Abstract
A continuous-flow porcine cecal bacterial culture has been used experimentally as treatment against enterotoxigenic Escherichia coli in weanling pigs. Periodically, the cultures must be started from frozen stock. Our results indicate that denaturing gradient gel electrophoresis can be applied as an indirect indication of culture similarity for each new batch generated from frozen stock.
Beneficial commensal gastrointestinal bacteria provide protection in young pigs against enteropathogens and present barriers to host cell invasion. Dietary changes and weaning impose stresses that may disrupt commensal populations and promote colonization with enterotoxigenic Escherichia coli (ETEC) (11). Colonization with ETEC can disrupt nutrient absorption accompanied by diarrhea. Antibiotic treatment presents the potential for the emergence of antibiotic-resistant organisms as well as the disruption of the normal digestive microflora (1, 3).
A porcine continuous-flow (PCF) culture been proven to be effective against ETEC in weanling pigs (2, 5-8). Periodically, new PCF cultures must be started from frozen stock. The objective of the current study was to examine the usefulness of denaturing gradient gel electrophoresis (DGGE) to determine similarities of PCF cultures derived at different times from frozen stock. In three separate trials, frozen stock from a previously established PCF culture was used as a starter culture for new PCF cultures. Triplicate samples were collected from each culture at 0, 4, 8, and 12 h and then at 24-h intervals from 0 h for the next 9 days. Samples were used to determine aerobic and anaerobic CFU/ml, pH, lactic acid, and volatile fatty acid (acetic, propionic, and butyric acids) concentrations (10) and DGGE profiles (9, 12).
Facultative bacterial populations at 0 h were 5.65, 5.72, and 5.72 log10 CFU/ml for trials 1, 2, and 3, respectively. Cell counts at 10 days were 6.28, 6.77, and 6.81 log10 CFU/ml in the three trials, respectively. Total anaerobe log10 CFU/ml increased from 6.61, 6.08, and 7.23 at 0 h to 8.38, 8.11, and 8.66, respectively, by 10 days. Other than indicating that the cultures are growing well, cell counts do not tell too much about the comparative qualities of culture in each trial. Culture pH at 1 day in trial 3 was 5.3 compared to 5.7 and 5.7, respectively, for trials 1 and 2. By 10 days, pH stabilized to 5.9, 5.8, and 6.1, respectively, for trials 1, 2, and 3.
Acetic acid levels were the highest of the three volatile fatty acids examined. Average levels for the three cultures ranged from 23.30 to 43.83 μmol/ml for the full 10 days. The propionic acid level was lower, and averages for the full 10 days for the three cultures ranged from 4.30 to 14.78 μmol/ml. Butyric acid averages in the three trials by 10 days ranged from 7.76 to 15.93 μmol/ml. Lactic acid concentrations for the three trials varied during the 10 days starting at 0 h at 21.7, 19.7, and 3.4 μmol/ml and stabilizing at 25.4, 27.1, and 2.8 μmol/ml by 10 days, respectively, for trials 1, 2, and 3. Interestingly, lactic acid levels were measured at 1 day, when the levels in trial 3 were 169 μmol/ml, while levels in the other two trials remained relatively modest. This difference alone hinted that the culture in trial 3 was somewhat different from those in the other trials.
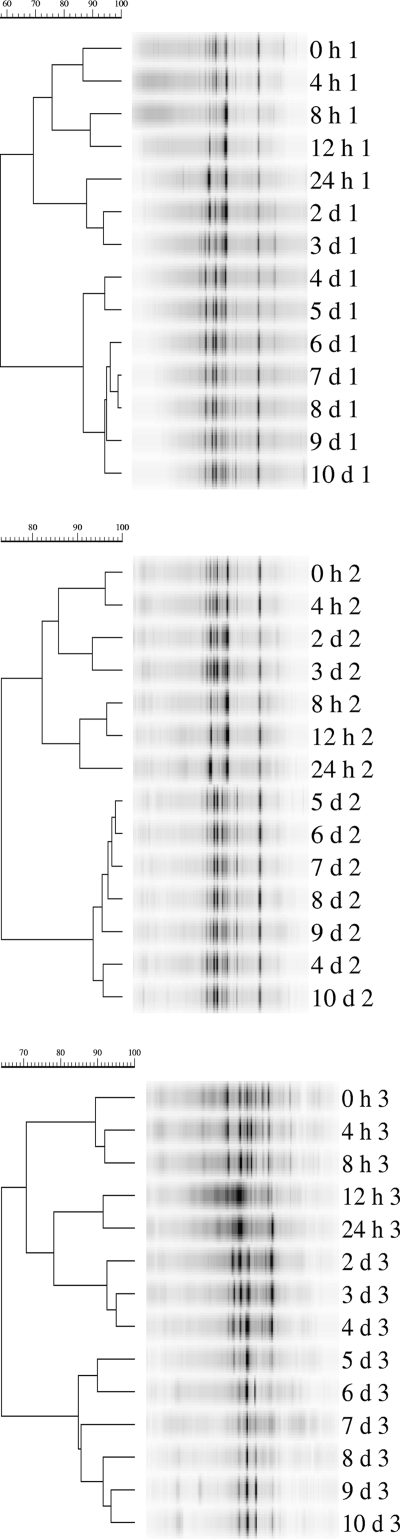
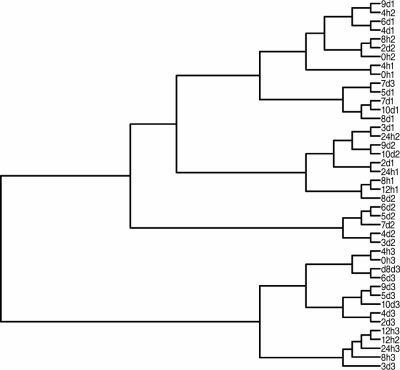
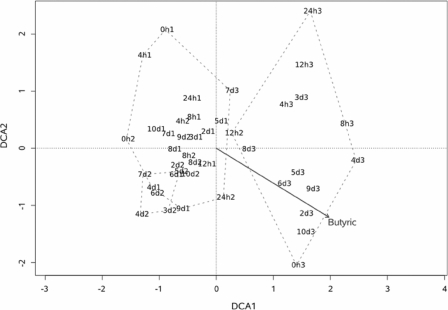
Genomic DNA for DGGE was isolated from 1 ml of each sample with a QIAamp DNA mini kit (Qiagen, Valencia, CA) according to the method described in the kit. DGGE was run according to the method of Muyzer et al. (12), with modification (9), using PCR primers to conserved regions flanking the variable V3 region of 16S ribosomal DNA (12). Fragment pattern relatedness was determined with molecular analysis fingerprinting software, version 1.6 (Bio-Rad Laboratories, Hercules, CA), based on the Dice similarity coefficient and the unweighted-pair group method using arithmetic averages for clustering (Fig. 1). The DGGE profiles were digitized and converted to a binary matrix using a set of scripts written in the Python programming language (Python Software Foundation, Hampton, NH). The data analysis was carried out with the R statistical software (Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org) using various R-packages, the similarity matrix was computed based on Sorensen's coefficient using the ade4 package (4), and clustering and correspondence analysis were carried out using the vegan package (J. Oksanen, R. Kindt, P. Legendre, B. O'Hara, M. Henry, and H. Stevens, Community ecology package, version 1.8-8; http://cran.r-project.org/ and http://r-forge.r-project.org/projects/vegan/). The significance of clustering was tested using TreeClimber (15). A cladogram was obtained from the similarity matrix, which was calculated according to the Sorensen coefficient, 2A/(2A + B + C), where A is the number of bands that occur in both lanes, B is the number of bands found only in the first lane, and C is the number of bands found only in the second lane. The resulting cluster is divided into three major clades: the first contains all of the samples from trial 3 and includes the 12-h sample from trial 2, the second contains samples for 2 to 7 days from trial 2, and the third contains all of the remaining samples from trials 1 and 2 and includes the 7-day sample from trial 3. To assess the reliability of the observed clusters, a parsimony test was performed using the TreeClimber software. The analysis showed that there were significant differences among the community structures of the three trial groups, while there were no significant differences observed among time groups. A detrended correspondence analysis was conducted to produce a two-dimensional plot of the major variation in the data set and displayed the results of clustering on the ordination space. Cluster 1 and cluster 2 overlap, and points in those groups are very close, which means that the profiles are very similar (Fig. 2). Profiles from trial 3 occupy a distinct space on the ordination diagram. The various environmental parameters were overlaid on the ordination space to determine if there was a correlation between the environmental parameters and DGGE profiles (Fig. 3). The most significant parameter was butyric acid. A Mantel test was conducted comparing the similarity matrix of DGGE profiles with the distance matrix of the environmental parameters. The trials showed no correlation with environmental parameters. Additionally, Mantel tests were conducted with each environmental parameter separately and analyses showed a significant correlation with butyric acid for trial 1 and for trial 3.
FIG. 1.
Dendrograms for trials 1 (top panel), 2 (middle panel), and 3 (bottom panel) for swine cecal bacterial continuous-flow chemostat cultures.
FIG. 2.
Cladogram from the similarity matrix calculated according to the Sorensen coefficient.
FIG. 3.
Detrended correspondence analysis (DCA) of the major variation in the data set and the results of clustering on the ordination space (The first number indicates the time in hours or days. The second number indicates trial 1, 2, or 3).
The results indicate that DGGE band analysis can be applied as an indirect indication of culture similarity for each new batch generated from frozen stock. Nisbet et al. (13, 14) introduced the idea of maintaining commensal probiotic cultures as continuous-flow cultures. Decreased batch-to-batch variation and the opportunity to identify culture microbial components made the technology more appropriate for continued use and efficacy than cultures freshly acquired from changing animal sources. Routine fluctuations in culture characteristics may be misinterpreted, and valuable time and resources may be sacrificed before a decision (possibly incorrect) can be made to terminate the culture. These results indicate that DGGE holds the potential to shorten the decision-making time between a putative contaminating event and halting production of a new continuous-flow subculture.
Acknowledgments
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Amezcua, R., R. M. Friendship, C. E. Dewey, C. Gyles, and J. M. Fairbrother. 2002. Presentation of postweaning Escherichia coli diarrhea in southern Ontario, prevalence of hemolytic E coli serogroups involved and their antimicrobial resistance patterns. Can. J. Vet. Res. 66:73-78. [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. C., L. H. Stanker, C. R. Young, S. A. Buckley, K. J. Genovese, R. B. Harvey, J. R. DeLoach, N. K. Keith, and D. J. Nisbet. 1999. Effect of competitive exclusion treatment on colonization of early-weaned pigs by Salmonella serovar Choleraesuis. J. Swine Health Prod. 7:155-160. [Google Scholar]
- 3.Casewell, M., C. Friis, E. Marco, P. McMullin, and I. Phillips. 2003. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 52:159-161. [DOI] [PubMed] [Google Scholar]
- 4.Chessel, D., A.-B. Dufour, and J. Thioulouse. 2004. The ade4 package. I. One-table methods. R News 4:5-10. [Google Scholar]
- 5.Genovese, K. J., R. C. Anderson, T. R. Callaway, T. L. Poole, T. S. Edrington, P. J. Fedorka-Cray, and D. J. Nisbet. 2003. Competitive exclusion of Salmonella from the gut of neonatal and weaned pigs. J. Food Prot. 66:1353-1359. [DOI] [PubMed] [Google Scholar]
- 6.Genovese, K. J., R. C. Anderson, R. B. Harvey, and D. J. Nisbet. 2000. Competitive exclusion treatment reduces the mortality and fecal shedding associated with entertoxigenic Escherichia coli infection in nursery-raised neonatal pigs. Can. J. Vet. Res. 64:204-207. [PMC free article] [PubMed] [Google Scholar]
- 7.Genovese, K. J., R. B. Harvey, R. C. Anderson, and D. J. Nisbet. 2001. Protection of suckling neonatal pigs against infection with an enterotoxigenic Escherichia coli expressing 987P fimbriae by the administration of a bacterial competitive exclusion culture. Microb. Ecol. Health Dis. 13:223-228. [Google Scholar]
- 8.Harvey, R. B., R. C. Anderson, K. J. Genovese, T. R. Callaway, and D. J. Nisbet. 2005. Use of competitive exclusion to control enterotoxigenic strains of Escherichia coli in weaned piges. J. Anim. Sci. 83(Suppl. E):E44-E47. [Google Scholar]
- 9.Hume, M. E., L. F. Kubena, T. S. Edrington, C. J. Donskey, R. W. Moore, S. C. Ricke, and D. J. Nisbet. 2003. Poultry digestive microflora biodiversity as indicated by denaturing gradient gel electrophoresis. Poult. Sci. 82:1100-1107. [DOI] [PubMed] [Google Scholar]
- 10.Hume, M. E., D. J. Nisbet, S. A. Buckley, R. L. Ziprin, R. C. Anderson, and L. H. Stanker. 2001. Inhibition of in vitro Salmonella typhimurium colonization in porcine cecal bacteria continuous-flow competitive exclusion cultures. J. Food Prot. 64:17-22. [DOI] [PubMed] [Google Scholar]
- 11.Kyriakis, S. C., V. K. Tsiloyiannis, J. Vlemmas, K. Sarris, A. C. Tsinas, C. Alexopoulos, and L. Jansegers. 1999. The effect of probiotic LSP 122 on the control of post-weaning diarrhea syndrome of piglets. Res. Vet. Sci. 67:223-228. [DOI] [PubMed] [Google Scholar]
- 12.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nisbet, D. J., D. E. Corrier, and J. D. DeLoach. 1993. Effect of mixed cecal microflora maintained in continuous culture and of dietary lactose on Salmonella Typhimurium colonization in broiler chicks. Avian Dis. 37:528-535. [PubMed] [Google Scholar]
- 14.Nisbet, D. J., D. E. Corrier, S. C. Ricke, M. E. Hume, J. A. Byrd II, and J. R. DeLoach. 1996. Maintenance of the biological efficacy in chicks of a cecal competitive-exclusion culture against Salmonella by continuous-flow fermentation. J. Food Prot. 59:1279-1283. [DOI] [PubMed] [Google Scholar]
- 15.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]