Abstract
We describe a cross-sectional study of the molecular epidemiology of Campylobacter jejuni in a dairy farmland environment, with the aim of elucidating the dynamics of horizontal transmission of C. jejuni genotypes among sources in the area. A collection of 327 C. jejuni isolates from cattle, wildlife, and environmental sources in a 100-km2 area of farmland in northwest England was characterized by multilocus sequence typing. A total of 91 sequence types and 18 clonal complexes were identified. Clonal complexes ST-21, ST-45, and ST-61, which have been frequently associated with human disease, were the most commonly recovered genotypes in this study. In addition, widely distributed genotypes as well as potentially host-associated genotypes have been identified, which suggests that both restricted and interconnecting pathways of transmission may be operating in the dairy farmland environment. In particular, the ST-61 complex and the ST-21 complex were significantly associated with cattle. In contrast, complex strains ST-45, ST-952, and ST-677 were isolated predominantly from wild birds, wild rabbits, and environmental water. A considerable number of novel sequence types have also been identified, which were unassigned to existing clonal complexes and were frequently isolated from wildlife and environmental sources. The segregated distribution of genotypes among samples from different sources suggests that their transmission to humans is perhaps via independent routes. Insight into the dynamics and interactions of C. jejuni populations between important animal reservoirs and their surrounding environment would improve the identification of sources of Campylobacter infection and the design of control strategies.
Campylobacteriosis is the most frequent explanation of acute bacterial gastroenteritis and is a zoonotically transmitted disease caused primarily by the pathogen Campylobacter jejuni via food-borne routes. Campylobacter infections have a substantial public health and socioeconomic impact (28, 31); over 46,000 cases have been reported in England and Wales in 2006 (http://www.hpa.org.uk/infections), and an estimated 2 million cases and 2,000 deaths in the United States are attributed to Campylobacter infections annually (33).
C. jejuni has a widespread distribution with a broad range of animal hosts and environmental reservoirs. Food-producing animals such as poultry (6), cattle, and sheep (22, 32) commonly harbor vast numbers of C. jejuni isolates in their gastrointestinal tracts and represent an important route through which organisms could enter the food chain. In addition, wildlife such as wild birds (2, 3, 34) and mammals (27) are also common hosts of the organism. C. jejuni isolates are also found in the environment, including in surface waters from rivers, streams, ponds, and agricultural runoffs (15, 17, 18) and in sand from bathing beaches (1), and have been implicated in outbreaks of campylobacteriosis (13, 26). According to a recent survey, Campylobacter spp. were implicated in 14% of waterborne infectious intestinal disease outbreaks from 1992 to 2003 in England and Wales, where the common sources were springs, wells, and boreholes (30).
The transmission dynamics of this sporadic disease are complex, and the relative contribution of different sources in C. jejuni infections has remained unclear, reflected in part by a lack of reliable and definitive characterization methods. Recently, multilocus sequence typing (MLST) has been proven to be a valuable and robust characterization tool for C. jejuni (5, 9, 22, 25) and has provided useful information in epidemiological and population dynamic studies of this important pathogen.
Following the recent MLST studies that have highlighted the importance of cattle in C. jejuni epidemiology (12, 22), we further present a cross-sectional study that is the final part of a detailed survey of C. jejuni isolates in a 100-km2 dairy farming area in Cheshire, United Kingdom (12), with the primary objective of detailing the molecular epidemiology of C. jejuni in dairy cattle and the surrounding farming environment in order to elucidate the significance of horizontal transmission among different sources. The distribution and diversity of C. jejuni genotypes among cattle, wildlife, and environmental sources were investigated from a population snapshot based on the systematic sampling of isolates within the area. Isolates included in the study originated from a wide range of sources, including cattle, sheep, wild birds and ducks, rabbits, badgers, and environmental sources such as groundwater, soil, and a drinking-water trough.
MATERIALS AND METHODS
Study site and sample collection.
A total of 100 km2 of rural dairy farming area in Cheshire, United Kingdom was systematically sampled for cattle and wildlife feces and for soil and water, where the regional geographical features, study design, and protocol for spatially structured sampling have been detailed previously (4, 12, 23). Of the C. jejuni isolates investigated in this study (n = 327), 247 were sampled between May and July 2000 from all sources, where the spatial analysis and modeling for 172 isolates have been detailed previously (12). In 2001, an additional 80 isolates were obtained from cattle by sampling from fresh fecal pats in the same locations that had been sampled in the year 2000 (see Table S1 in the supplemental material).
Bacterial growth conditions and species differentiation.
Fecal samples from cattle and wildlife (1 ml), soil samples, and water-sample filters (0.2-μm pores; Nalge Nunc International, Rochester, NY) were added to 9 ml of Campylobacter enrichment broth (Lab M, Bury, United Kingdom) and incubated at 42°C for 24 h under microaerophilic conditions. The broth was inoculated onto Campylobacter blood-free agar (modified charcoal cefoperazone deoxycholate agar) containing an antibiotic supplement (cefoperazone and amphotericin; catalogue no. X112 and X212; Lab M) and was further incubated for 48 h under the same conditions. Three to four colonies of bacterial growth were subcultured onto Columbia blood agar (Lab M) and incubated for 24 to 48 h. On the basis of growth and colony morphology on agar under the described conditions, oxidase and catalase tests, Gram staining, and cell shape and size, Campylobacter spp. were presumptively identified and were stored frozen at −70°C in a cryogenic preservative (Microbank; Pro-Lab Diagnostics, Neston, United Kingdom) until required for culture. Bacterial cells from subsequent culture were heated and lysed to release DNA for PCR. Campylobacter species were identified by performing single-reaction PCRs with previously described primers and conditions (14, 24).
MLST.
Characterization of isolates by MLST was performed as per the published scheme for C. jejuni isolates (10). Briefly, fragments of seven gene targets for each isolate were amplified using the following published primers and reaction conditions: aspA (aspartase A), glnA (glutamine synthase), gltA (citrate synthase), glyA (serine hydroxymethyltransferase), pgm (phosphoglucomutase), tkt (transketolase), and uncA (ATP synthase alpha subunit). Amplicons were purified with the MultiScreen PCR filter plate and MultiScreen vacuum manifold (Millipore Corporation) according to the manufacturer's instructions. Dideoxy termination sequencing reactions were performed at least once on each of the amplified forward and reverse DNA strands in 10-μl volumes containing a 10-μM primer, BigDye reaction mix version 3.1, and 5× sequencing buffer (PE Applied Biosystems). The reaction conditions were 30 cycles at 96°C for 20 s, 50°C for 20 s, and 60°C for 4 min. Sequencher software version 4.0 (Gene Codes Corporation) was used for sequence editing and assembly on a computer. The Campylobacter MLST database (http://pubmlst.org/campylobacter) (16) was used for sequence typing and the clonal complex assignment of C. jejuni isolates.
Statistical and gene flow analysis.
The association between C. jejuni isolate genotypes and isolation sources was analyzed with statistical tests using SPSS software version 14.0, where pairwise comparisons of clonal complex distributions between isolation sources using the Yates' continuity-corrected χ2 test or Fisher's exact two-tailed test were performed. P values of ≤0.05 were the criteria chosen for statistical significance. Isolation sources and clonal complexes with a small number of isolates in the data set were not included.
Estimates of the gene flow between C. jejuni populations from different sources were carried out using DnaSP software (29). Gene flow is measured by an FST value that falls between 0 and 1, where a value of 0 indicates that the two populations are indistinguishable, and a value of 1 indicates that the two populations are genetically distinct.
RESULTS
C. jejuni genotypes in the farmland environment.
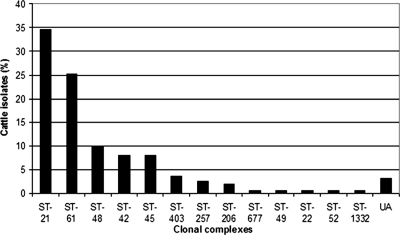
C. jejuni isolates (n = 327) from different animal and environmental sources within the dairy farmland sampling area were characterized by MLST. Among the 91 sequence types identified, the most prevalent sequence types were ST-45 (n = 44; 13.5%), ST-21 (n = 35; 10.7%), and ST-61 (n = 31; 9.5%), which collectively represented 33.7% of the entire data set, while 16.8% of the data set was represented by 55 sequence types which appeared only once. Fifty-nine sequence types (n = 283; 86.5%) were grouped into 18 clonal complexes, while the remaining 32 sequence types (n = 44; 13.5%) were unassigned to any clonal complexes (Fig. 1; Table 1).
FIG. 1.
Occurrence of clonal complexes of 327 C. jejuni isolates from within a farmland environment. UA, unassigned sequence types.
TABLE 1.
Distribution of C. jejuni among sources from 327 isolates
| Clonal complex | Sequence type | % of cattle in:
|
% of isolates from:
|
No. of isolates from:
|
Total no. of isolates | Total % of isolates | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2000 | 2001 | Both yrs combined (avg) | Rabbit | Bird | Water | Soil | Trough | Badger | Sheep | Duck | ||||
| ST-21 | ST-21 | 15.9 | 13.8 | 14.8 | 4.8 | 3.1 | 5.7 | 3 | 2 | 35 | 10.7 | |||
| ST-19 | 6.1 | 5.0 | 5.6 | 2.4a | 10 | 3.1 | ||||||||
| ST-522 | 8.5 | 2.5 | 5.6 | 9.5 | 13 | 4.0 | ||||||||
| ST-53 | 1.2 | 1.3 | 1.2 | 7.1 | 3.1 | 7 | 2.1 | |||||||
| ST-262 | 2.4 | 5.0 | 3.7 | 6 | 1.8 | |||||||||
| ST-266 | 2.4 | 1.2 | 2 | 0.6 | ||||||||||
| ST-797 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-806 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-104 | 1.3 | 0.6a | 1 | 0.3 | ||||||||||
| ST-956 | 1.3 | 0.6a | 1 | 0.3 | ||||||||||
| Total | 39.0 | 30.0 | 34.6 | 23.8 | 6.3 | 5.7 | 77 | 23.5 | ||||||
| ST-45 | ST-45 | 6.1 | 1.3 | 3.7 | 19.0 | 9.4 | 37.1 | 2 | 8 | 1 | 44 | 13.5 | ||
| ST-583 | 2.4 | 1.2 | 6.3 | 5.7 | 8 | 2.4 | ||||||||
| ST-137 | 1.3 | 0.6a | 4.8 | 4.7 | 1 | 7 | 2.1 | |||||||
| ST-334 | 1.3 | 0.6a | 4.7 | 4 | 1.2 | |||||||||
| ST-230 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-233 | 4.7 | 3 | 0.9 | |||||||||||
| ST-792 | 1 | 1 | 0.3 | |||||||||||
| ST-97 | 1.6a | 1 | 0.3 | |||||||||||
| ST-11 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-241 | 1.3 | 0.6a | 31.3 | 42.9 | 1 | 0.3 | ||||||||
| Total | 11.0 | 5.0 | 8.0 | 23.8 | 71 | 21.7 | ||||||||
| ST-61 | ST-61 | 14.6 | 18.8 | 16.7 | 4.8 | 1.6a | 2.9a | 31 | 9.5 | |||||
| ST-618 | 2.4 | 5.0 | 3.7 | 4.8 | 1 | 9 | 2.8 | |||||||
| ST-352 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-628 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-820 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-848 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-864 | 2.5 | 1.2 | 2 | 0.6 | ||||||||||
| ST-852 | 1.3 | 0.6a | 1 | 0.3 | ||||||||||
| ST-955 | 1.3 | 0.6a | 1 | 0.3 | ||||||||||
| ST-948 | 1.6a | 1 | 0.3 | |||||||||||
| Total | 22.0 | 28.8 | 25.3 | 9.5 | 3.1 | 2.9 | 49 | 15.0 | ||||||
| ST-42 | ST-42 | 6.1 | 6.3 | 6.2 | 7.1 | 3.1 | 2.9a | 1 | 17 | 5.2 | ||||
| ST-447 | 1.2 | 1.3 | 1.2 | 1.6a | 1 | 4 | 1.2 | |||||||
| ST-957 | 1.3 | 0.6 | 1 | 0.3 | ||||||||||
| Total | 7.3 | 8.8 | 8.0 | 7.1 | 4.7 | 2.9 | 22 | 6.7 | ||||||
| ST-48 | ST-48 | 1.2 | 10.0 | 5.6 | 9 | 2.8 | ||||||||
| ST-38 | 2.4 | 2.5 | 2.5 | 5.7 | 6 | 1.8 | ||||||||
| ST-798 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-804 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-205 | 1.3 | 0.6a | 1 | 0.3 | ||||||||||
| Total | 6.1 | 13.8 | 9.9 | 5.7 | 18 | 5.5 | ||||||||
| ST-677 | ST-677 | 1.2 | 0.6a | 9.5 | 3.1 | 8.6 | 10 | 3.1 | ||||||
| ST-794 | 3.1 | 2 | 0.6 | |||||||||||
| Total | 1.2 | 0.6 | 9.5 | 6.3 | 8.6 | 12 | 3.7 | |||||||
| ST-952 | ST-799 | 5.7 | 2 | 0.6 | ||||||||||
| ST-808 | 3.1 | 2 | 0.6 | |||||||||||
| ST-807 | 2.9a | 1 | 0.3 | |||||||||||
| ST-809 | 2.4a | 1 | 0.3 | |||||||||||
| ST-949 | 2.9a | 1 | 0.3 | |||||||||||
| ST-952 | 2.4a | 1 | 0.3 | |||||||||||
| Total | 4.8 | 3.1 | 11.4 | 8 | 2.4 | |||||||||
| ST-403 | ST-55 | 1.2 | 3.8 | 2.5 | 4 | 1.2 | ||||||||
| ST-270 | 2.4 | 1.2 | 2 | 0.6 | ||||||||||
| Total | 3.7 | 3.8 | 3.7 | 6 | 1.8 | |||||||||
| ST-257 | ST-257 | 2.4 | 2.5 | 2.5 | 4 | 1.2 | ||||||||
| ST-206 | ST-795 | 1.2 | 0.6a | 1 | 0.3 | |||||||||
| ST-206 | 2.5 | 1.2 | 2 | 0.6 | ||||||||||
| Total | 1.2 | 2.5 | 1.9 | 3 | 0.9 | |||||||||
| ST-1287 | ST-945 | 2.4a | 1.6a | 2.9a | 3 | 0.9 | ||||||||
| ST-49 | ST-49 | 1.2 | 0.6a | 3.1 | 3 | 0.9 | ||||||||
| ST-508 | ST-508 | 3.1 | 2 | 0.6 | ||||||||||
| ST-22 | ST-22 | 1.2 | 0.6a | 1 | 0.3 | |||||||||
| ST-283 | ST-267 | 1 | 1 | 0.3 | ||||||||||
| ST-52 | ST-796 | 1.2 | 0.6a | 1 | 0.3 | |||||||||
| ST-177 | ST-563 | 1.6a | 1 | 0.3 | ||||||||||
| ST-1332 | ST-696 | 1.2 | 0.6a | 1 | 0.3 | |||||||||
| UAb | ST-448 | 4.8 | 4.7 | 5 | 1.5 | |||||||||
| ST-810 | 2.4a | 4.7 | 4 | 1.2 | ||||||||||
| ST-793 | 1.6a | 2.9a | 2 | 0.6 | ||||||||||
| ST-800 | 3.1 | 2 | 0.6 | |||||||||||
| ST-801 | 1.6a | 2.9a | 2 | 0.6 | ||||||||||
| ST-812 | 3.1 | 2 | 0.6 | |||||||||||
| ST-953 | 2.4a | 1.6a | 2 | 0.6 | ||||||||||
| ST-586 | 1.2 | 0.6a | 1 | 0.3 | ||||||||||
| ST-704 | 1.3 | 0.6a | 1 | 0.3 | ||||||||||
| ST-802 | 2.4a | 1 | 0.3 | |||||||||||
| ST-803 | 2.9a | 1 | 0.3 | |||||||||||
| ST-805 | 2.4a | 1 | 0.3 | |||||||||||
| ST-811 | 1.6a | 1 | 0.3 | |||||||||||
| ST-833 | 2.9a | 1 | 0.3 | |||||||||||
| ST-834 | 1.6a | 1 | 0.3 | |||||||||||
| ST-835 | 2.9a | 1 | 0.3 | |||||||||||
| ST-836 | 1.6a | 1 | 0.3 | |||||||||||
| ST-837 | 1.6a | 1 | 0.3 | |||||||||||
| ST-838 | 1 | 1 | 0.3 | |||||||||||
| ST-839 | 2.9a | 1 | 0.3 | |||||||||||
| ST-840 | 1.6a | 1 | 0.3 | |||||||||||
| ST-841 | 1.6a | 1 | 0.3 | |||||||||||
| ST-851 | 1.3 | 0.6a | 1 | 0.3 | ||||||||||
| ST-853 | 1.3 | 0.6a | 1 | 0.3 | ||||||||||
| ST-865 | 1.3 | 0.6a | 1 | 0.3 | ||||||||||
| ST-944 | 1.6a | 1 | 0.3 | |||||||||||
| ST-946 | 1.6a | 1 | 0.3 | |||||||||||
| ST-947 | 2.4a | 1 | 0.3 | |||||||||||
| ST-950 | 1 | 1 | 0.3 | |||||||||||
| ST-951 | 1.6a | 1 | 0.3 | |||||||||||
| ST-954 | 1.6a | 1 | 0.3 | |||||||||||
| ST-959 | 2.4a | 1 | 0.3 | |||||||||||
| Total | 1.2 | 5.0 | 3.1 | 19.0 | 35.9 | 17.1 | 2 | 44 | 13.5 | |||||
Sequence types represented by a single isolate within a source.
UA, unassigned sequence types.
Sixty percent of the data set belonged to three predominant clonal complexes, namely, the ST-21 complex (n = 77; 23.5%), the ST-45 complex (n = 71; 21.7%), and the ST-61 complex (n = 49; 15.0%). Each of the 15 remaining clonal complexes was represented by 6.7% or fewer isolates in the data set. Nine clonal complexes were represented by just one sequence type, five of which occurred only once. From 25 isolates, 21 sequence types (ST-848, ST-851 to ST-853, ST-864 to ST-865, ST-944 to ST-957, and ST-959, inclusive) were newly described in this study.
Ruminant isolates.
A total of 162 C. jejuni isolates from cattle were analyzed in this study. Forty-nine sequence types were identified, from which 30 sequence types appeared only once. The most prevalent sequence types were ST-61 (n = 27; 16.7%) and ST-21 (n = 24; 14.8%), and each of the 47 remaining sequence types was represented by up to 6.2% of isolates in the cattle data set (Table 1). All isolates were grouped into 13 clonal complexes, except for five unassigned sequence types (3.1%), three of which were described for the first time in this study (ST-851, ST-853, and ST-865).
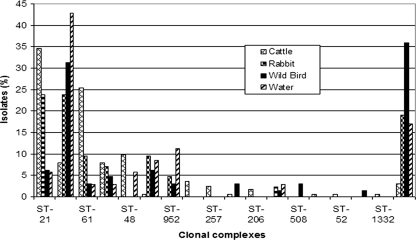
The majority of the cattle data set (n = 139; 85.8%) belonged to five clonal complexes (complexes ST-21, ST-61, ST-48, ST-42, and ST-45), and 60% (n = 97) belonged to either the ST-21 complex or the ST-61 complex (Fig. 2). Each of the eight remaining clonal complexes was represented by 3.7% or fewer cattle isolates. Six clonal complexes were represented by only one sequence type, from which five complexes appeared only once in the data set.
FIG. 2.
Distribution of clonal complexes of 303 C. jejuni isolates recovered from cattle, rabbits, wild birds, and environmental waters. UA, unassigned sequence types.
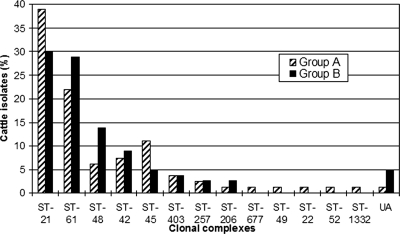
The cattle data set was derived from samples obtained 12 months apart, designated group A (n = 82; collected in 2000) and group B (n = 80; collected in 2001) (Table 1). Consequently, 34 sequence types and 13 clonal complexes were identified in group A, while 29 sequence types and 8 clonal complexes were identified in group B. A number of genotypes overlapped between the two groups; 14 sequence types were shared, and all clonal complexes identified in group B also appeared in group A (Fig. 3). Sequence types and clonal complexes which did not overlap were represented by no more than two isolates in each group. In addition, C. jejuni clonal complexes (complexes ST-21, ST-61, ST-48, ST-42, and ST-45) were predominant in both groups and collectively represented 85.4% and 86.4% of isolates in group A and group B, respectively. Furthermore, there appeared to be a comparable prevalence between group A and group B for clonal complexes ST-21 (39.0% and 30.0%, respectively), ST-61 (22.0% and 28.8%, respectively), and ST-42 (7.3% and 8.8%, respectively).
FIG. 3.
Distribution of C. jejuni clonal complexes among two groups of cattle isolates from the same geographical area collected 12 months apart. UA, unassigned sequence types.
Only five isolates from sheep were characterized, with three isolates belonging to the ST-21 complex and two belonging to the ST-45 complex.
Wildlife isolates.
From the wild bird isolates (n = 64), 31 sequence types were identified, of which 12 appeared only once. No obvious dominant sequence type was observed, and the most commonly encountered sequence type, ST-45, was represented by only six isolates (9.4%) (Table 1). Nineteen sequence types (n = 41; 64.1%) were grouped into 10 clonal complexes, and 12 sequence types (n = 23; 35.9%) in the data set were unassigned to clonal complexes.
The ST-45 complex was predominant in the wild bird data set, represented by 20 isolates (31.3%), while all other nine lineages were individually represented by 6.3% or fewer wild bird isolates (Fig. 2). Five clonal complexes were represented by only one sequence type, and two appeared only once in the data set. In addition, two isolates were collected from ducks, both of which were of the ST-21 genotype.
Among the 21 sequence types identified from rabbit isolates (n = 42), ST-45 was the most frequently identified sequence type (n = 8; 19.0%), while 8 other sequence types were represented by two to four (4.8% to 9.5%) isolates, and 12 sequence types appeared only once in the rabbit data set (Table 1). Thirteen sequence types (n = 34, 81.0%) in the rabbit data set were grouped into seven clonal complexes, and 8 sequence types (n = 8; 19.0%) were not assigned to clonal complexes. The ST-45 complex and the ST-21 complex were equally dominant and were represented by 10 isolates each (23.5%) (Fig. 2). All other clonal complexes were individually represented by four or fewer rabbit isolates. Three clonal complexes were represented by one sequence type, one of which was represented by a single isolate.
Of the 11 isolates obtained from badgers, the majority belonged to the ST-45 complex (n = 9), with the remaining two isolates belonging to clonal complexes ST-42 and ST-283.
Environmental isolates.
Sixteen sequence types were identified from 35 C. jejuni isolates obtained from environmental water samples, with 9 sequence types having occurred only once (Table 1). ST-45 was the dominant sequence type (n = 13; 37.1%), with all remaining sequence types being represented by three or fewer isolates.
Eleven sequence types (n = 29; 82.9%) were grouped into eight clonal complexes, and 6 sequence types (n = 6, 17.1%) were unassigned to clonal complexes. The ST-45 clonal complex was the predominant genotype (42.9%), while the ST-957 complex (11.4%) and the ST-677 complex (8.6%) were the next most common (Fig. 2). The five remaining clonal complexes were represented by two or fewer isolates. Three clonal complexes were represented by one sequence type, all of which were represented by a single isolate.
Isolates obtained from soil samples (n = 5) belonged to the ST-45 complex (n = 2) and the ST-42 complex (n = 1), and two sequence types (ST-838 and ST-950) were not assigned to a clonal complex. A single isolate that originated from a drinking-water trough belonged to the ST-61 complex.
Association between genotypes and sources.
The frequency distribution of C. jejuni clonal complexes described appeared to be nonrandom among sources. Consequently, pairwise comparisons of the clonal complex distribution between isolation sources using the χ2 tests were performed to investigate possible associations (Table 2). However, certain isolation sources and clonal complexes with a small number of isolates in the data set were not included. Accordingly, isolates which originated from cattle, wild birds, wild rabbits, and environmental sources (water, soil, and water trough, collectively), isolates which belonged to clonal complexes ST-21, ST-45, ST-61, ST-42, ST-48, ST-677, and ST-952, and those with unassigned sequence types were analyzed.
TABLE 2.
Summary of P values of significance in the pairwise comparison for the differences in the distribution of C. jejuni clonal complexes between isolation sources
| Source of isolation |
P value for cattle with indicated ST
|
P value for wild bird (ST-21 complex) | P value for rabbit (ST-21 complex) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ST-61 | ST-45 | ST-21 | ST-48 | ST-677 | ST-952 | Unassigned STs | |||
| Wild bird | <0.0005 | <0.0005 | <0.0005 | 0.024 | <0.0005 | ||||
| Rabbit | 0.047 | 0.011 | 0.007 | 0.007 | 0.042 | 0.001 | 0.020 | ||
| Environment | 0.008 | <0.0005 | <0.0005 | 0.047 | 0.028 | 0.001 | 0.001 | 0.032 | |
The proportion of isolates which belonged to the ST-61 complex was found to be significantly higher in cattle than in wild birds (P < 0.0005), wild rabbits (P = 0.047), and environmental sources (P = 0.008), but there were no significant differences in the prevalence of the ST-61 complex within these sources.
In contrast, the prevalence of the ST-45 complex was significantly higher in wild birds (P < 0.0005), rabbits (P = 0.011), and environmental sources (P < 0.0005) than in cattle, but there were no significant differences in the prevalence of the ST-45 complex among these sources.
A significantly higher prevalence of the ST-21 complex was observed in both cattle and rabbits than in wild birds (P < 0.0005 and P = 0.020, respectively) and environmental isolates (P < 0.0005 and P = 0.032, respectively), but no significant difference in the prevalence of the ST-21 complex was observed between cattle and rabbits.
The prevalence of the ST-42 complex was less variable among sources, where no significant differences were found. The presence of the ST-48 complex was detected only in cattle and environmental sources, and the difference was significant between cattle and wild bird (P = 0.007) and rabbit populations (P = 0.047).
The ST-677 complex was significantly associated with wild birds (P = 0.024), rabbits (P = 0.007), and environmental sources (P = 0.028), but no significant differences were found among these sources.
Similarly, the ST-952 complex was absent in cattle, significantly associated with rabbits (P = 0.042) and environmental sources (P = 0.001), and marginally associated with wild birds (P = 0.079), but no significant differences were found between these sources.
The proportion of isolates with unassigned sequence types was also significantly higher in wild birds (P < 0.0005), rabbits (P = 0.001), and environmental sources (P = 0.001) than in cattle, although there were no significant differences among these sources.
Gene flow analysis between isolation sources.
Based on the concatenated sequences of all seven housekeeping loci of C. jejuni isolates from cattle, wild birds, wild rabbits, and the environment, a gene flow analysis between these sources was performed.
The FST values generated from six pairwise comparisons between isolate sources ranged from 0.009 to 0.199, reflecting the presence of both genetically similar and moderately different C. jejuni isolate populations (Table 3). In particular, higher FST values were observed for isolates from cattle and birds (0.199) and cattle and environmental sources (0.175), indicating limited genetic exchange between these C. jejuni isolate populations. In contrast, lower FST values for the remaining four comparisons, ranging from 0.009 between wild birds and the environment to 0.083 between cattle and rabbits, suggest that these C. jejuni isolates represent a genetically indistinguishable population.
TABLE 3.
Pairwise FST values of the gene flow analysis of C. jejuni isolates between sources
| Source of isolation |
FST value for isolates from:
|
||
|---|---|---|---|
| Cattle | Wild bird | Rabbit | |
| Wild bird | 0.199 | ||
| Rabbit | 0.080 | 0.032 | |
| Environment | 0.175 | 0.009 | 0.022 |
DISCUSSION
A better understanding of the epidemiology of C. jejuni is clearly necessary as it continues to be the most common bacterial cause of human gastroenteritis. Indeed, the routes of disease transmission and the relative significance of infection sources to the burden on human disease have largely remained elusive. Nonetheless, several previous studies have greatly improved the understanding of C. jejuni epidemiology by investigating C. jejuni isolate populations from a range of animal hosts using geographically and temporally diverse isolate collections which were characterized by MLST (5, 9, 22, 25). However, the dynamics and interactions of C. jejuni isolate populations between animal reservoirs and their immediate environment have not been established. Given the important role of cattle in C. jejuni epidemiology (22, 32), this study addresses the molecular epidemiology of C. jejuni isolate populations in a dairy farmland environment, with the aim of elucidating the dynamics of horizontal transmission of C. jejuni genotypes among different sources within a defined area, specifically whether cattle would acquire C. jejuni isolates from the environment and vice versa. Insight into such interactions would advance the knowledge of C. jejuni epidemiology to inform on the relative importance of potential reservoirs for human infection.
This study has demonstrated that clonal complexes ST-21, ST-45, and ST-61 were the most common C. jejuni genotypes in the dairy farming environment under study, comprising 60% of the entire data set (Table 1; Fig. 1). This finding may have significant implications for disease control and prevention, as not only do these strains have the capacity to cause disease, they are also the most frequently isolated genotypes in humans (9). Moreover, seven further clonal complexes identified have also been associated with human infections, albeit on a smaller scale (9). These observations clearly highlight the need to recognize that cattle and their associated environment could act as important reservoirs for human disease.
The genotypic composition of the C. jejuni population in cattle was found to be consistent with that delineated from a sizeable number of isolates in a previous longitudinal study (22), where nine clonal complexes overlapped between studies. Clonal complexes ST-21 and ST-61 were highly prevalent, and the ST-48 complex was moderately common in both cases, although a relatively lower number of ST-42 complex isolates and higher number of ST-45 complex isolates were observed in this study, whereas strains which did not overlap accounted for less than 4% of the isolates in each data set. These findings therefore confirmed the major C. jejuni genotypes found in cattle from the longitudinal study; they further reinforce the hypothesis that ST-61 complex isolates may be from a cattle-adapted C. jejuni lineage, especially in the United Kingdom, as suggested in previous studies (5, 9, 12, 22, 25). Further, since all but one of the clonal complexes (ST-1332) identified in cattle in this study have been associated with human infections in the past, there is evidence to suggest that cattle may serve as a major reservoir for C. jejuni infections. A longitudinal aspect was also included in the cattle isolates in this study, where isolates obtained on two occasions separated by 12 months had similar genotype distributions (Fig. 3). This indicates that C. jejuni genotypes in dairy cattle were largely stable over time, which would concur with the finding that there were minimal seasonal patterns in C. jejuni genotypes in cattle from the previous longitudinal study (22).
The ST-45 complex was found to be the most prevalent (31.3%) and the only dominant genotype among wild birds in this study, which was apparent by comparing the ST-45 complex prevalence to that of the next most common lineages, the ST-21 complex (6.3%) and the ST-677 complex (6.3%) (Table 1). Despite this, however, the ST-45 lineage was found to be highly diverse and comprised of various sequence types with similar prevalence. Likewise, the distribution of isolates among sequence types of other lineages was fairly even throughout the data set, while the overall population structure of C. jejuni isolates in wild birds appeared to be highly diverse (Table 1). This feature was somewhat unique to the wild bird data set, since it has been observed that C. jejuni isolates from other sources often belong to dominant sequence types within clonal lineages, including the ST-45 complex. However, this observation may support the suggestion that the relatively high body temperature of birds could provide an optimal growth environment for campylobacters (19), and therefore, isolates may be genetically more diverse (25). Adding to this observation was the presence of a large group of genetically distinct isolates with novel sequence types that were unassigned to clonal complexes (35.9%) (Fig. 2), which contributed to 52% of all such isolates of the entire data set. This observation may also be a reflection of the high diversity of C. jejuni isolates in wild birds but warrants further investigation. The ST-45 complex (5, 9, 25) and unassigned sequence types (25) were found to be highly prevalent in poultry sources in previous studies, and the results of this study suggest that this may also be true in wild birds. The coincidence of such findings in poultry and wild birds could suggest that similar mechanisms may exist in the gastrointestinal tract of avian animals to facilitate the niche adaptation of similar C. jejuni strains.
Given the association of the ST-45 complex between poultry and human infections (9, 25), the high prevalence of the ST-45 complex observed in this data set strongly suggests that wild birds may play a part in the role of disease transmission and should also be regarded as a potential reservoir for C. jejuni infections. Additionally, seven other complex strains with lower prevalence in the wild bird data set have also caused human gastroenteritis in the past. However, these observations are in contrast to previous studies that demonstrated a significantly limited degree of overlap between C. jejuni genotypes found in certain wild bird species and those found in human clinical disease using pulsed-field gel electrophoresis (2, 3). Possible explanations for inconsistencies may include geographical differences, typing methods used, and the difference in bird taxa investigated between studies, where it has been demonstrated that C. jejuni strains could differ according to species and/or specific feeding habits and ecology (3).
The C. jejuni genotypes found in environmental water closely resembled those found in the wild bird data set, where the ST-45 complex and unassigned sequence types were predominant. Unlike in birds, however, these C. jejuni isolates appeared to be less diverse, as the ST-45 complex only included two sequence types and presented fewer unassigned sequence types. This may indicate that, while environmental water is contaminated with C. jejuni isolates from wild animals, only a proportion of genotypes were more adapted to survive and persist in the environment. Further, the low diversity of sequence types observed in water may also be, in part, a reflection of the fact that Campylobacter spp. do not replicate outside hosts (21), hence the lack of means by which genetic variation could be generated following contamination.
There are implications for human infection from the finding that six of the eight clonal complexes isolated from environmental water in this area matched those that have caused disease in humans and that geographical areas such as the location studied are used for recreational activities such as water sports, camping, and picnicking. This suggestion, however, also conflicts with two previous studies that were conducted in New Zealand, which have concluded that genotypes found in environmental water did not overlap with those that cause human disease, although different characterization methods were used in these studies (7, 11).
The wild mammal isolates included in this study consisted mainly of rabbit and a number of badger samples, where the ST-45 complex was predominant, although the ST-21 clonal complex and unassigned sequence types were also highly prevalent in rabbits. Interestingly, rabbit was the only source where there was comparable prevalence between a genotype that was found to be dominant in cattle (the ST-21 complex) and genotypes that were found to be dominant in wild birds and water (ST-45 and unassigned sequence types). In addition, similar to that found in water samples, the ST-45 complex from these sources was also less diverse and had fewer unassigned sequence types than that found in wild birds. The most prevalent genotypes in the wild mammal data set were also the most relevant to human infections, in particular, clonal complexes ST-45, ST-21, ST-61, ST-42 (9), and ST-677 (20), which collectively represented 73.7% of rabbit isolates.
The associations of clonal complexes between isolation sources were compared, and distinct as well as overlapping genotypes were identified among different sources within the study area. For example, the ST-61 complex was significantly associated with cattle, while the ST-21 complex was also found in significantly higher numbers compared to that found in other sources except rabbits. A set of common genotypic characteristics among wildlife and environmental isolates that was distinct from cattle isolates was also apparent, which was the high prevalence of clonal complexes ST-45, ST-677, and ST-952 and unassigned sequence types.
The finding that ST-45 complex isolates were significantly associated with sources from wildlife and the environment is in contrast with the suggestion from previous studies that it is predominantly a poultry-adapted strain. This may indicate that the ST-45 complex was also widespread in the natural environment, or it may be a reflection of contamination of the environment from animal sources. The latter, however, may be a more plausible explanation; wild birds could be a likely source of environmental contamination considering the hypothesis that, as previously discussed, the ST-45 complex may be an avian-adapted C. jejuni strain (5, 25), although additional studies would be necessary to form robust conclusions. Nonetheless, the high prevalence of the ST-45 complex observed in both wild birds and in environmental water suggests that this strain is capable of withstanding marked differences between the high temperatures of birds and the ambient temperatures of environmental waters.
In addition, despite the relatively lower prevalence, the ST-677 and ST-952 complex strains were predominantly isolated from wildlife and environmental water sources, and only one isolate belonging to the ST-677 complex was found in cattle. While the ST-952 complex was a newly identified lineage in this study, in a previous study, infection with the ST-677 complex was found to be associated with drinking unchlorinated water or water from natural sources (20). An increase in the sample size may reveal a higher number of these strains from water and wildlife sources.
A considerable number of uncommon and genetically distinct sequence types, many of which were newly identified in this study, have been observed from wildlife and environmental sources. This observation is in-line with a previous study, where infections with novel and unassigned sequence types were associated with swimming in natural bodies of water (20). Therefore, it is suspected that these isolates may in fact represent a group of closely related isolates for which the genetic links are yet to be identified due to the small numbers recovered to date. Further studies on a larger scale may be able to identify the relationship between these strains and genotypes that are commonly found in the environment, such as the ST-952 complex, or as new emerging C. jejuni clonal groups associated with the environment.
The finding of low numbers of the ST-21 complex from wildlife and environmental sources was somewhat surprising, since it has been considered to be a stable cluster of C. jejuni isolates that is ubiquitous and has adapted to a wide range of hosts and environments (5, 9, 25). However, due to the limited number of environmental isolates that has been investigated using MLST to date, the question of whether the ST-21 complex is indeed less widespread in the natural environment deserves further research.
The associations between clonal complexes and sources were confirmed by performing a gene flow analysis based on 3,309-bp-long concatenated nucleotide sequences from all seven housekeeping loci (Table 3). Considering that an FST value of 0.932 was found between the two Campylobacter species (C. jejuni and C. coli) in a previous study (8), the FST values of 0.199 and 0.175 found within C. jejuni isolates are noteworthy and are indicative of a limited gene flow between C. jejuni isolate populations in cattle with those in wild birds and the environment, respectively. This is in agreement with the segregated distribution of clonal complexes observed between these sources, suggesting that transmission from cattle to humans is perhaps through nonenvironmental routes, that is, through the food chain.
Our primary objective was to investigate the potential role of wildlife and the environment as a source of C. jejuni infection in cattle by comparing the distribution of genotypes from diverse sources. These observations have demonstrated that C. jejuni isolate populations in wildlife and environmental samples were notably distinct from those in cattle within this geographical area. The finding that certain clonal complexes were predominantly shared among isolates from wildlife and environmental sources is indicative of common or interconnecting horizontal transmission pathways of C. jejuni isolates within these sources. However, the dissemination of genotypes between cattle and the environment and wildlife appeared to be considerably restricted, and we suggest that C. jejuni isolates from the environment have limited significance as reservoirs for infection of the cattle population currently under study.
From the public health viewpoint, the finding that the majority of the C. jejuni isolate genotypes identified in this study have been associated with human infections emphasizes the need to recognize cattle and their associated environment as important potential reservoirs for human disease, in particular for clonal complex strains ST-21, ST-45, and ST-61. Furthermore, the distinct distribution of these genotypes among samples from different sources observed in this study would imply that their transmission to humans is perhaps via independent routes. This situation would have implications in the design of control strategies.
Investigations that have addressed the importance of the potential transmission of C. jejuni isolates between cattle and environmental sources have been scarce. Based on the systematic sampling of isolates in this study, a population snapshot that mirrored the dynamics of C. jejuni isolates within a dairy farmland microcosm was explored with MLST, a characterization tool proven to be valuable for discerning the molecular epidemiology of C. jejuni. This has offered further insight into C. jejuni epidemiology, where both widely distributed genotypes and potentially host-associated genotypes have been identified. The added knowledge and understanding of such relationships would further our ability to recognize the sources of campylobacteriosis. To corroborate these findings, and given the high degree of significance of the C. jejuni genotypes identified in this study with respect to human disease, further studies involving a larger number of isolates and focusing on wildlife and environmental samples should be an essential research priority.
Supplementary Material
Acknowledgments
The funding of this work was supported by the Health Protection Agency North West, United Kingdom.
This study made use of the Campylobacter jejuni multilocus sequence typing website (http://pubmlst.org/campylobacter/) developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford (15). The development of this site has been funded by the Wellcome Trust.
Footnotes
Published ahead of print on 27 June 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bolton, F. J., S. B. Surman, K. Martin, D. R. A. Wareing, and T. J. Humphrey. 1999. Presence of Campylobacter and Salmonella in sand from bathing beaches. Epidemiol. Infect. 122:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broman, T., H. Palmgren, S. Bergström, M. Sellin, J. Waldenström, M. L. Danielsson-Tham, and B. Olsen. 2002. Campylobacter jejuni in black-headed gulls (Larus ridibundus): prevalence, genotypes, and influence on C. jejuni epidemiology. J. Clin. Microbiol. 40:4594-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broman, T., J. Waldenström, D. Dahlgren, I. Carlsson, I. Eliasson, and B. Olsen. 2004. Diversities and similarities in PFGE profiles of Campylobacter jejuni isolated from migrating birds and humans. J. Appl. Microbiol. 96:834-843. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. E., O. F. Christensen, H. E. Clough, P. J. Diggle, C. A. Hart, S. Hazel, R. Kemp, A. J. H. Leatherbarrow, A. Moore, J. Sutherst, J. Turner, N. J. Williams, E. J. Wright, and N. P. French. 2004. Frequency and spatial distribution of environmental Campylobacter spp. Appl. Environ. Microbiol. 70:6501-6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colles, F. M., K. Jones, R. M. Harding, and M. C. J. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corry, J. E. L., and H. I. Atabay. 2001. Poultry as a source of Campylobacter and related organisms. Symp. Ser. Soc. Appl. Microbiol. 30:96S-114S. [DOI] [PubMed] [Google Scholar]
- 7.Devane, M. L., C. Nicol, A. Ball, J. D. Klena, P. Scholes, J. A. Hudson, M. G. Baker, B. J. Gilpin, N. Garrett, and M. G. Savill. 2005. The occurrence of Campylobacter subtypes in environmental reservoirs and potential transmission routes. J. Appl. Microbiol. 98:980-990. [DOI] [PubMed] [Google Scholar]
- 8.Dingle, K. E., F. M. Colles, D. Falush, and M. C. J. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. A. Wareing, and M. C. J. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eyles, R. F., H. J. L. Brooks, C. R. Townsend, G. A. Burtenshaw, N. C. K. Heng, R. W. Jack, and P. Weinstein. 2006. Comparison of Campylobacter jejuni PFGE and Penner subtypes in human infections and in water samples from the Taieri River catchment of New Zealand. J. Appl. Microbiol. 101:18-25. [DOI] [PubMed] [Google Scholar]
- 12.French, N., M. Barrigas, P. Brown, P. Ribiero, N. Williams, H. Leatherbarrow, R. Birtles, E. Bolton, P. Fearnhead, and A. Fox. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116-1126. [DOI] [PubMed] [Google Scholar]
- 13.Furtado, C., G. K. Adak, J. M. Stuart, P. G. Wall, H. S. Evans, and D. P. Casemore. 1998. Outbreaks of waterborne infectious intestinal disease in England and Wales, 1992-5. Epidemiol. Infect. 121:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, I., K. A. Grant, P. T. Richardson, S. F. Park, and M. D. Collins. 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson, J. A., C. Nicol, J. Wright, R. Whyte, and S. K. Hasell. 1999. Seasonal variation of Campylobacter types from human cases, veterinary cases, raw chicken, milk and water. J. Appl. Microbiol. 87:115-124. [DOI] [PubMed] [Google Scholar]
- 16.Jolley, K. A., M. S. Chan, and M. C. J. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, K. 2001. Campylobacters in water, sewage and the environment. J. Appl. Microbiol. 90:S68-S79. [DOI] [PubMed] [Google Scholar]
- 18.Jones, K., M. Betaieb, and D. R. Telford. 1990. Thermophilic campylobacters in surface waters around Lancaster, UK: negative correlation with Campylobacter infections in the community. J. Appl. Bacteriol. 69:758-764. [DOI] [PubMed] [Google Scholar]
- 19.Kapperud, G., and O. Rosef. 1983. Avian wildlife reservoir of Campylobacter fetus subsp. jejuni, Yersinia spp., and Salmonella spp. in Norway. Appl. Environ. Microbiol. 45:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kärenlampi, R., H. Rautelin, D. Schönberg-Norio, L. Paulin, and M. L. Hänninen. 2007. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 73:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 22.Kwan, P. S. L., A. Birtles, F. J. Bolton, N. P. French, S. E. Robinson, L. S. Newbold, M. Upton, and A. J. Fox. 2008. Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74:3626-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leatherbarrow, A. J. H., C. A. Hart, R. Kemp, N. J. Williams, A. Ridley, M. Sharma, P. J. Diggle, E. J. Wright, J. Sutherst, and N. P. French. 2004. Genotypic and antibiotic susceptibility characteristics of a Campylobacter coli population isolated from dairy farmland in the United Kingdom. Appl. Environ. Microbiol. 70:822-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 25.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millson, M., M. Bokhout, J. Carlson, L. Spielberg, R. Aldis, A. Borczyk, and H. Lior. 1991. An outbreak of Campylobacter jejuni gastroenteritis linked to meltwater contamination of a municipal well. Can. J. Public Health 82:27-31. [PubMed] [Google Scholar]
- 27.Petersen, L., E. M. Nielsen, J. Engberg, S. L. W. On, and H. H. Dietz. 2001. Comparison of genotypes and serotypes of Campylobacter jejuni isolated from Danish wild mammals and birds and from broiler flocks and humans. Appl. Environ. Microbiol. 67:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, J. A., P. Cumberland, P. N. Sockett, J. Wheeler, L. C. Rodrigues, D. Sethi, and P. J. Roderick. 2003. The study of infectious intestinal disease in England: socio-economic impact. Epidemiol. Infect. 130:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 30.Smith, A., M. Reacher, W. Smerdon, G. K. Adak, G. Nichols, and R. M. Chalmers. 2006. Outbreaks of waterborne infectious intestinal disease in England and Wales, 1992-2003. Epidemiol. Infect. 134:1141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sockett, P. 1993. Social and economic aspects of food-borne disease. Food Policy 18:110-119. [Google Scholar]
- 32.Stanley, K., and K. Jones. 2003. Cattle and sheep farms as reservoirs of Campylobacter. J. Appl. Microbiol. 94:104S-113S. [DOI] [PubMed] [Google Scholar]
- 33.Tauxe, R. V. 1992. Epidemiology of Campylobacter jejuni infections in the United States and other industrial nations, p. 9-12. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins (ed.), Campylobacter jejuni: current and future trends. American Society for Microbiology, Washington, DC.
- 34.Waldenström, J., T. Broman, I. Carlsson, D. Hasselquist, R. P. Achterberg, J. A. Wagenaar, and B. Olsen. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.