Abstract
A membrane-bound protein purified from Gluconobacter oxydans M5 was confirmed to be a pyrroloquinoline quinone-dependent d-sorbitol dehydrogenase. Gene disruption and complementation experiments demonstrated that this enzyme is responsible for the oxidation of 1-(2-hydroxyethyl) amino-1-deoxy-d-sorbitol (1NSL) to 6-(2-hydroxyethyl) amino-6-deoxy-l-sorbose (6NSE), which is the precursor of an antidiabetic drug, miglitol.
Gluconobacter strains are able to oxidize many sugar alcohols incompletely to produce the corresponding aldehydes, ketones, and organic acids, e.g., l-ascorbic acid, d-gluconic acid, ketogluconic acids, and dihydroxyacetone. Their responsible polyol dehydrogenases have been confirmed (1, 4, 15). It was reported that Gluconobacter oxydans strain M5 can catalyze the oxidation of 1-(2-hydroxyethyl) amino-1-deoxy-d-sorbitol (1NSL) to 6-(2-hydroxyethyl) amino-6-deoxy-l-sorbose (6NSE), a precursor of the antidiabetic drug miglitol (11, 20). However, which dehydrogenase is involved has not been revealed yet. The aim of this work was to find out the responsible enzyme.
The strains and plasmids used in this study are listed in Table 1. G. oxydans M5 was grown in a medium containing 20 g of d-sorbitol, 3 g of yeast extract, 10 g of polypeptone, 1 g of KH2PO4, and 0.2 g of MgSO4-7H2O in 1 liter of deionized water. The different fractions of the cells were prepared by supercentrifugation at 100,000 × g for 60 min. The activities of 1NSL oxidation were measured in the presence of 200 mM substrate, 1NSL (8). The substrate and product were analyzed by silica gel thin-layer chromatography (TLC), with ethanol-methanol-ammonia (1.5:1:1 [vol/vol/vol]) as its eluant, and then dyed with iodine. The major part of the activities was found in the membrane fraction, but little activity could be found in the cytoplasmic fraction (Fig. 1). A second experiment showed phenazine methosulfate (PMS)-dependent 1NSL dehydrogenase activity but not NAD(P)-dependent 1NSL dehydrogenase activity in the membrane fraction. On the contrary, the cytoplasmic fraction had only NAD(P)-dependent 1NSL dehydrogenase activity. In cell extracts, the total activity of PMS-dependent 1NSL dehydrogenase was some 28-fold higher than that of the NAD(P)-dependent 1NSL dehydrogenase (data not shown). These results suggested that the proteins responsible for 1NSL oxidation in G. oxydans are located mainly in the cytoplasmic membrane.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| G. oxydans M5 | Wild strain producing 6NSE from 1NSL | Our laboratory |
| G. oxydans M6 | G. oxydans M5 derivative; sldA::kan | This study |
| G. oxydans M7 | G. oxydans M6 containing pBBR1MCS5-sldAB | This study |
| E. coli JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ [traD36 proAB+lacIqlacZ::M15] | |
| BW25113 | lacIqrrnBT14 ΔlacZwj16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 3 |
| Plasmids | ||
| pKD46 | Red recombinase expression plasmid; Ampr | 3 |
| pKD46RCm | pKD46 derivative; Cmr; red recombinase expression plasmid | Our laboratory |
| pKD4 | Knr gene cassette | 3 |
| pGEM-sldAB | pGEM-T carrying sldAB; Ampr | This study |
| pGEM-sldB-sldA::kan | pGEM-sldAB carrying a modified sldA gene; Ampr Knr | This study |
| pBBR1MCS-5 | Broad-host-range cloning vector; Gmr | 10 |
| pBBR1MCS5-sldAB | pBBR1MCS-5 carrying sldAB; Gmr | This study |
Cmr, chloramphenicol resistant; Knr, kanamycin resistant; Ampr, ampicillin resistant; Gmr, gentamicin resistant; cat, chloramphenicol resistance gene; kan, kanamycin resistance gene.
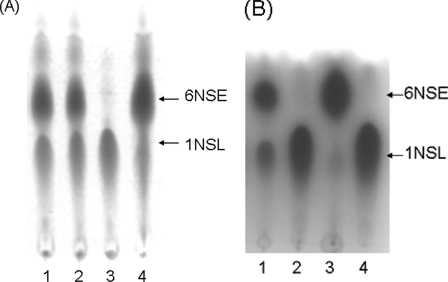
FIG. 1.
TLC analysis of the 6NSE product. (A) TLC analysis of the 6NSE products of various cell fractions or resting cells of G. oxydans M5 catalyzing the 1NSL reaction. Lanes: 1, resting cell reaction mixture; 2, membrane fraction reaction mixture; 3, soluble fraction reaction mixture; 4, purified enzyme reaction mixture. (B) TLC analysis of the 6NSE product of various mutant resting cells catalyzing the 1NSL reaction. Lanes 1, wild-type G. oxydans M5 reaction mixture; 2, G. oxydans M6 defective sldA reaction mixture; 3, G. oxydans M7 complementation of the sldAB genes reaction mixture; 4, standard of substrate 1NSL.
Gluconobacter strains contain a large number of dehydrogenases, which can be classified into two major groups (15). The first group is formed by cytoplasmic-soluble NAD(P)-dependent polyol dehydrogenases, which were believed to participate in the synthesis of precursors and are obviously involved in the maintenance of cells in the stationary growth phase (12, 15). The second group possesses various membrane-bound dehydrogenases, which are shown to be responsible for the rapid oxidation of some important substrates (16, 17). According to our previous work, the oxidation of 1NSL to 6NSE by G. oxydans M5 was fast, with almost 95% of the conversion rate occurring after 8 h (9). Therefore, the responsible dehydrogenase is thought to be compatible with the first type of dehydrogenase.
Purification and identification were performed to reveal the amino acid sequence of the 1NSL dehydrogenase. The membrane fraction was suspended in a 20 mM sodium acetate buffer (pH 6.0) containing 1% Tween-20, stirred, and centrifuged at 10,000 rpm for 30 min. The resulting supernatant was purified with a carboxymethyl (CM) cellulose column, eluted with the same buffer containing 0.1 M KCl, and put on a Sephacryl HR 400 column for further purification (Table 2). The purified enzyme migrated as a single protein band with a molecular mass of about 80 kDa, using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2). The 80-kDa band was isolated from the SDS-PAGE preparation and digested with trypsin. The product was subjected to mass mapping by using matrix-assisted laser desorption ionization-time of flight tandem mass spectrometry, with a model 4700 Proteomics Analyzer. De novo sequences from six polypeptides covering a total of 72 amino acid residues were determined, and the data showed there was a 100% match to the large subunit of a putative pyrroloquinoline quinone (PQQ)-dependent membrane-bound d-sorbitol dehydrogenase (SLDH) encoded by sldAB (GeneID accession no. gox: 0854-0855) in G. oxydans ATCC 621H (17) and a gluconate/polyol dehydrogenase (EBI accession no. AJ577472) of G. suboxydans IFO12528 (18). Then, the sldAB genes were amplified from the G. oxydans M5 chromosome, using the oligonucleotide primers sldAB-KpnI-fwd (GTTTACGATGGTACCGGTTCTGG) and sldAB-EcoRI-rev (GCTTCCCACCCGAATTCTGGAAAAAACG) in GC buffer, with an annealing temperature of 54°C and purified, and cloned into the pGEM-T plasmid, resulting in the pGEM-sldAB plasmid. The sldA gene encodes a polypeptide of 743 residues with a molecular mass of 81,730 Da, which is compatible with the molecular mass of about 80 kDa of the protein band from the SDS-PAGE sample. These results confirmed that the purified enzyme was the product of the sldA gene carried by G. oxydans M5. Interestingly, the small subunit of 126 residues with the molecular mass of 13,860 Da encoded by sldB was not present in the SDS-PAGE result. The reason for its absence was not clear. However, according to previous research (21), the product of the sldB gene is necessary for the SLDH activity in vivo.
TABLE 2.
Purification of 1NSL dehydrogenase of G. oxydans M5
| Purification step | Total activity (units)a | Total protein (mg) | Sp act (U/mg of protein)a | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Membrane purification | 2,016 | 1,588 | 1.3 | 100 | 1 |
| Solubilization | 1,360 | 108 | 13 | 67 | 10 |
| CM-cellulose chromatography | 1,172 | 31 | 38 | 58 | 30 |
| Sephacryl HR 400 chromatography | 589 | 9.4 | 62 | 29 | 49 |
Standard deviations of the determination of specific enzyme activities were less than 20%.
FIG. 2.
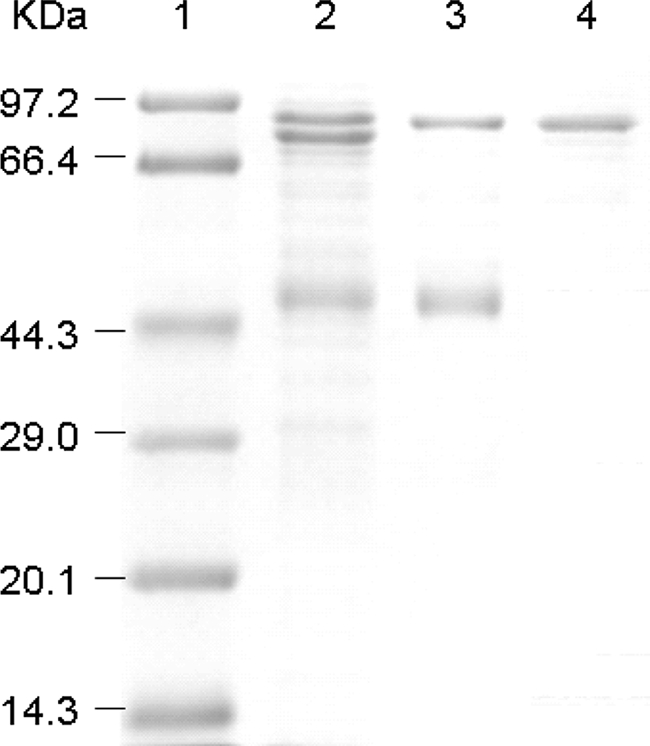
SDS gel electrophoresis of purified 1NSL dehydrogenase. The SDS-15% polyacrylamide gel was stained with Coomassie brilliant blue R-250, and the molecular mass of 1NSL dehydrogenase was measured. Lanes: 1, standard marker proteins (kDa); 2, solubilization from membrane fraction; 3, pooled fraction after CM-column chromatography; 4, pooled fraction after Sephacryl HR 400 column chromatography.
Previously, a membrane-bound PQQ-dependent SLDH from G. suboxydans IFO3255 was encoded by the same genes, sldAB (GenBank accession no. AB065091) (7). The two SLDH genes and their corresponding amino acid residues had 81% and 85% identity, respectively. The quinoproteins SLDH, d-arabitol dehydrogenase, and d-gluconate dehydrogenase from various Gluconobacter strains were confirmed to be identical (13). It is suggested that SLDH is responsible for the oxidation of the substrates, with Bertrand-Hudson configuration (6). Therefore, it is reasonable that a 1NSL substrate with a similar configuration can be oxidized by SLDH.
Disruption of the sldA gene and complementation were carried out to reveal its function in vivo. To generate a gene replacement vector for the inactivation of sldA, the modified highly efficient recombineering-based strategy described by Cotta-de-Almeida et al. was used (2, 3). Several kanamycin-resistant colonies were purified and cultivated to prepare the plasmids. The modified plasmids were confirmed by PCR with the primers KnP1 (GTGTAGGCTGGAGCTGCTTCG) and KnP2 (CATATGAATATCCTCCTTAGT) and resulted in the pGEM-sldB-sldA::kan plasmid. For the construction of an sldAB expression vector, the broad-host-range vector pBBR1MCS-5 (10) and the amplification product were digested with the restriction endonucleases KpnI and EcoRI and ligated, resulting in the pBBR1MCS5-sldAB plasmid. The cloned products were sequenced through a commercial service offered by Invitrogen (Shanghai, China). General molecular biological techniques were performed according to Sambrook et al. (19).
The knockout vector and the expression vector were electrotransferred into G. oxydans cells essentially as described by Mostafa et al. (14). For screening potential sldA-deficient mutants, colonies were transferred to glucose-calcium carbonate agar (5) containing the appropriate selective antibiotics and incubated at 30°C for 3 days. The colonies of mutants were screened with the phenotype of Knr Amps and a clear zone, and the gene disruption was confirmed by PCR. To construct an overexpression strain, the culture plate was incubated for 36 h until gentamicin-resistant colonies appeared.
The mutant was cultivated in complex medium containing 0.5% yeast extract, 0.3% tryptone, and 5% d-glucose (pH 6.0). When mutant cells were used as the resting-cell-reaction system in a mixture containing 200 mM of 1NSL, the disruptant cells did not produce 6NSE, in contrast to that produced by the parental strain at any pH (Fig. 1B, lane 2). This result indicated that the mutant strain has no 1NSL oxidation activity. When the mutant was reconstituted with the recombination plasmid pBBR1MCS5-sldAB, the 1NSL oxidation activity was restored (Fig. 1B, lane 3). These results clearly showed that the quinoprotein SLDH encoded by sldAB is involved in the process of 6NSE production. From these experiments, it can be concluded that the PQQ-dependent membrane-bound SLDH is the primary source enzyme responsible for the oxidation of 1NSL in G. oxydans M5.
Acknowledgments
We thank C. Jin, Institute of Microbiology, Chinese Academy of Science, and S. S.-H. Yang, China Agricultural University, for providing the pKD46 and pBBR1MCS-5 plasmids. We also thank J. Y. Zhang, College of Medicine, East China University of Science and Technology, for membrane protein purification and Y. S.-H. Ma and Q. Y. Ma for their advice on this work.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Adachi, O., D. Moonmangmee, H. Toyama, M. Yamada, E. Shinagawa, and K. Matsushita. 2003. New developments in oxidative fermentation. Appl. Microbiol. Biotechnol. 60:643-653. [DOI] [PubMed] [Google Scholar]
- 2.Cotta-de-Almeida, V., S. Schonhoff, T. Shibata, A. Leiter, and S. B. Snapper. 2003. A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 13:2190-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deppenmeier, U., M. Hoffmeister, and C. Prust. 2002. Biochemistry and biotechnological applications of Gluconobacter strains. Appl. Microbiol. Biotechnol. 60:233-242. [DOI] [PubMed] [Google Scholar]
- 5.Gupta, A., V. Verma, and G. N. Qazi. 1997. Transposon induced mutation in Gluconobacter oxydans with special reference to its direct-glucose oxidation metabolism. FEMS Microbiol. Lett. 147:181-188. [DOI] [PubMed] [Google Scholar]
- 6.Hann, R. M., E. B. Tilden, and C. S. Hudson. 1938. The oxidation of sugar alcohols by Acetobacter suboxydans. J. Am. Chem. Soc. 60:1201-1203. [Google Scholar]
- 7.Hoshino, T., T. Sugisawa, M. Shinjoh, N. Tomiyama, and T. Miyazaki. 2003. Membrane-bound d-sorbitol dehydrogenase of Gluconobacter suboxydans IFO 3255—enzymatic and genetic characterization. Biochim. Biophys. Acta 1647:278-288. [DOI] [PubMed] [Google Scholar]
- 8.Kagan, F., M. A. Rebenstorf, and R. V. Heinzelman. 1957. The preparation of glycamines. J. Am. Chem. Soc. 79:3541-3544. [Google Scholar]
- 9.Keliang, G., and W. Dongzhi. 2006. Asymmetric oxidation by Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 70:135-139. [DOI] [PubMed] [Google Scholar]
- 10.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibioticresistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 11.Landis, B. H., J. K. McLaughlin, R. Heeren, R. W. Grabner, and P. T. Wang. 2002. Bioconversion of N-butylglucamine to 6-deoxy-6-butylamino sorbose by Gluconobacter oxydans. Org. Process Res. Dev. 6:547-552. [Google Scholar]
- 12.Matsushita, K., H. Toyama, and O. Adachi. 1994. Respiratory chains and bioenergetics of acetic acid bacteria. Adv. Microb. Physiol. 36:247-301. [DOI] [PubMed] [Google Scholar]
- 13.Matsushita, K., Y. Fujii, Y. Ano, H. Toyama, M. Shinjoh, N. Tomiyama, T. Miyazaki, T. Sugisawa, T. Hoshino, and O. Adachi. 2003. 5-keto-d-Gluconate production is catalyzed by a quinoprotein glycerol dehydrogenase, major polyol dehydrogenase, in Gluconobacter species. Appl. Environ. Microbiol. 69:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mostafa, H. E., K. J. Heller, and A. Geis. 2002. Cloning of Escherichia coli lacZ and lacY genes and their expression in Gluconobacter oxydans and Acetobacter liquefaciens. Appl. Environ. Microbiol. 68:2619-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muynck, C. D., C. S. S. Pereira, M. Naessens, S. Parmentier, W. Soetaert, and E. J. Vandamme. 2007. The genus Gluconobacter oxydans: comprehensive overview of biochemistry and biotechnological applications. Crit. Rev. Biotechnol. 27:147-171. [DOI] [PubMed] [Google Scholar]
- 16.Pronk, J. T., P. R. Levering, W. Olijve, and J. P. Vandijken. 1989. Role of NADP-dependent and quinoprotein glucose dehydrogenases in gluconic acid production by Gluconobacter oxydans. Enzyme Microb. Technol. 11:160-164. [Google Scholar]
- 17.Prust, C., M. Hoffmeister, H. Liesegang, A. Wiezer, W. F. Fricke, A. Ehrenreih, G. Gottschalk, and U. Deppenmeier. 2005. Complete genome sequence of the acetic bacterium Gluconobacter oxydans. Nat. Biotechnol. 23:195-200. [DOI] [PubMed] [Google Scholar]
- 18.Salusjärvi, T., M. Povelainen, N. Hvorslev, E. V. Eneyskaya, A. A. Kulminskaya, K. A. Shabalin, K. N. Neustroev, N. Kalkkinen, and A. N. Miasnikov. 2004. Cloning of a gluconate/polyol dehydrogenase gene from Gluconobacter suboxydans IFO 12528, characterization of the enzyme and its use for the production of 5-ketogluconate in a recombinant Escherichia coli strain. Appl. Microbiol. Biotechnol. 65:306-314. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Schedel, M. 2000. Regioselective oxidation of aminosorbitol with Gluconobacter oxydans, a key reaction in the industrial synthesis of 1-deoxynojirimycin, p. 296-308. In D. R. Kelly (ed.), Biotechnology, vol. 8b, biotransformations I. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 21.Shinjoh, M., N. Tomiyama, T. M Iyazaki, and T. Hoshino. 2002. Main polyol dehydrogenase of Gluconobacter suboxydans IFO 3255, membrane-bound d-sorbitol dehydrogenase, that needs product of upstream gene, sldB, for activity. Biosci. Biotechnol. Biochem. 66:2314-2322. [DOI] [PubMed] [Google Scholar]