Abstract
Environmental remediation efforts often utilize either biodegradative microbes or surfactants, but not in combination. Coupling both strategies holds the potential to dramatically increase the rate and extent of remediation because surfactants can enhance the bioavailability of contaminants to microbes. However, many surfactants permeabilize bacterial cell membranes and are effective disinfectants. An important goal then is to find or genetically modify microorganisms that possess both desirable degradative capabilities and the ability to thrive in the presence of surfactants. The guts of some marine invertebrates, particularly deposit feeders, have previously been shown to contain high levels of biosurfactants. Our primary aim was to mine these natural, surfactant-rich habitats for surfactant-resistant bacteria. Relative to sediment porewaters, the gut contents of two polychaete deposit feeders, Nereis succinea and Amphitrite ornata, exhibited a significantly higher ratio of bacteria resistant to both cationic and anionic surfactants. In contrast, bacteria in the gut fluids of a holothuroid, Leptosynapta tenuis, showed surfactant susceptibility similar to that of bacteria from sediments. Analyses of 16S rRNA gene sequences revealed that the majority of surfactant-resistant isolates were previously undescribed species of the genus Vibrio or were of a group most closely related to Spongiobacter spp. We also tested a subset of resistant bacteria for the production of biosurfactants. The majority did produce biosurfactants, as demonstrated via the oil-spreading method, but in all cases, production was relatively weak under the culture conditions employed. Novel surfactant-resistant, biosurfactant-producing bacteria, and the habitats from which they were isolated, provide a new source pool for potential microorganisms to be exploited in the in situ bioremediation of marine sediments.
In situ bioremediation strategies may involve the stimulation of native bacterial strains or the introduction of nonnative microorganisms. These microbes then transform contaminants into non- or less-hazardous chemicals. Despite such efforts, the prolonged persistence of hydrophobic organic pollutants in the environment is common due to their solubilization-limited bioavailability. A wide array of bacteria have been isolated that are capable of mineralizing otherwise persistent toxic chemicals in situ (24). However, the efficacy of microbe-driven bioremediation is limited by at least three factors: (i) the bioavailability of the toxic substrate to be degraded, (ii) the ease with which the toxic substrate can diffuse or be taken up by the degrading microorganisms, and (iii) the capability of the degrading microorganisms to survive in the polluted environment (24).
A possible means to enhance the availability of contaminants is the application of surfactants. Surfactants are amphipathic molecules with both hydrophilic and hydrophobic moieties that partition preferentially at the interface between the fluid phases of different polarities, such as oil/water. These properties render surfactants capable of reducing surface and interfacial tension and lead to the formation of microemulsions in which hydrocarbons can solubilize in water. Surfactants can increase the surface area of hydrophobic, water-insoluble growth substrates, increasing their bioavailability by raising their solubility or desorbing them from surfaces (24). At a sufficient concentration, known as the critical micelle concentration (CMC), surfactants form spherical aggregations of molecules in solution. Micelles can surround and sequester hydrocarbons and other hydrophobic compounds, increasing their solubility in water (11). However, a prerequisite for surfactant-enhanced biodegradation is that the degradative microorganisms not be adversely affected by the surfactant. Because of their amphipathic nature, many surfactants permeabilize bacterial cell membranes and are effective disinfectants (9), although particular bacterial strains can employ a variety of mechanisms (e.g., cell impermeability, efflux pumps, and surfactant degradation) to counter these negative effects (23). Thus, the net effect of surfactants on biodegradation is variable (2, 24).
An important goal then is to find bacteria with both the desired biodegradative capabilities and the ability to thrive in the presence of surfactants. Alternatively, if resistant bacteria do not possess the desired degradative abilities, genetic engineering can be used to add biodegradative capabilities (or surfactant resistance genes could be added to degraders). The guts of some marine invertebrates, particularly deposit feeders, from shallow waters off the coasts of Maine and Washington have previously been shown to contain high levels of biosurfactants (19). The functions of these gut surfactants are unknown, but they likely contribute to digestion by aiding cell lysis or through the desorption of organics from sedimentary particles.
Biosurfactants have several advantages over the chemically synthesized surfactants typically used for bioremediation. Some benefits include lower toxicity, higher biodegradability, and high selectivity and specific activity at extreme temperatures, pHs, and salinities (11). Although it has been documented that deposit-feeder guts are surfactant rich (18), the source of these surfactants is unclear (1). It is possible that the deposit feeders themselves secrete the surfactants, but another potential source is symbiotic gut bacteria.
The primary aim of the present study was to mine natural, surfactant-rich marine habitats for surfactant-resistant bacteria in order to isolate bacteria applicable in bioremediation. We also tested resistant bacteria for the production of biosurfactants.
MATERIALS AND METHODS
Sample collection and preparation.
During three sampling periods, Dec 2005/Jan 2006, June/July 2006, and Jan 2007, two species of deposit feeders were collected and sampled: Amphitrite ornata Leidy (Polychaeta: Terebellidae) and Nereis succinea Frey and Leuckart (Polychaeta: Nereididae). Leptosynapta tenuis Verrill (Holothuroidea: Synaptidae) was also collected during the June 2006 sampling period. A. ornata and N. succinea were gathered at Grice Cove, adjacent to the Grice Marine Laboratory, Charleston Harbor, SC (32°45.1′N, 79°53.9′W). L. tenuis was collected at Breach Inlet, between Sullivan's Island and Isle of Palms, SC (32°46.6′N, 79°48.7′W). All invertebrates were collected by digging with a garden fork or shovel during low tide. Groups of deposit feeders were kept alive in seawater-filled buckets until after transport to the laboratory for dissection. In addition, surficial (<0.5-cm depth) sediment samples were collected with sterile spatulas from the animal collection sites on all sampling dates.
Under a stereomicroscope, the midsection of each animal was opened with a shallow incision. Gut fluid was withdrawn directly from the alimentary canal using a hypodermic needle and syringe. Approximately 20-μl volumes of gut fluid samples, typically obtained from two or three animals, were placed in sterile 1.5-ml microcentrifuge tubes. Both the gut and sediment samples were centrifuged for 5 min at 1,500 rpm (<200 × g) to accumulate fluids from the sides of the microcentrifuge tubes and to separate the fluid from the sediment pellet.
Surfactant concentrations.
Both the gut fluid and sediment interstitial fluid samples were screened for surfactant concentration using the drop-collapse assay previously described by Van Der Vegt and coworkers (31). Briefly, the assay detects the decrease in surface tension caused by the presence of surfactants by measuring the degree to which the shape of a droplet of liquid placed on a nonpolar surface is flattened. A 5-μl drop of each gut and sediment fluid sample was pipetted onto a glass slide wrapped in Parafilm. The drop profiles were captured from above using a Model 1000 VersaDoc imaging system (Bio-Rad Laboratories, Hercules, CA) and a mirror placed at a 45° angle to the glass slide or were photographed from the side using a 6.1-megapixel Kodak EasyShare DX7630 digital camera. The relative surfactant levels were determined by measuring each droplet's height and radius using the ruler function in Adobe Photoshop 7.0 for Mac OS X (Adobe Systems, San Jose, CA) and comparing the height-to-radius (h/r) ratios.
Enumeration of surfactant-resistant bacteria.
Both gut and sediment interstitial fluid samples were serially diluted to 10−2 and 10−3 with autoclaved seawater. Duplicate 100-μl aliquots of each dilution were spread on plates containing nonselective marine peptone-yeast extract (PY) medium (0.1% yeast extract, 0.01% peptone, and 1.5% agar in sterile seawater) and on selective PY medium plates containing added surfactant. The plates were incubated aerobically at 22 to 24°C.
The three surfactants added to the separate selective plates were the cationic surfactant cetyltrimethylammonium bromide (CTAB) at concentrations of 100 μM and 2 mM, the anionic surfactant sodium dodecyl sulfate (SDS) at concentrations of 500 μg/ml and 2.5 mg/ml, and the nonionic surfactant Triton X-100 at 0.1, 1, and 5%. In January 2006, only the CTAB (both concentrations) and Triton X-100 (0.1 and 1%) plates were employed. These three surfactants were chosen to represent a common synthetic surfactant from each of the three surfactant types: cationic, anionic, and nonionic. The particular concentrations were chosen such that the lower concentration was below the CMC and the higher was above the CMC.
Identification of surfactant-resistant bacteria.
The major colony morphologies on selective and nonselective plates were characterized with regard to shape, color, size, and margin. The most common colony morphologies (colonies on surfactant plates typically appeared to be monocultures), in addition to select unique colonies, were picked and streaked three times in succession on nonselective PY plates to ensure purity. Broth cultures (1% peptone, 0.5% yeast extract, dissolved in seawater) were inoculated with single colonies of the purified strains. Genomic DNA was extracted from the cultures using the standard phenol-chloroform method and resuspended in Tris-EDTA buffer, as previously described by Neumann et al. (22).
Utilizing PCR primers 1492r and 530f, an approximately 900-bp segment of the 16S rRNA genes of the DNA extracts was amplified by PCR. The PCR protocol has been previously described in greater detail (25). Sequencing of the PCR products was completed using a Beckman Coulter dye terminator cycle quick start kit. Following the cycle sequencing, DNA samples were sequenced by capillary electrophoresis on a Beckman Coulter CEQ 8000 automated DNA sequencer. For the presumptive identification of the isolated bacteria, the sequences of the PCR products were compared to those in GenBank using the BLAST function of the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov).
A phylogenetic tree was generated using the neighbor-joining method with MEGA 4 (30), based on alignments from CLUSTAL W (8).
Biosurfactant production assays.
A subset of bacterial isolates were grown in PY broth at 25°C on a shaker table (∼100 rpm). After 24 h, aliquots of the broth were withdrawn and tested for the production of biosurfactants using the measurement of the h/r ratio of the culture supernatants (as described above) or employing the oil-spreading technique described by Youssef et al. (34). In the oil-spreading method, 25 ml of distilled water was poured into a 15-mm by 100-mm petri dish, and then 30 μl of decane reference oil (Acros Organics, Morris Plains, NJ) was added onto the water surface. Next, 10 μl of culture supernatant was delicately pipetted onto the center of the oil surface. Around each drop of supernatant, the diameter of the cleared, halo region on the oil surface was measured. Each isolate was tested in triplicate. Biosurfactant production was measured as a decrease in surface tension, as indicated by an increase in the diameter of the clear zone on the oil surface around the culture supernatant compared to that around the uninoculated broth control.
Data analysis.
t tests or one-way analyses of variance were used to compare the surfactant levels and the abundance of surfactant-resistant bacteria in gut fluid versus sediment and to test for biosurfactant production.
Nucleotide sequence accession numbers.
Sequences were deposited in GenBank under accession numbers EU709812 to EU709817 and EU797578 to EU797596.
RESULTS
Surfactant levels.
Compared with the Grice Cove sediment porewaters, the gut fluid samples from both A. ornata and N. succinea flattened and spread out more when placed on the nonpolar Parafilm surface (P values of <0.001 with respect to both species and season; Table 1), indicating that the gut contents of both species had significantly greater surfactant levels than the porewaters. A. ornata also had significantly greater surfactant levels than N. succinea (P < 0.001). A significant seasonal effect was noted in the A. ornata gut samples, with higher surfactancy (lower h/r ratio) in winter (P = 0.025), whereas seasonal differences were not significant for N. succinea fluids or sediments (P values of 0.481 and 0.152, respectively). In contrast to the other two deposit feeders, L. tenuis gut fluid did not exhibit higher surfactancy than sediment porewater (P = 0.160 compared to Breach Inlet sediment; Table 1).
TABLE 1.
Mean h/r ratios (± standard errors of the means) of 5-μl drops of gut and sediment fluid on Parafilma
| Sample source | Mean h/r ratio (±SEM)
|
|
|---|---|---|
| Winter 2006 | Summer 2006 | |
| N. succinea gut | 0.42 (0.05) | 0.55 (0.02) |
| A. ornata gut | 0.17 (0.01) | 0.32 (0.01) |
| Grice Cove sediment | 1.13 (0.01) | 1.18 (0.03) |
| L. tenuis gut | ND | 0.71 (0.02) |
| Breach Inlet sediment | ND | 0.80 (0.06) |
Lower ratios indicate higher surfactancy. ND, not determined; SEM, standard error of the mean.
Surfactant-resistant bacteria in guts and sediments.
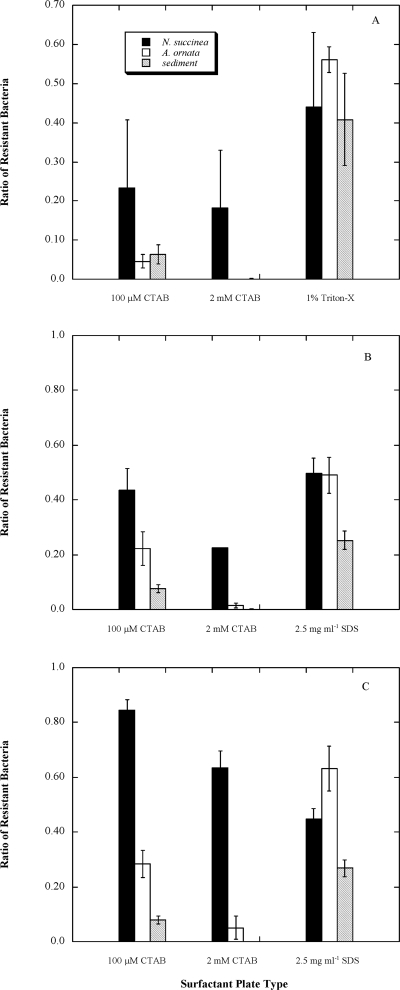
The numbers of bacteria forming colonies on the surfactant-laden plates were clearly reduced relative to the numbers on the nonselective plates with the winter 2006 samples (Fig. 1A). High levels (2 mM) of CTAB had the most deleterious effect, with an almost complete inhibition of the growth of bacteria from the sediment and A. ornata samples. However, a significantly higher proportion of bacteria in the N. succinea gut fluids were capable of growth on these plates, compared to the proportion in the sediment porewater samples (P = 0.047). Although patterns appeared to be qualitatively similar on the 100-μM CTAB plates, numerical comparisons between the colony counts from the deposit feeders and the sediment porewaters were not significant (P values of 0.507 and 0.632 for N. succinea and A. ornata, respectively), due at least in part to high variability and low sample sizes (n = 3). The least inhibition was observed on the Triton X-100 plates, and the ratios of resistant bacteria were no different among gut fluids or sediments (P values of >0.400 for all such comparisons; Fig. 1A).
FIG. 1.
Mean ratios (± standard errors of the means) of surfactant-resistant bacteria in N. succinea guts, A. ornata guts, and Grice Cove sediment during winter 2006 (A), summer 2006 (B), and winter 2007 (C). The ratios were calculated by dividing the number of colonies enumerated on each type of surfactant-laden plates by the number enumerated on nonselective plates.
On the subsequent dates (summer 2006 and winter 2007), both the plates with the weak and strong concentrations (up to 5%) of Triton X-100 failed to consistently reduce the number of bacteria that could be enumerated from either the gut or sediment samples relative to the nonselective plates. Similarly, the low-concentration SDS plates (500 μg/ml) also failed to reduce the number of CFU relative to those of the nonselective plates. Therefore, neither the Triton X-100 nor the low-concentration SDS plates were used to screen for surfactant resistance later in the study.
In the summer, the gut fluids of deposit-feeding polychaetes generally had significantly greater ratios of surfactant-resistant bacteria than the sediment porewater (Fig. 1B). N. succinea gut fluids contained higher levels of resistant bacteria relative to the levels in the sediment porewater samples on the 100-μM CTAB, 2-mM CTAB, and 2.5-mg/ml SDS plates (P values of <0.001 for all three comparisons). A. ornata gut fluids likewise contained higher levels of resistant bacteria on the 100-μM CTAB (P = 0.029) and 2.5-mg/ml SDS plates (P = 0.004). However, A. ornata gut fluid on 2 mM-CTAB-laden plates did not show a significantly different ratio of surfactant resistance than the sediment porewater (P = 0.163).
The same trends were seen for samples taken during the winter of 2007, although the higher proportions of resistant bacteria in the gut fluids of N. succinea were more pronounced (Fig. 1C). With one exception, deposit-feeder gut fluid samples had significantly greater ratios of surfactant-resistant bacteria than sediment fluid (N. succinea fluids on 100-μM CTAB, 2-mM CTAB, and 2.5-mg/ml SDS plates and A. ornata on 100-μM CTAB and 2.5-mg/ml SDS plates; P values of <0.002 for all). At higher CTAB concentrations (2-mM CTAB plates), A. ornata gut fluid did not show a significantly different ratio of surfactant resistance than the sediment porewater (P = 0.221).
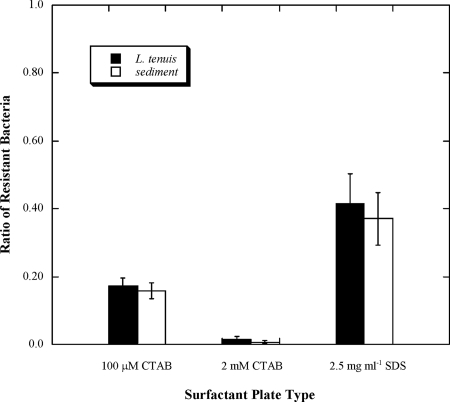
There was no significant difference in the proportions of surfactant-resistant bacteria in the L. tenuis gut contents and the sediment porewater (P values of 0.617, 0.226, and 0.433 for the 100-μM CTAB, 2-mM CTAB, and 2.5-mg/ml SDS plates, respectively; Fig. 2).
FIG. 2.
Mean ratios (± standard errors of the means) of surfactant-resistant bacteria in L. tenuis guts and Breach Inlet sediment, collected in June 2006. The ratios were calculated by dividing the number of colonies enumerated on each type of surfactant-laden plates by the number enumerated on nonselective plates.
16S rRNA gene sequence analysis.
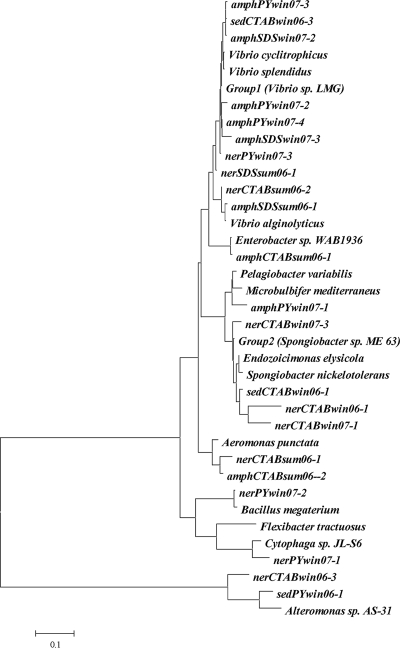
All but 2 of the 42 sediment and gut isolates were gammaproteobacteria. The two exceptions were isolated from nonselective plates and represented Cytophaga-Flexibacter-Bacteroides and Firmicutes phyla, respectively. The isolates showed similarities ranging from 84 to 100% with GenBank reference strains (Table 2). The majority differed (<98% sequence similarity) from any previously sequenced bacterial species, although one large group of 13 isolates was an exception. This group formed a single operational taxonomic unit (OTU) (>99% similarity) and also showed high similarity to Vibrio sp. strain LMG (Table 1). Although most colonies on surfactant plates looked so similar as to appear to be a monoculture, in fact, these colonies consisted of numerous OTUs. However, the majority, especially in the winter samples, did derive from either a closely related group of vibrios or a group of Spongiobacter-like isolates (Fig. 3). Broadly speaking, these two taxonomic groups dominated on surfactant plates, regardless of the surfactant type, the animal from which they were isolated, or the season. However, a subtle seasonal effect was noted in that different Vibrio taxa, more aligned with V. alginolyticus, dominated in the summer (Fig. 3). Additionally, during the winter, N. succinea had both Vibrio and Spongiobacter-like species in its gut lumen, while A. ornata appeared to contain only Vibrio species.
TABLE 2.
Summary of the phylogenetic diversity of isolates based on 16S rRNA gene sequences
| Season and isolate | Closest relative (accession no.) | Sequence similarity (%) |
|---|---|---|
| Winter 2006 | ||
| nerCTABwin06-1 | Uncultured Spongiobacter sp. clone ME19 (DQ917863) | 84 |
| nerCTABwin06-2 | Vibrio sp. strain ACH-24 (DQ408387) | 87 |
| nerCTABwin06-3 | Uncultured gammaproteobacterium clone C23 (DQ917863) | 92 |
| nerCTABwin06-4 | Uncultured Spongiobacter sp. clone ME63 (DQ917871) | 97 |
| sedPYwin06-1 | Alteromonas sp. strain AS-331 (AJ391192) | 94 |
| nerCTABwin06-5 | Uncultured Spongiobacter sp. clone ME63 (DQ917871) | 97 |
| nerCTABwin06-6 | Uncultured Spongiobacter sp. clone ME63 (DQ917871) | 97 |
| sedCTABwin06-1 | Uncultured Spongiobacter sp. clone ME63 (DQ917871) | 96 |
| sedCTABwin06-2 | Vibrio sp. strain LMG 23856 (EF599163) | 95 |
| amphCTABwin06-1 | Vibrio splendidus (AJ874364) | 94 |
| amphCTABwin06-2 | Vibrio sp. strain LMG 23856 (EF599163) | 95 |
| sedCTABwin06-3 | Vibrio sp. strain Y511 (AM778457) | 96 |
| sedCTABwin06-4 | Vibrio sp. strain LMG 23856 (EF599163) | 97 |
| Summer 2006 | ||
| nerCTABsum06-1 | Uncultured bacterium clone aaa96f09 (DQ817157) | 89 |
| nerCTABsum06-2a | Vibrio sp. strain HS1 (EU086102) | 99 |
| nerSDSsum06-1 | Vibrio rumoiensis (DQ530289) | 99 |
| amphSDSsum06-1 | Vibrio alginolyticus (EF050431) | 95 |
| amphCTABsum06-1 | Enterobacter sp. strain WAB1936 (AM184275) | 96 |
| amphCTABsum06-2a | Aeromonas sp. strain ATCC 15467 (AB235956) | 87 |
| Winter 2007 | ||
| nerCTABwin07-1a | Uncultured Spongiobacter sp. clone ME19 (DQ917863) | 89 |
| nerCTABwin07-2a | Uncultured Spongiobacter sp. clone ME63 (DQ917871) | 98 |
| nerSDSwin07-1a | Vibrio sp. strain LMG 23856 (EF599163) | 98 |
| nerSDSwin07-2a | Vibrio sp. strain LMG 23856 (EF599163) | 99 |
| nerSDSwin07-3a | Vibrio sp. strain LMG 23856 (EF599163) | 99 |
| nerCTABwin07-3a | Uncultured Spongiobacter sp. clone ME149 (DQ917896) | 93 |
| amphCTABwin07-1a | Vibrio sp. strain LMG 23856 (EF599163) | 99 |
| amphCTABwin07-2a | Vibrio sp. strain LMG 23856 (EF599163) | 100 |
| amphCTABwin07-3a | Vibrio sp. strain LMG 23856 (EF599163) | 99 |
| amphSDSwin07-1a | Vibrio sp. strain LMG 23856 (EF599163) | 99 |
| amphSDSwin07-2a | Vibrio sp. strain Y320 (AM778460) | 99 |
| amphSDSwin07-3a | Vibrio sp. strain LMG 23856 (EF599163) | 94 |
| amphCTABwin07-4a | Vibrio sp. strain VS6 (AJ1332988) | 99 |
| amphCTABwin07-5a | Vibrio sp. strain LMG 23856 (EF599163) | 99 |
| nerSDSwin07-4a | Uncultured Spongiobacter sp. clone ME63 (DQ917871) | 98 |
| nerCTABwin07-4a | Uncultured Spongiobacter sp. clone ME63 (DQ917871) | 98 |
| amphPYwin07-1 | Microbulbifer sp. strain A4B17 (EF207053) | 92 |
| nerPYwin07-1 | Uncultured CFB group clone MERTZ 0CM 94 (AF425755) | 95 |
| amphPYwin07-2 | Vibrio sp. strain LMG 23856 (EF599163) | 96 |
| amphPYwin07-3 | Vibrio sp. strain Y320 (AM778460) | 94 |
| amphPYwin07-4 | Vibrio cyclitrophicus (AM422804) | 96 |
| nerPYwin07-2 | Bacillus megaterium (EU169176) | 95 |
| nerPYwin07-3 | Vibrio tasmaniensis (AM422801) | 95 |
Positive biosurfactant production by oil-spreading method (winter 2006 not tested).
FIG. 3.
Phylogenetic relationships among 16S rRNA gene sequences of bacterial isolates and reference sequences from GenBank. The tree was constructed using the neighbor-joining method. The bar indicates the number of base changes per nucleotide position. Isolates are named by source (amph, Amphitrite ornata gut; ner, Nereis succinea gut; sed, sediment), type of medium used for isolation (PY, peptone-yeast, nonselective; SDS, PY plus 2.5 mg/ml SDS; CTAB, PY plus 100 μM or 2 mM CTAB), and season of sample collection (sum, summer of 2006; win, winter of 2006 or 2007). OTUs (>99% similarity) that were found more than once were classified as groups. Group 1 consists of the following 13 isolates with close sequence similarity to Vibrio sp. strain LMG: sedCTABwin06-2, amphCTABwin06-1, amphCTABwin06-2, sedCTABwin06-4, nerSDSwin07-1, nerSDSwin07-2, nerSDSwin07-3, amphCTABwin07-1, amphCTABwin07-2, amphCTABwin07-3, amphSDSwin07-1, amphCTABwin07-4, and amphCTABwin07-5. Group 2 consists of the following six isolates closest to Spongiobacter sp. clone ME 63: nerCTABwin06-4, nerCTABwin06-5, nerCTABwin06-6, nerCTABwin07-2, nerSDSwin07-4, and nerCTABwin07-5.
Biosurfactant production.
No significant differences in h/r ratios were detected between the culture supernatants and the controls (P values of >0.100 for all). However, using the more sensitive oil-spreading method, biosurfactant production was detected in a majority of the surfactant-resistant isolates. In total, 26 summer isolates were tested, 15 of which produced surfactants. The diameter of the cleared halo region on the oil surface was from 109 to 127% greater than that produced by samples from uninoculated controls (P values of <0.047 for all such comparisons). Of the six resistant strains that were characterized by DNA sequencing, only two produced detectible levels of biosurfactants (Table 2). In contrast, all 16 surfactant-resistant isolates tested from the winter 2007 samples produced measurable levels (halos ranging from 116 to 137% relative to the sterile medium) of biosurfactants (P values between 0.009 and 0.052; Table 2).
DISCUSSION
Our finding of high surfactant concentrations in the guts of the two polychaete worms, but not in the sea cucumber, is in agreement with prior investigations. Mayer et al. (18) found that the deposit-feeding polychaetes Arenicola marina and Nereis virens had very strong surfactant activities in their gut fluids, while the gut contents of the holothuroid Leptosynapta clarki exhibited relatively weak activity. Subsequent studies reinforced the finding of lower surfactant levels in echinoderms relative to polychaetes (13, 19).
The function and composition of surfactants in the guts of deposit-feeding organisms has not been conclusively established (1). It has been speculated that surfactants assist in the digestive process by either solubilizing hydrophobic food particles or preventing adsorptive attrition of digestive enzymes and dissolved organic matter onto sediment particles ejected from the gut lumen (1). Deposit-feeding polychaetes and holothuroids have markedly different digestive strategies. In comparison with polychaetes, echinoderms are generally slower moving and have lower metabolic demands (16), leading to lower luminal enzyme activities, larger gut volumes, and longer gut residency times. The slower digestive rate of echinoderms may demand lower surfactant concentrations than those required by faster-digesting polychaetes (18).
The hypothesis that a larger proportion of surfactant-resistant bacteria would be found in habitats with higher surfactant levels was supported. Comparatively high abundances of surfactant-resistant bacteria were isolated from the gut lumen of the polychaete worms N. succinea and A. ornata. In contrast, L. tenuis had lower surfactant activity and harbored no more surfactant-resistant bacteria than did ambient sediment. These results suggest that only the guts of specific marine deposit feeders are potential sources of novel surfactant-resistant bacteria. Previous studies have screened sediment sites subject to anthropogenic surfactant contamination and found elevated numbers of biosurfactant-producing and emulsifier-tolerant species of bacteria. For instance, Gaze et al. (14) found higher ratios of CTAB-resistant bacteria in soils contaminated with quaternary ammonium compounds from a wool finishing mill than in agricultural soils.
Elevated numbers of bacteria resistant to both anionic (SDS) and cationic (CTAB) surfactants were found in the guts of our two model polychaetes, whereas the proportions of bacteria on nonselective and nonionic surfactant (Triton X-100) plates were similar in guts and sediment. These findings agree with the general notion that cationic surfactants are most toxic to bacteria, anionic ones are less so, and nonionic ones are nontoxic (32). Little is known about the structure of digestive surfactants in deposit-feeding invertebrates, with the exception of those employed by Arenicola marina. Because a mixture of anionic surfactants has been identified in the gut of A. marina (29), greater resistance to this type of surfactant (i.e., SDS) was anticipated. However, the polychaetes studied here may contain different, or additional, surfactants compared to A. marina. Alternatively, the mechanisms that allow bacteria to thrive in the presence of surfactants (e.g., membrane impermeability or efflux pumps; 12, 23) might perform equally well with anionic and cationic surfactants.
The nonrandom sampling scheme used for the isolation of bacteria precludes a comprehensive view of bacterial community structure; therefore, comparisons between sample types must be treated with caution. However, our observed difference between the winter and summer bacterial community compositions was not unexpected. Rajendran and Nagatomo (26) and Rooney-Varga et al. (27) found seasonal variation in microbial community composition in coastal marine sediments using phospholipid ester-linked fatty acid and genetic analyses, respectively. In temperate estuaries, temperature has been suggested as the prime regulator of seasonal variations in the microbial community (4) and likely accounts for the seasonal influence on bacterial composition of the polychaete gut lumen.
Variation in the composition and diversity of bacteria between surfactant and nonselective plates was also noted. A relatively small set of gammaproteobacteria dominated on surfactant plates, whereas a wider variety of bacteria, including three distinct phyla, was represented on nonselective plates. Two groups, the vibrios, especially a group similar to Vibrio sp. strain LMG, and a Spongiobacter-like group, dominated on the surfactant plates. However, DNA sequencing demonstrated that these same groups (e.g., the two groups resembling Vibrio sp. strain LMG and Spongiobacter strain ME 63) could be found in sediment samples. It is therefore unlikely that these surfactant-resistant bacteria are strict gut symbionts.
The present study identified a variety of biosurfactant-producing bacteria, many of which do not appear to belong to any known genus (<95% 16S rRNA gene sequence similarity; 3). Those surfactant-positive strains that could be assigned to known genera belonged to Vibrio, Spongiobacter, or Bacillus. The list of biosurfactant-producing bacterial genera is diverse and extensive (7), including the particularly well-represented and well-studied genera Pseudomonas, Bacillus, Flavobacterium, and Streptomyces (32). The Vibrio spp. and Spongiobacter-like isolates found in this study appear to be new additions to the list of genera known to produce biosurfactants.
Biosurfactant production by many of the isolated surfactant-resistant strains suggests that the resident gut bacteria could be the source of the digestive surfactants in the deposit feeders. However, the surfactant levels produced by bacterial cultures were much lower than those measured in gut fluid. As in our study, Youssef et al. (34) detected “weak” biosurfactant production in bacterial strains, which tested negative using the drop-collapse method but exhibited significant surfactancy by the more sensitive oil-spreading technique. In accord with these parallel findings, and based on similar clear zone diameters, our cultures appear to have produced surfactants at similar, low levels (50 to 63 mg/liter; 34). It is possible that the growth conditions used in this study were not optimal for isolates to produce natural levels of biosurfactants. In studies demonstrating high yields of biosurfactants, bacteria were grown under specialized conditions (e.g., specific medium type, temperature, and duration) designed to optimize biosurfactant production by a particular strain. For example, Joshi et al. (17) increased the biosurfactant yield from Bacillus licheniformis by a factor of 10 after optimizing the components in the growth medium.
The deposit feeders themselves have previously been proposed as the source of the strong gut surfactant activity (29). Given demonstrated biosurfactant production by gut isolates, further investigation into whether the deposit-feeder hosts or the associated bacteria produce the surfactants is warranted. One means to determine the source would be to determine the chemical structure of the gut surfactants (e.g., by mass and infrared spectrometry) for comparison to those produced by cultured bacteria.
The findings reported here relate to bioremediation efforts in several ways. First, it appears that passage through the guts of deposit feeders, especially polychaetes, selects for bacteria that are resistant to surfactants and are often biosurfactant producers. Previous studies have demonstrated a “worm effect,” wherein hydrocarbon-contaminated sediments were more readily remediated in the presence of sediment-reworking invertebrates (6, 20). Two leading hypotheses for this effect are (i) the enhanced oxygenation of sediments due to bioturbative activities and (ii) the increased bioavailability of polycyclic aromatic hydrocarbons and other hydrocarbons to hydrocarbonoclastic bacteria due to solubilizers in the animal gut (33). An alternative notion is that gut passage selects for hydrocarbonoclastic bacteria, thus increasing their prevalence in contaminated sediments. Cuny et al. (10) noted distinct changes in community composition, including an increase in Alcanivorax spp., in sediments with added Blend Arabian light oil and the deposit feeder Nereis diversicolor compared to uncontaminated sediments or those with added oil alone. Alcanivorax spp. are well known alkane-degrading, surfactant-producing bacteria thought to be central to the natural cleansing of oil-polluted marine ecosystems (15). Second, previously unknown bacteria capable of producing surfactants were isolated. Biosurfactants are often superior to commercial surfactants at solubilizing pollutants and are more easily biodegraded (5). The microorganisms isolated in this study could well be sources of novel biosurfactants. Last, the guts of deposit-feeding invertebrates have been shown to be a source of surfactant-resistant bacteria. Bioremediation efforts typically employ either biodegradative bacteria or surfactants separately. Combining the two methods can greatly improve the efficiency and extent of remediation (21, 28). Exploiting recombinant surfactant-resistant transformants for biodegradation seems feasible. The inherent surfactant resistance of these isolates would enhance the application of an engineered strain for sustained in situ bioremediation.
Acknowledgments
Funding for this work was provided by NSF REU site award DBI-0552828, the Department of Defense ASSURE Program, and a College of Charleston summer research grant, contribution no. 328 of the Grice Marine Laboratory, College of Charleston, Charleston, SC.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Ahrens, M. J., J. H. Hertz, E. M. Lamoureux, G. R. Lopez, A. E. McElroy, and B. J. Brownawell. 2001. The role of digestive surfactants in determining bioavailability of sediment-bound hydrophobic organic contaminants to 2 deposit-feeding polychaetes. Mar. Ecol. Prog. Ser. 212:145-157. [Google Scholar]
- 2.Allen, C. C. R., D. R. Boyd, F. Hempenstall, M. J. Larkin, and N. D. Sharma. 1999. Contrasting effects of a nonionic surfactant on the biotransformation of polycyclic aromatic hydrocarbons to cis-dihydrodiols by soil bacteria. Appl. Environ. Microbiol. 65:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apple, J. K., P. A. del Giorgi, and W. M. Kemp. 2006. Temperature regulation of bacterial production, respiration, and growth efficiency in a temperate salt-marsh estuary. Aquat. Microb. Ecol. 43:243-254. [Google Scholar]
- 5.Banat, I. M. 1995. Characterization of biosurfactants and their use in pollution removal—state of the art. Acta Biotechnol. 15:251-267. [Google Scholar]
- 6.Bauer, J. E., R. P. Kerr, M. F. Bautista, C. J. Decker, and D. G. Capone. 1988. Stimulation of microbial activities and polycyclic aromatic hydrocarbon degradation in marine sediments inhabited by Capitella capitata. Mar. Environ. Res. 25:63-84. [Google Scholar]
- 7.Bodour, A. A., K. P. Drees, and R. M. Maier. 2003. Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid southwestern soils. Appl. Environ. Microbiol. 69:3280-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, H. Y., T. Tsuchido, H. Ono, and M. Takano. 1990. Cell death of Bacillus subtilis caused by surfactants at low concentrations results from induced cell autolysis. J. Ferment. Bioeng. 70:11-14. [Google Scholar]
- 10.Cuny, P., G. Miralles, V. Cornet-Barthaux, M. Acquaviva, G. Stora, V. Grossi, and F. Gilbert. 2007. Influence of bioturbation by the polychaete Nereis diversicolor on the structure of bacterial communities in oil contaminated coastal sediments. Mar. Pollut. Bull. 54:452-459. [DOI] [PubMed] [Google Scholar]
- 11.Desai, J. D., and I. M. Banat. 1997. Microbial production of surfactants and the commercial potential. Microbiol. Mol. Biol. Rev. 61:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar, R., and E. Bibi. 1997. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 179:2274-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaskell, P. N., A. C. Brooks, and L. Maltby. 2007. Variation in the bioaccumulation of a sediment-sorbed hydrophobic compound by benthic macroinvertebrates: patterns and mechanisms. Environ. Sci. Technol. 41:1783-1789. [DOI] [PubMed] [Google Scholar]
- 14.Gaze, W. H., N. Abdouslam, P. M. Hawkey, and E. M. H. Wellington. 2005. Incidence of class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrob. Agents Chemother. 49:1802-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harayama, S., Y. Kasai, and A. Hara. 2004. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 15:205-214. [DOI] [PubMed] [Google Scholar]
- 16.Jangoux, M., and J. M. Lawrence. 1982. Echinoderm nutrition. CRC Press, Rotterdam, The Netherlands.
- 17.Joshi, S., S. Yadav, A. Nerurkar, and A. J. Desai. 2007. Statistical optimization of medium components for the production of biosurfactant by Bacillus licheniformis K51. J. Microbiol. Biotechnol. 17:313-319. [PubMed] [Google Scholar]
- 18.Mayer, L. M., L. L. Schick, R. F. L. Self, P. A. Jumars, R. H. Findlay, Z. Chen, and S. Sampson. 1997. Digestive environments of benthic macroinvertebrate guts: enzymes, surfactants, and dissolved organic matter. J. Mar. Res. 55:785-812. [Google Scholar]
- 19.Mayer, L. M., D. P. Weston, and M. J. Bock. 2001. Benzo[a]pyrene and zinc solubilization by digestive fluids of benthic invertebrates—a cross-phyletic study. Environ. Toxicol. Chem. 20:1890-1900. [PubMed] [Google Scholar]
- 20.McElroy, A. E., J. W. Farrington, and J. M. Teal. 1990. Influence of mode of exposure and the presence of a tubiculous polychaete on the fate of benz[a]anthracene in the benthos. Environ. Sci. Technol. 24:1648-1655. [Google Scholar]
- 21.Mulligan, C. N., R. N. Yong, and B. F. Gibbs. 2001. Surfactant-enhanced remediation of contaminated soil: a review. Eng. Geol. 60:371-380. [Google Scholar]
- 22.Neumann, B., A. Pospiech, and H. U. Schairer. 1992. Rapid isolation of genomic DNA from gram-negative bacteria. Trends Genet. 8:332-333. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382-388. [DOI] [PubMed] [Google Scholar]
- 24.Pieper, D. H., and W. Reineke. 2000. Engineering bacteria for bioremediation. Curr. Opin. Biotechnol. 11:262-270. [DOI] [PubMed] [Google Scholar]
- 25.Plante, C. J., and S. B. Wilde. 2004. Biotic disturbance, recolonization, and early succession of bacterial assemblages in intertidal sediments. Microb. Ecol. 48:154-166. [DOI] [PubMed] [Google Scholar]
- 26.Rajendran, N., and Y. Nagatomo. 1999. Seasonal changes in sedimentary microbial communities of two eutrophic bays as estimated by biomarkers. Hydrobiologia 393:117-125. [Google Scholar]
- 27.Rooney-Varga, J. N., R. Devereux, R. S. Evans, and M. E. Hines. 1997. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl. Environ. Microbiol. 63:3895-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singer, A. C., E. S. Gilbert, E. Luepromcai, and D. E. Crowley. 2000. Bioremediation of polychlorinated biphenyl-contaminated soil using carvone and surfactant-grown bacteria. Appl. Microbiol. Biotechnol. 54:838-843. [DOI] [PubMed] [Google Scholar]
- 29.Smoot, J. C., L. M. Mayer, M. J. Bock, P. C. Wood, and R. H. Findlay. 2003. Structures and concentrations of surfactants in the gut fluid of the marine polychaete Arenicola marina. Mar. Ecol. Prog. Ser. 258:161-169. [Google Scholar]
- 30.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 31.Van Der Vegt, W., H. C. Vander Mei, J. Noordmans, and H. J. Busscher. 1991. Assessment of bacterial biosurfactant production through axisymmetric drop shape analysis by profile. Appl. Microbiol. Biotechnol. 35:766-770. [Google Scholar]
- 32.Van Hamme, J. D., A. Singh, and O. P. Ward. 2006. Physiological aspects. Part 1 in a series of papers devoted to surfactants in microbiology and biotechnology. Biotechnol. Adv. 24:604-620. [DOI] [PubMed] [Google Scholar]
- 33.Weston, D. P., and L. M. Mayer. 1998. In vitro digestive fluid extraction as a measure of the bioavailability of sediment-associated polycyclic aromatic hydrocarbons: sources of variation and implications of partitioning models. Environ. Toxicol. Chem. 17:820-829. [Google Scholar]
- 34.Youssef, N. H., K. H. Duncan, D. P. Nagle, K. N. Savage, R. M. Knapp, and M. J. McInerney. 2004. Comparison of methods to detect biosurfactant production by diverse microorganisms. J. Microbiol. Methods 56:339-347. [DOI] [PubMed] [Google Scholar]