. Glucose induces the expression of gcd (27), and with active Gcd present, glucose is rapidly oxidized to gluconate and 2-ketogluconate, resulting in a swift downregulation of gcd. However, without active Gcd, glucose persists in the medium, resulting in increased levels of gcd mRNA. So, with an intact gcd gene and associated RBS present in the evolved strain, and apparently even increased transcription levels, the absence of active Gcd must be attributed to some posttranslational effect. The amino acid sequence of Gcd shows that the protein is excreted and that it contains four transmembrane regions (not shown), which is consistent with the periplasmic oxidation of sugars. It raises the possibility that malfunctions appear in the Gcd translocation machinery, leading to faulty localization of the enzyme, improper folding, or inadequate anchoring to the inner membrane. The exact cause of the inactivity of Gcd remains to be investigated.
In conclusion, a P. putida S12 strain was obtained that efficiently utilizes the three most-abundant sugars in lignocellulose, glucose, xylose, and arabinose, as sole carbon sources. The applied evolutionary approach proved to be a powerful method to optimize the initial inefficient xylose-utilizing strain. Transcriptome and proteome analyses, as well as metabolic flux analysis, are currently being performed to identify the changes in the metabolism of the evolved xylose-utilizing strain. The insight gained into the molecular background of the efficient pentose utilization will be employed to incorporate this property into optimized substitute-aromate-producing P. putida S12-derived strains, thereby contributing to the economical feasibility of the production of such biochemicals from renewable feedstock.
Acknowledgments
We thank Nicole van Luijk for developing the markerless gene disruption system in P. putida S12, Hendrik Ballerstedt for performing qPCR experiments, Maaike Westerhof for practical assistance, and Jan Wery for helpful discussions.
This project was financially supported by The Netherlands Ministry of Economic Affairs and the B-Basic partner organizations (www.b-basic.nl) through B-Basic, a public-private NWO-ACTS (Advanced Chemical Technologies for Sustainability) program.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.de Bont, J. A. 1998. Solvent-tolerant bacteria in biocatalysis. Trends Biotechnol. 16:493-499. [Google Scholar]
- 2.del Castillo, T., J. L. Ramos, J. J. Rodríguez-Herva, T. Fuhrer, U. Sauer, and E. Duque. 2007. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J. Bacteriol. 189:5142-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eliasson, A., E. Boles, B. Johansson, M. Österberg, J. M. Thevelein, I. Spencer-Martins, H. Juhnke, and B. Hahn-Hägerdal. 2000. Xylulose fermentation by mutant and wild-type strains of Zygosaccharomyces and Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 53:376-382. [DOI] [PubMed] [Google Scholar]
- 4.Fuhrer, T., E. Fischer, and U. Sauer. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187:1581-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao, Q., M. Zhang, J. D. McMillan, and D. S. Kompala. 2002. Characterization of heterologous and native enzyme activity profiles in metabolically engineered Zymomonas mobilis strains during batch fermentation of glucose and xylose mixtures. Appl. Biochem. Biotechnol. 98-100:341-355. [DOI] [PubMed] [Google Scholar]
- 6.Hahn-Hägerdal, B., K. Karhumaa, M. Jeppsson, and M. F. Gorwa-Grauslund. 2007. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108:147-177. [DOI] [PubMed] [Google Scholar]
- 7.Hardy, G. P., M. J. Teixeira de Mattos, and O. M. Neijssel. 1993. Energy conservation by pyrroloquinoline quinol-linked xylose oxidation in Pseudomonas putida NCTC 10936 during carbon-limited growth in chemostat culture. FEMS Microbiol. Lett. 107:107-110. [DOI] [PubMed] [Google Scholar]
- 8.Hartmans, S., J. P. Smits, M. J. van der Werf, F. Volkering, and J. A. M. de Bont. 1989. Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl. Environ. Microbiol. 55:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmans, S., M. J. van der Werf, and J. A. de Bont. 1990. Bacterial degradation of styrene involving a novel flavin adenine dinucleotide-dependent styrene monooxygenase. Appl. Environ. Microbiol. 56:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawaguchi, H., M. Sasaki, A. A. Vertes, M. Inui, and H. Yukawa. 27 October 2007. Engineering of an l-arabinose metabolic pathway in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. doi: 10.1007/s00253-007-1244-x. [DOI] [PubMed]
- 11.Kawaguchi, H., A. A. Vertès, S. Okino, M. Inui, and H. Yukawa. 2006. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 72:3418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuyper, M., H. R. Harhangi, A. K. Stave, A. A. Winkler, M. S. Jetten, W. T. de Laat, J. J. den Ridder, H. J. Op den Camp, J. P. van Dijken, and J. T. Pronk. 2003. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 4:69-78. [DOI] [PubMed] [Google Scholar]
- 13.Kuyper, M., M. J. Toirkens, J. A. Diderich, A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2005. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 5:925-934. [DOI] [PubMed] [Google Scholar]
- 14.Lee, J. 1997. Biological conversion of lignocellulosic biomass to ethanol. J. Biotechnol. 56:1-24. [DOI] [PubMed] [Google Scholar]
- 15.Lessie, T. G., and P. V. Phibbs, Jr. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38:359-388. [DOI] [PubMed] [Google Scholar]
- 16.Liu, Q., J. R. Kirchhoff, C. R. Faehnle, R. E. Viola, and R. A. Hudson. 2006. A rapid method for the purification of methanol dehydrogenase from Methylobacterium extorquens. Protein Expr. Purif. 46:316-320. [DOI] [PubMed] [Google Scholar]
- 17.Nijkamp, K., N. van Luijk, J. A. de Bont, and J. Wery. 2005. The solvent-tolerant Pseudomonas putida S12 as host for the production of cinnamic acid from glucose. Appl. Microbiol. Biotechnol. 69:170-177. [DOI] [PubMed] [Google Scholar]
- 18.Nijkamp, K., R. G. Westerhof, H. Ballerstedt, J. A. de Bont, and J. Wery. 2007. Optimization of the solvent-tolerant Pseudomonas putida S12 as host for the production of p-coumarate from glucose. Appl. Microbiol. Biotechnol. 74:617-624. [DOI] [PubMed] [Google Scholar]
- 19.Pastinen, O., K. Visuri, H. E. Schoemaker, and M. Leisola. 1999. Novel reactions of xylose isomerase from Streptomyces rubiginosus. Enzyme Microb. Technol. 25:695-700. [Google Scholar]
- 20.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 21.Richard, P., M. H. Toivari, and M. Penttilä. 2000. The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol. Lett. 190:39-43. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., T. Maniatis, and E. Fritsch. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 23.Sauer, B., and N. Henderson. 1988. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 85:5166-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song, S., and C. Park. 1998. Utilization of D-ribose through D-xylose transporter. FEMS Microbiol. Lett. 163:255-261. [DOI] [PubMed] [Google Scholar]
- 25.Sternberg, N., and D. Hamilton. 1981. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 150:467-486. [DOI] [PubMed] [Google Scholar]
- 26.Verhoef, S., H. J. Ruijssenaars, J. A. de Bont, and J. Wery. 2007. Bioproduction of p-hydroxybenzoate from renewable feedstock by solvent-tolerant Pseudomonas putida S12. J. Biotechnol. 132:49-56. [DOI] [PubMed] [Google Scholar]
- 27.Vicente, M., and J. L. Cánovas. 1973. Regulation of the glucolytic enzymes in Pseudomonas putida. Arch. Microbiol. 93:53-64. [DOI] [PubMed] [Google Scholar]
- 28.Wery, J., B. Hidayat, J. Kieboom, and J. A. de Bont. 2001. An insertion sequence prepares Pseudomonas putida S12 for severe solvent stress. J. Biol. Chem. 276:5700-5706. [DOI] [PubMed] [Google Scholar]
- 29.Wierckx, N. J., H. Ballerstedt, J. A. de Bont, and J. Wery. 2005. Engineering of solvent-tolerant Pseudomonas putida S12 for bioproduction of phenol from glucose. Appl. Environ. Microbiol. 71:8221-8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wisselink, H. W., M. J. Toirkens, M. D. Franco Berriel, A. A. Winkler, J. P. van Dijken, J. T. Pronk, and A. J. van Maris. 2007. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl. Environ. Microbiol. 73:4881-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, M., C. Eddy, K. Deanda, M. Finkelstein, and S. Picataggio. 1995. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240-243. [DOI] [PubMed] [Google Scholar]