Abstract
A 1,026-bp open reading frame sharing significant similarity with queA, which encodes a predicted S-adenosylmethionine:tRNA ribosyltransferase-isomerase responsible for queosine modification of tRNAs, was found immediately 5′ of the gene for the transcriptional activator (ArcR) of the arginine deiminase system (ADS) operon of Streptococcus gordonii. The role of QueA in bacterial physiology is enigmatic, but loss of QueA has been shown to compromise stationary-phase survival or virulence in certain enteric bacteria. Interestingly, S. gordonii appears to be unique among ADS-positive bacteria in the linkage of queA with the ADS genes. A putative σ70 promoter (pqueA; TTGCCA-N21-TATAAT) was mapped 5′ of queA by primer extension, and queA and arcR were shown to be cotranscribed. The expression from pqueA was found to be constitutive under all conditions tested, but the expression of parcA, which drives the expression of the arc structural genes, was enhanced in stationary phase and could be induced by low pH and arginine. QueA and CcpA acted repressively on arc transcription, but neither QueA-deficient strains nor CcpA-deficient strains showed significant differences in arginine deiminase enzyme activities compared with the wild-type strain. The growth rate of a QueA-deficient strain did not differ significantly from that of the parental strain, but the QueA-deficient strain did not compete well with the wild-type during serial passage. In addition to the finding that ADS expression can be regulated separately by growth phase and pH, a significant linkage between the ADS, translational efficiency modulated by QueA, and post-exponential-phase survival of S. gordonii was found.
The arginine deiminase system (ADS) consists of arginine deiminase (AD), ornithine carbamyltransferase, and carbamate kinase. The ADS catalyzes the conversion of arginine to ornithine, ammonia, and CO2 with concomitant production of ATP (11). The ADS has a number of important physiological functions; it contributes to pH homeostasis, it protects cells from lethal acidification, and it provides ATP for growth and maintenance (8, 22, 30, 39). In addition, recent studies have linked the ADS with the pathogenicity of some organisms (12, 44). In the oral cavity, the ADS is one of two major ammonia-generating pathways (6), and several studies have demonstrated that there is an inverse relationship between arginine metabolism and dental caries (6, 34). A thorough analysis of the regulation and function of the ADS in oral biofilms is needed to understand the contribution of arginine catabolism to oral health and to establish a foundation for designing new strategies for caries prevention that capitalize on moderating dental plaque acidification (4, 6).
The ADS is widely distributed among prokaryotes, and the primary structures of the enzymes in the pathway have been conserved throughout evolution. In contrast, diversity in the gene organization and regulation of operons encoding the ADS have been revealed by physiologic and genetic studies (46). Most microorganisms studied so far have ADS genes that are organized in one cluster. The arcA, arcB, and arcC genes are conserved in the arc operons and encode AD, ornithine carbamyltransferase, and carbamate kinase, respectively. Additional genes are often associated with the ADS, including arcD, which encodes an arginine:ornithine antiporter, and arcT, which encodes a putative peptidase (45) that may release arginine from internalized peptides. In all cases examined thus far, regulation of the ADS in bacteria is tightly controlled, but the mode and mechanisms of control vary widely. For instance, both Pseudomonas aeruginosa and Bacillus licheniformis utilize the ADS exclusively under anaerobic conditions, and expression can be further enhanced in the presence of arginine (28, 29). In some lactic acid bacteria, such as Streptococcus sanguis and Lactobacillus sakei, the expression of the arc operon is under the control of carbon catabolite repression (CCR) and is inducible by arginine (18, 45). The ADS of S. sanguis and Streptococcus rattus can also be repressed by aeration (5).
We previously reported that the arc operon of Streptococcus gordonii DL1 is arranged as follows: arcABCDT (15). In addition to the genes encoding enzymes involved in arginine utilization, two other genes were shown to regulate arc operon expression. The first of these genes is flp (Fnr-like protein), which is located 5′ of arcA (Fig. 1) and encodes an activator of arc transcription that responds to oxygen availability (14). The second gene, arcR, is located 3′ of arcT and is transcribed in the opposite direction (Fig. 1). ArcR is an activator of the arc operon that governs induction of expression by arginine (15). We also showed that the expression of the arc operon in S. gordonii was subject to CCR through the catabolite control protein A (CcpA)-catabolite response element pathway (14).
FIG. 1.
Schematic diagram of the arc operon, arcR, queA, and flp of S. gordonii DL1, showing the gene order and arrangement. The size of each ORF (in nucleotides [nt]) is indicated. Transcription initiation sites upstream of arcA (14) and queA are indicated by arrows. A putative rho-independent terminator between flp and arcA is indicated by a loop.
In this study, we observed that a 1,026-bp open reading frame (ORF) is located 5′ of arcR and is transcribed in the same direction, with only 15 bp separating the genes. This ORF encodes a predicted protein with significant similarity to S-adenosylmethionine:tRNA ribosyltransferase-isomerase (QueA). Previous biochemical analysis of Escherichia coli revealed that the function of QueA was to modify tRNA with queuosine. Introduction of queuosine into tRNAs is initiated when a transglycosylase, encoded by the tgt gene, exchanges the guanine base at the first position of the GUN anticodon in tRNAs with preQ1, a precursor of the base (queuine) and queuosine. QueA then attaches a substituted cyclopentenyl group to preQ1 to produce epoxyqueuosine, which is followed by a B12-dependent step in which the epoxide in epoxyqueuosine is reduced, yielding queuosine (20, 24, 25, 33, 36, 37).
Although our understanding of the biochemistry of queuosine modification of tRNAs is increasing (21, 40, 41), it is not yet clear what the impact of this modification on gene expression and cellular physiology is. A connection between queuosine modification of tRNA and the efficiency and fidelity of translation has been reported (32). However, there have been only two cases in which the effect of queuosine modification of tRNAs on a bacterial phenotype was documented. In one case, it was shown that the pathogenic potential of Shigella flexneri is diminished in a queA mutant (17). In the second case, E. coli mutants lacking QueA exhibited an apparently normal growth phenotype during favorable conditions, but upon entry into stationary phase the wild-type strain survived better than a queuosine-deficient strain (19, 31). QueA has been identified in a variety of other organisms by genomic and genetic studies, but functional analyses have not been undertaken yet. In this paper, a unique linkage of queA with genes of the ADS is described and new information implicating QueA and the ADS in post-exponential-phase homeostasis is presented.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains were grown in Luria-Bertani medium supplemented with ampicillin (100 μg ml−1), kanamycin (Km) (40 μg ml−1), erythromycin (Em) (300 μg ml−1), spectinomycin (250 μg ml−1), or chloramphenicol (20 μg ml−1), where indicated. S. gordonii DL1 was maintained in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, MI) at 37°C in 5% CO2-95% air. Depending on the experiment, S. gordonii strains were cultured in tryptone-yeast extract (TY) medium (43) or the chemically defined medium FMC (38). All recombinant S. gordonii DL1 strains carrying a promoterless chloramphenicol acetyltransferase (CAT) gene (cat) fused to the arcA (parcA-cat) or queA (pqueA-cat) promoter cloned into pQY1 were selected and maintained on BHI agar supplemented with 5 μg ml−1 Em. The QueA-deficient strains were selected on BHI agar with Km (250 μg ml−1). The CcpA-deficient strains constructed previously (14) were maintained on BHI agar with Em (10 μg ml−1). Preparation of competent cells and transformation of S. gordonii were done as previously described (27).
RNA methods.
S. gordonii was grown to mid-exponential phase (optical density at 600 nm [OD600], ≃0.5 to 0.6) in TY medium containing 10 mM galactose and 50 mM arginine. RNA was extracted as previously described (10). For reverse transcription (RT)-PCR, the first-strand cDNA was amplified from total RNA with random hexamers. Then the first-strand cDNA was amplified by PCR at high stringency with primer SqueA (5′-TGCCTTTTCGACCAATTTCCACTTACC-3′), which contained the sense sequence of queA located 151 bp 5′ of the queA stop codon, and primer AsarcR (5′-AACAATAACGCCATTGGCTTCAAGGAG-3′), which contained the antisense sequence of arcR located 110 bp 3′ of the arcR start codon. Primer extension was carried out as previously described (3), with primer annealing and reverse transcription performed at 42°C. An oligonucleotide designated EXTQueA (5′-AAGAGGAGTTTGGGCAATAAGCTCCTCTGGTAAGTG-3′), which contained the antisense sequence of queA from 30 to 65 bases 3′ of the queA start codon, was used in primer extension reactions. Primer extension products were analyzed alongside DNA sequencing products generated with the same primer.
Construction of wild-type and CcpA-deficient strains of S. gordonii carrying a pqueA-cat fusion.
A 277-bp BamHI-SstI fragment immediately 5′ of the start codon of queA was amplified by PCR with primers pqueA5′ (5′-CTTTTCTCTAATATTATTGAGCTCTATAGACCAATTTTTG-3′) and pqueA3′ (5′-ATCGGCAGTGTTCATGGATCCTCCTTAAACAGTCCAT-3′). To facilitate fusion construction, SstI and BamHI recognition sequences (indicated by bold type) were included in primers pqueA5′ and pqueA3′, respectively. The product harboring pqueA was fused with the promoterless cat gene derived from pC194 (23) on pGEM-Zf3(+). The fusion was constructed so that translation was driven from the queA ribosome binding site. To facilitate integration of the pqueA-cat fusion, the integration vector pYQ1 was constructed. Plasmid pYQ1 is a derivative of pMJB8 (9) in which the ΩKm cassette was replaced with an Em cassette (42). Plasmid pYQ1 allows insertion of foreign DNA into the gtfG gene (encoding glucosyltransferase) with concomitant acquisition of an Em resistance phenotype. Plasmid pYQ1 carrying pqueA-cat was then used to transform the wild-type and CcpA-deficient strains of S. gordonii (14) in order to construct strains WT/pqueA-cat and CcpA−/pqueA-cat.
Construction of QueA-deficient strains of S. gordonii carrying a parcA-cat fusion.
A 1.26-kbp EcoRI-XbaI fragment containing the S. gordonii queA gene was amplified by recombinant PCR to introduce a unique BamHI site 515 bp 3′ of the start codon of queA. A BamHI fragment containing a nonpolar Km (26) resistance cassette was subsequently cloned into the BamHI site in the PCR product. The resulting plasmid was used to transform S. gordonii in order to generate nonpolar QueA-deficient mutants via double-crossover recombination. The correct configuration of integration was confirmed by PCR. A parcA-cat fusion, described elsewhere (14), was cloned into pYQ1 and used to transform the QueA-deficient and wild-type S. gordonii strains to construct QueA−/parcA-cat and WT/parcA-cat, respectively.
Growth phase regulation of pqueA and parcA expression.
The expression from pqueA and parcA as a function of growth phase was monitored by measuring the CAT activity in early-exponential-phase (OD600, ≃0.25 to 0.3), mid-exponential-phase (OD600, ≃0.5 to 0.6), early-stationary-phase (OD600, ≃1.0 to 1.1), late-stationary-phase (3 h after cultures entered stationary phase), and overnight cultures grown in FMC containing 20 mM glucose with 20 mM arginine.
Relationship of pqueA to CCR.
Expression from pqueA was monitored by measuring CAT activities of WT/pqueA-cat and CcpA−/pqueA-cat cultured in TY medium with 20 mM arginine and 20 mM galactose or glucose. Cells were collected at mid-exponential phase (OD600, ≃0.5 to 0.6).
Chemostat cultivation to monitor gene expression.
To determine potential effects of pH and arginine induction without inducing changes in the growth rate or growth phase, WT/pqueA-cat, WT/parcA-cat, QueA-deficient strains, and CcpA-deficient strains carrying a parcA-cat fusion were grown in a Biostat i twin-controller chemostat (B. Braun Biotech, Inc., Allentown, PA) in TY medium (43) supplemented with 10 mM glucose at a dilution rate of 0.3 h−1. The pH of the cultures was maintained at 5.9 or 7.0 by addition of 2 M KOH. Cultures were sampled when the cells reached steady state, which was achieved after 10 generations under particular growth conditions (43) or 1 h after pulsing with arginine.
Biochemical assays.
CAT activity was measured as previously described (10). Briefly, cells were washed once with 10 mM Tris-HCl (pH 7.8), and cell pellets were quickly frozen in an ethanol-dry ice bath and stored at −80°C until assays were performed. Cells were disrupted with a Bead Beater (Biospec Products, Inc., Bartlesville, OK) for a total of 40 s at 4°C. The cell lysates were centrifuged at 18,000 × g for 5 min, and each soluble fraction was recovered and used to measure the CAT activity by the method of Shaw (35). The concentration of protein was determined by using a Bradford protein assay (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard. AD activity was measured by monitoring citrulline production from arginine, as previously described (2). Cells were harvested by centrifugation, washed once with 10 mM Tris-maleate buffer (pH 6.8), and resuspended using 1/10 the original culture volume in the same buffer. The cells were permeablized by vortexing them with toluene and were collected by centrifugation at 18,000 × g. The supernatant fluid was discarded, and the pellet was resuspended in 10 mM Tris-maleate buffer and used to measure AD activity in a reaction mixture containing 20 mM arginine, 10 mM hexanoic acid, and 50 mM Tris-maleate buffer (pH 6.0). The concentration of protein used in each assay was determined as described above.
Real-time PCR.
Levels of arcA expression in the wild-type, QueA-deficient, and CcpA-deficient strains carrying the parcA-cat fusion were quantified by real-time PCR. Cells were cultured in TY media with 20 mM galactose and 20 mM arginine to mid-exponential phase. The primers used were arcA antisense (5′-GACCGCGAACCAATTCACTTCC-3′) and arcA sense (5′-CGCTCCAGGTGTTGTTGTTGTG-3′). Extraction of RNA, RT-PCR, and real-time RT-PCR were performed as previously described (1).
Competition-persistence comparison.
Overnight cultures of the wild-type and Km-resistant QueA-deficient strains of S. gordonii were diluted 1:100 in TY medium containing 0.2% glucose or TY medium containing 0.2% galactose and 10 mM arginine. When the OD600 of the cultures reached 0.5 to 0.6, equal volumes of the wild-type and QueA-deficient strains were mixed, and the culture was diluted 1:100 with fresh medium (zero subculture). The cultures were grown for 4 days, and samples were removed each day to determine the number of CFU/ml of culture. The levels of the wild-type and QueA-deficient S. gordonii strains in the cultures were determined by plating dilutions on BHI agar and BHI agar with Km (250 μg ml−1).
RESULTS AND DISCUSSION
queA and arcR are cotranscribed from a promoter 5′ of queA.
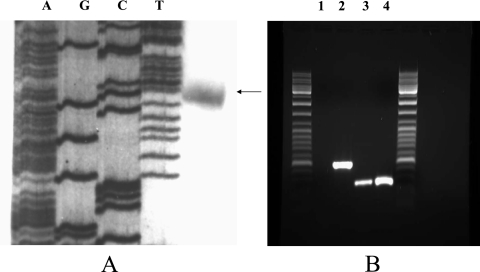
To locate the transcriptional initiation site of the queA gene, primer extension was performed. A single product, which was 181 bases 5′ of the queA start codon and started with a G residue, was observed. A putative σ70 promoter, TTGCCA-N21-TATAAT, was identified 8 bases 5′ of the transcription initiation site (Fig. 2A). The results of the primer extension analysis were further confirmed by 5′ random amplification of cDNA ends (data not shown). It is unlikely that a functional promoter is present in the 310 bp 5′ of the arcR start codon, since when we tried to complement an ArcR-deficient strain (13) with the entire arcR structural gene and 310 bp of DNA upstream of the arcR start codon delivered on plasmid pDL278 (16), arginine-dependent activation of ADS expression could not be restored (data not shown). However, when the promoter for the urease operon of Streptococcus salivarius, which has been demonstrated to be an efficient promoter for gene expression in S. gordonii (9), was fused with the entire arcR gene and cloned onto pDL278, the resulting construct was able to restore the AD activity to wild-type levels (data not shown).
FIG. 2.
Cotranscription of queA with arcR and primer extension analysis of S. gordonii queA. (A) Transcription initiation site of S. gordonii queA analyzed by primer extension. The arrow indicates the transcription initiation site at a G residue 181 bases upstream of the queA start codon. (B) RT-PCR analysis of mRNA from S. gordonii grown in TY medium containing 10 mM galactose with 50 mM arginine. Primers specific for the arcR and queA intergenic regions were used to amplify cDNA. Lane 1, negative control, in which the reaction mixture contained mRNA from S. gordonii but no RT was performed; lane 2, positive control, in which the control RNA from the RT reaction kit was used as the template to perform RT-PCR as recommended by the supplier; lanes 3 and 4, arcR-queA intergenic region, in which mRNA from S. gordonii and chromosomal DNA, respectively, were used as the templates for RT-PCR.
By searching the S. gordonii genome at http://ncbi.nlm.nih.gov, a 1,026-bp ORF that was 15 bp 5′ of, and was transcribed in the same orientation as, arcR was identified. Based on BLAST searches, this ORF was predicted to code for an S-adenosylmethionine:tRNA ribosyltransferase-isomerase involved in queosine modification of tRNAs. The overall similarity of the deduced amino acid sequences of the product of this ORF and QueA of other bacteria was high (Table 1). Thus, we designated this gene in S. gordonii queA. The ORF immediately 5′ of the queA gene in S. gordonii, whose product was revealed by BLAST searches to be a conserved hypothetical protein, was transcribed in the orientation opposite that of queA transcription.
TABLE 1.
Homologies of the deduced amino acid sequence of S. gordonii QueA with the QueA sequences of other bacteria
| Species |
S. gordonii QueA homology
|
|
|---|---|---|
| % Similarity | % Identity | |
| S. pneumoniae | 92 | 85 |
| S. pyogenes | 90 | 83 |
| E. faecalis | 88 | 76 |
| B. subtilis | 78 | 65 |
| E. coli | 67 | 48 |
| S. flexneri | 67 | 48 |
The queA gene was preceded by a putative Shine-Dalgarno sequence and began with an ATG codon. No putative rho-independent terminator could be located between queA and arcR. To determine whether queA and arcR were cotranscribed, RT-PCR using primers that were internal to queA and arcR was employed, as described in Materials and Methods. A PCR product of the expected size was detected after RT-PCR, indicating that a continuous queA-arcR transcript was present (Fig. 2B).
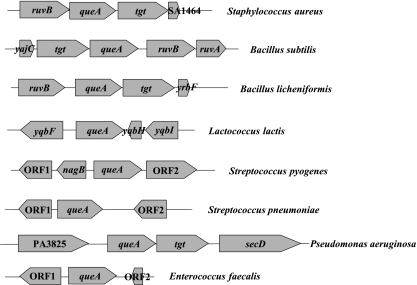
We examined other known bacterial arc operons and their flanking regions in sequenced bacterial genomes to determine whether it was common to find queA and arcR genetically linked. We found queA genes in Streptococcus pyogenes, Enterococcus faecalis, Bacillus subtilis, Lactococcus lactis, B. licheniformis, P. aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae, but none of them were linked to the arc operons (Fig. 3). We also searched for the tgt gene, which encodes the tRNA-guanine transglycosylase that is needed for the first step in queosine modification of tRNA. An apparent tgt gene was found in all cases and was linked to queA, except in the streptococci, E. faecalis, and L. lactis (the latter two organisms were previously members of the genus Streptococcus). In the S. gordonii genome, tgt was not linked to queA or the ADS genes. Thus, low-G+C-content gram-positive cocci show substantial divergence in the genomic organization of the tgt and queA genes, and S. gordonii is unique in the linkage of queA and the arc operon among organisms for which such sequence information is available.
FIG. 3.
Genetic arrangement of queA in ADS-positive bacteria. In S. aureus, ruvB encodes a holiday junction resolveasome helicase subunit and SA1464 encodes a hypothetical protein. In B. subtilis, yrbF encodes a hypothetical protein, tgt encodes queuine tRNA ribosyltransferase, ruvB encodes a holiday junction resolveasome helicase subunit, and ruvA encodes a holiday junction DNA helicase. In L. lactis, yqbF encodes a hypothetical protein, yqbH encodes a transcriptional regulator, and yqbI encodes a hypothetical protein. In S. pyogenes, ORF1 encodes a putative pseudouridylate synthetase, nagB encodes a putative N-acetylglucosamine-6-phosphate isomerase, and ORF2 encodes a hypothetical protein. In S. pneumoniae, ORF1 encodes glucosamine-6-phosphate isomerase and ORF2 encodes a surface protein. In B. licheniformis, ruvB encodes a holiday junction resolveasome helicase subunit, tgt encodes queuine tRNA ribosyltransferase, and yrbF encodes a preprotein translocase subunit. In P. aeruginosa, PA3825 encodes a hypothetical protein, tgt encodes queuine tRNA ribosyltransferase, and secD encodes a preprotein translocase subunit. In E. faecalis, ORF1 encodes a glyoxylase family protein and ORF2 encodes a transcriptional regulator belonging to the Cro/CI family.
Expression of pqueA and parcA.
S. gordonii WT/pqueA-cat and WT/parcA-cat were grown in FMC containing 20 mM glucose with 20 mM arginine. The expression level of pqueA, which drives the expression of the arc operon regulator arcR, and the expression level of parcA, which drives the expression of the ADS genes (14, 15), were examined in different growth phases. The data showed that there was little difference in expression of pqueA as a function of growth phase, but parcA expression was significantly higher in cells that had entered stationary phase than in exponential-phase cells (Fig. 4A). Also, AD enzyme assays showed that ADS expression was upregulated during stationary phase (Fig. 4B). The pH of the culture medium dropped from 6.6 in early exponential phase to 4.4 in overnight cultures (Fig. 4). Consequently, enhanced ADS expression during stationary phase could benefit S. gordonii not only by providing ATP but also by moderating acidification of the environment. Caldelari et al. previously reported greater AD enzyme activity as cells entered stationary phase (7), although in their study, no effort was made to control pH and cells were cultivated in the presence of glucose. As a result, it was not possible to attribute increases in AD activity measured in stationary-phase cells solely to growth phase, since a decrease in the pH and alleviation of CCR in stationary phase may have contributed to the observations.
FIG. 4.
CAT specific activities (A) and AD activities (B) of cells of WT/pqueA-cat and WT/parcA-cat in different growth phases. Cells were grown in FMC containing 20 mM glucose with 20 mM arginine to the early exponential, mid-exponential, early stationary, and late stationary phases and overnight and were used to measure the CAT specific activity and AD activity. The final pH values of the cultures were 6.6 at early exponential phase, 5.8 at mid-exponential phase, 5.2 at early stationary phase, 5.18 at stationary phase, and 4.4 in overnight cultures. The values are the means and standard deviations of three independent experiments.
Since the expression of the arcA operon of S. gordonii is under the control of CCR (13), we examined whether the queA-arcR operon is also CCR sensitive. Wild-type and CcpA-deficient S. gordonii/pqueA-cat strains were cultured in TY medium with 20 mM arginine and 20 mM galactose (nonrepressing conditions) or glucose (repressing conditions) to mid-exponential phase, and pqueA expression was examined. No significant differences in CAT activities were detected between the wild type and the CcpA mutant (Table 2), indicating that there was no modulation of queA-arcR transcription by CCR.
TABLE 2.
CAT specific activities of wild-type and CcpA-deficient pqueA-cat strains of S. gordonii grown in TY medium with 20 mM arginine and 20 mM galactose or glucose
| Strain | CAT sp acta
|
|
|---|---|---|
| With galactose | With glucose | |
| WT/pqueA-cat | 61.73 ± 11.21 | 64.73 ± 9.31 |
| CcpA−/pqueA-cat | 70.13 ± 19.72 | 76.28 ± 15.74 |
Cells were collected at mid-exponential phase (OD600, 0.5 to 0.6). The data are the means ± standard deviations of three independent experiments and are expressed in nanomoles of chloramphenicol acetylated per minute per milligram of total protein. All CAT assays were done in triplicate.
To examine the effects of acidic conditions on ADS expression and to compare the expression of pqueA and parcA in response to pH and arginine, S. gordonii WT/pqueA-cat and WT/parcA-cat were grown in a chemostat at pH 7.0 and 5.9 to a steady state following a 10 mM arginine pulse for 60 min. The use of pH 5.9 was dictated by the observation that S. gordonii could not be maintained at steady state at lower pH values under the conditions tested because of washout. As noted with batch cultivation, no effects of arginine or low pH on pqueA expression were observed (Table 3). However, parcA expression was 1.5-fold higher at pH 5.9 than at pH 7.0 and was induced 1.3-fold by addition of arginine (Table 3). For both the WT/pqueA-cat and WT/parcA-cat strains, threefold upregulation of arginine deminase enzyme activity was observed under acidic conditions compared to the activity at neutral pH (Table 3). While the increase was consistent with arcA transcriptional data, the greater increase in AD activity than in reporter gene activity raises the possibility that there is posttranscriptional regulation of AD expression at low pH. The expression level of pqueA was around 10-fold lower than that of parcA, as determined from comparisons of the levels of expression of cat in various culture conditions. We are presently exploring how other factors, such as lower oxygen tension and growth rate effects, could contribute to ADS upregulation in stationary phase.
TABLE 3.
CAT specific activities and AD activities of wild-type, CcpA-deficient, and QueA-deficient parcA-cat S. gordonii strains grown in chemostats containing TY medium at pH 7.0 and 5.9 separately with 10 mM glucose at a dilution rate of 0.3h−1a
| Strain | pH | CAT activityb
|
AD activityc
|
||
|---|---|---|---|---|---|
| Before arginine pulse | After arginine pulse | Before arginine pulse | After arginine pulse | ||
| WT/pqueA-cat | 7.0 | 28.98 ± 0.93d | 26.80 ± 0.82d | 56.10 ± 6.41 | 102.10 ± 8.57 |
| 5.9 | 30.12 ± 0.46d | 30.72 ± 1.60d | 138.00 ± 27.04 | 190.81 ± 23.77 | |
| WT/parcA-cat | 7.0 | 216.70 ± 2.12 | 380.99 ± 9.46 | 58.35 ± 6.71 | 116.60 ± 16.69 |
| 5.9 | 301.24 ± 12.88 | 450.25 ± 10.24 | 126.95 ± 12.73 | 241.45 ± 44.35 | |
| CcpA−/parcA-cat | 7.0 | 540.96 ± 3.90e | 532.92 ± 30.21e | 60.95 ± 22.22 | 94.83 ± 12.02 |
| 5.9 | 1,025.26 ± 5.12e | 1,004.00 ± 6.70e | 155.13 ± 27.57 | 160.52 ± 14.20 | |
| QueA−/parcA-cat | 7.0 | 680.95 ± 102.55e | 1,041.87 ± 119.35e | 68.81 ± 3.39 | 106.51 ± 4.19 |
| 5.9 | 1,071.10 ± 168.17e | 1,694.36 ± 109.52e | 175.12 ± 33.48 | 168.34 ± 2.87 | |
Cells were cultured to steady state and then pulsed with 10 mM arginine for 60 min.
The data are the means ± standard deviations of three independent experiments and are expressed in nanomoles of chloramphenicol acetylated per minute per milligram of total protein. All CAT assays were done in triplicate.
The data are the means ± standard deviations of three independent experiments and are expressed in micrograms of citrulline per minute per milligram of total protein. All AD assays were done in triplicate.
The CAT activities of WT/pqueA-cat differed significantly (P < 0.05) from those of WT/parcA-cat when cells were cells were cultured in the same environmental conditions, including pH and arginine concentration.
The CAT activities of the CcpA−/parcA-cat and QueA−/parcA-cat strains differed significantly (P < 0.05) from those of WT/parcA-cat when cells were cultured in the same environmental conditions, including pH and arginine concentration.
Analysis of QueA effects on parcA.
Very little is known about the physiological consequences of queuosine modification of tRNA in bacteria, although work with E. coli and S. flexneri suggested a possible role in modulation of translational efficiency, virulence, and post-exponential-phase survival (17, 19, 31). We posited that a possible role for QueA in ADS regulation is to modulate the translational efficiency of ArcR based on a comparison of the expression of the ADS operon promoter (parcA) of the wild-type strain and the expression of the ADS operon promoter of the QueA-deficient strain. The expression of the arcA promoter in the QueA-deficient strain was around threefold higher than that in the wild-type strain at both pH 7.0 and pH 5.9 in chemostat cultures (Table 3). These results were consistent with real-time PCR data showing that the arcA transcript was 14-fold more abundant in the QueA-deficient strain than in the wild-type strain (Fig. 5). These results support the hypothesis that QueA has a role in modulation of ADS gene expression, perhaps exerting a negative effect on the translation of arc regulatory proteins. Interestingly, no significant differences in AD activity were detected between the wild-type and QueA-deficient strains, in spite of the fact that transcription of arcA was increased in the queA mutant. These results imply that there is tight posttranscriptional control over the levels of AD activity in the cells, which is probably critical for maintaining appropriate arginine pools to support protein synthesis. It is also noteworthy that the queA transcript contains a large leader mRNA, raising the possibility that the efficiency of queA translation may be regulated or perhaps autoregulated. We are also investigating the possibility that there is a small protein that is expressed from the mRNA immediately upstream of queA.
FIG. 5.
Quantitative real-time PCR of arcA gene expression. After RT from 1 μg of total RNA from wild-type, QueA-deficient, and CcpA-deficient strains of S. gordonii, the amount of arcA gene cDNA was determined by real-time PCR using SYBR green. The data are means ± standard deviations, which were obtained using three different RNA preparations and RT reactions. An asterisk indicates that the copy number of the arcA mRNA of the QueA-deficient or CcpA-deficient strain differs significantly from that of the wild-type strain of S. gordonii (P < 0.05, as determined by the Student t test).
CcpA is not required for low-pH induction of the ADS.
CcpA appears to have primary control of CCR of the arc operon of S. gordoniii (14). Our chemostat studies revealed that the expression from the arcA promoter in the CcpA-deficient strain was about 2.5-fold higher than that in the wild-type strain at pH 7.0 and was 3-fold higher at pH 5.9. Real-time PCR demonstrated that arcA mRNA levels were 21-fold higher in the CcpA-deficient strain than in the wild-type background (Fig. 5), confirming that CcpA represses arc transcription (14). AD enzyme activity was also measured to test whether CcpA could affect the translation of arcA. As noted for the queA mutant, however, a substantial increase in arcA promoter activity was not accompanied by an increase in AD activity, further supporting the idea that there is tight posttranslational control over the amount of AD enzyme that can accumulate in cells, possibly as a result of differential mRNA stability and translational efficiency.
The results of the chemostat experiments clearly showed that a low pH and arginine could increase arc operon expression in wild-type S. gordonii (Table 3). Importantly, our results indicate that neither QueA nor CcpA is involved in the responses of S. gordonii to pH or arginine. It is fairly well established (14) that ArcR is the primary control circuit for arginine induction of the operon, but the factor(s) responsible for pH induction, as well as stationary-phase gene expression, remains undefined. While the importance of acid tolerance in persistence and virulence of oral streptococci is well documented, the molecular mechanisms of regulation of genes by pH in these organisms remain largely unexplored. The ADS will undoubtedly prove to be valuable for identifying factors responsible for activation or derepression of genes in response to acidic conditions.
Growth of S. gordonii and QueA-deficient strains.
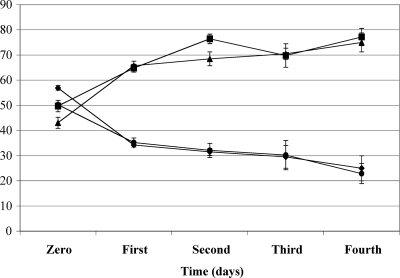
An E. coli mutant lacking QueA exhibited a decreased ability to survive in stationary phase compared to the wild-type strain (19, 31). To determine if the QueA-deficient S. gordonii strain has a growth deficiency compared to the wild-type strain in stationary phase, we examined the growth rates of the wild-type and queA mutant strains of S. gordonii. No significant differences were detected in the growth rates of the wild-type and queA mutant strains of S. gordonii in FMC containing glucose or galactose with arginine (data not shown). Therefore, a persistence experiment was carried out by using serial passage of a mixture of the wild-type and mutant strains for 4 days in TY medium containing 0.2% glucose or TY medium containing 0.2% galactose with 10 mM arginine (Fig. 6). Such competition experiments can sensitively detect minor changes in the fitness of a mutant strain. From day 1 to day 4 in both media, the proportion of QueA-deficient cells declined (Fig. 5), suggesting that the lack of queuosine modification in S. gordonii may adversely affect the competitive fitness of the organism. Although the final pH of the glucose culture was 0.5 pH unit lower than that of the galactose-arginine culture, no significant differences in the proportions in the cultures were detected (Fig. 5). Thus, the poorer competition associated with the deficiency of the QueA mutant did not correlate closely with pH and arginine metabolism, so the underlying basis for the defect remains to be determined.
FIG. 6.
Persistence experiment using a mixed inoculum containing wild-type S. gordonii and a nonpolar QueA-deficient strain. The proportions of the wild type (▴ and ▪) and the QueA-deficient strain (⧫ and •) in TY medium containing 0.2% glucose (⧫ and ▴) and TY medium containing 0.2% galactose (• and ▪) with 10 mM arginine are shown. The values are the means and standard deviations (error bars) of three independent experiments.
Summary.
In summary, the findings obtained in this study enhance our understanding of the mode and mechanisms of regulation of the ADS of S. gordonii and highlight the complexity underlying differential expression of arc genes. In addition, the results not only revealed a novel association between QueA, the arc operon, and post-exponential-phase gene expression but also demonstrated that ADS gene expression, in addition to being highly regulated by carbohydrate and arginine, is sensitive to pH and is upregulated in stationary phase independent of CCR or pH. Given that the nutrient source, the growth rate, pH, oxygen, and other influences and stresses that regulate ADS expression also have a profound impact on oral biofilm physiology and the expression of virulence, the ADS continues to be an excellent model for studying gene regulation and physiology in oral streptococci. In addition, there is tremendous potential to exploit the ADS and arginine catabolism in preventive strategies against dental caries and other oral diseases (6), so this exploration provided considerable practical knowledge for developing strategies to optimize ADS expression in oral biofilms.
Acknowledgments
This work was supported by Public Health Service grant DE10362 from the National Institute of Dental and Craniofacial Research.
We thank L. Zeng for critical evaluation of the manuscript.
Footnotes
Published ahead of print on 13 June 2008.
REFERENCES
- 1.Ahn, S. J., J. A. Lemos, and R. A. Burne. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J. Bacteriol. 187:3028-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archibald, R. M. 1944. Determination of citrulline and allantoin and demonstration of citrulline in blood plasma. J. Biol. Chem. 156:121-142. [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 4.Bowden, G. H., and I. R. Hamilton. 1987. Environmental pH as a factor in the competition between strains of the oral streptococci Streptococcus mutans, S. sanguis, and “S. mitior” growing in continuous culture. Can. J. Microbiol. 33:824-827. [DOI] [PubMed] [Google Scholar]
- 5.Burne, R. A., D. T. Parsons, and R. E. Marquis. 1991. Environmental variables affecting arginine deiminase expression in oral streptococci, p. 276-280. In P. P. Dunny, G. M. Cleary, and L. L. McKay (ed.), Genetics and molecular biology of streptococci, lactococci, and enterococci. American Society for Microbiology, Washington, DC.
- 6.Burne, R. A., and R. E. Marquis. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1-6. [DOI] [PubMed] [Google Scholar]
- 7.Caldelari, I., B. Loeliger, H. Langen, M. P. Glauser, and P. Moreillon. 2000. Deregulation of the arginine deiminase (arc) operon in penicillin-tolerant mutants of Streptococcus gordonii. Antimicrob. Agents Chemother. 44:2802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casiano-Colon, A., and R. E. Marquis. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 54:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y. Y., M. J. Betzenhauser, and R. A. Burne. 2002. cis-acting elements that regulate the low-pH-inducible urease operon of Streptococcus salivarius. Microbiology 148:3599-3608. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. Y., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degnan, B. A., J. M. Palmer, T. Robson, C. E. Jones, M. Fischer, M. Glanville, G. D. Mellor, A. G. Diamond, M. A. Kehoe, and J. A. Goodacre. 1998. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect. Immun. 66:3050-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, Y. 2004. The arginine deiminase system of Streptococcus gordonii DL-1. Ph.D. dissertation. University of Florida, Gainesville.
- 14.Dong, Y., Y. Y. Chen, and R. A. Burne. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong, Y., Y. Y. Chen, J. A. Snyder, and R. A. Burne. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl. Environ. Microbiol. 68:5549-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durand, J. M., N. Okada, T. Tobe, M. Watarai, I. Fukuda, T. Suzuki, N. Nakata, K. Komatsu, M. Yoshikawa, and C. Sasakawa. 1994. vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of Escherichia coli K-12. J. Bacteriol. 176:4627-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferro, K. J., G. R. Bender, and R. E. Marquis. 1983. Coordinately repressible arginine deiminase system in Streptococcus sanguis. Curr. Microbiol. 9:145-150. [Google Scholar]
- 19.Frey, B., G. Janel, U. Michelsen, and H. Kersten. 1989. Mutations in the Escherichia coli fnr and tgt genes: control of molybdate reductase activity and the cytochrome d complex by fnr. J. Bacteriol. 171:1524-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frey, B., J. McCloskey, W. Kersten, and H. Kersten. 1988. New function of vitamin B12: cobamide-dependent reduction of epoxyqueuosine to queuosine in tRNAs of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 170:2078-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaur, R., and U. Varshney. 2005. Genetic analysis identifies a function for the queC (ybaX) gene product at an initial step in the queuosine biosynthetic pathway in Escherichia coli. J. Bacteriol. 187:6893-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartmann, R., H. D. Sickinger, and D. Oesterhelt. 1980. Anaerobic growth of halobacteria. Proc. Natl. Acad. Sci. USA 77:3821-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwata-Reuyl, D. 2003. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg. Chem. 31:24-43. [DOI] [PubMed] [Google Scholar]
- 25.Kinzie, S. D., B. Thern, and D. Iwata-Reuyl. 2000. Mechanistic studies of the tRNA-modifying enzyme QueA: a chemical imperative for the use of AdoMet as a “ribosyl” donor. Org. Lett. 2:1307-1310. [DOI] [PubMed] [Google Scholar]
- 26.Kremer, B. H., M. van der Kraan, P. J. Crowley, I. R. Hamilton, L. J. Brady, and A. S. Bleiweis. 2001. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J. Bacteriol. 183:2543-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeBlanc, D. J., and F. P. Hassell. 1976. Transformation of Streptococcus sanguis Challis by plasmid deoxyribonucleic acid from Streptococcus faecalis. J. Bacteriol. 128:347-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, C. D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maghnouj, A., T. F. de Sousa Cabral, V. Stalon, and C. Vander Wauven. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor argR. J. Bacteriol. 180:6468-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquis, R. E., G. R. Bender, D. R. Murray, and A. Wong. 1987. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 53:198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noguchi, S., Y. Nishimura, Y. Hirota, and S. Nishimura. 1982. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J. Biol. Chem. 257:6544-6550. [PubMed] [Google Scholar]
- 32.Okada, N., S. Noguchi, H. Kasai, N. Shindo-Okada, T. Ohgi, T. Goto, and S. Nishimura. 1979. Novel mechanism of post-transcriptional modification of tRNA. Insertion of bases of Q precursors into tRNA by a specific tRNA transglycosylase reaction. J. Biol. Chem. 254:3067-3073. [PubMed] [Google Scholar]
- 33.Okada, N., S. Noguchi, S. Nishimura, T. Ohgi, T. Goto, P. F. Crain, and J. A. McCloskey. 1978. Structure determination of a nucleoside Q precursor isolated from E. coli tRNA: 7-(aminomethyl)-7-deazaguanosine. Nucleic Acids Res. 5:2289-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 36.Slany, R. K., M. Bosl, P. F. Crain, and H. Kersten. 1993. A new function of S-adenosylmethionine: the ribosyl moiety of AdoMet is the precursor of the cyclopentenediol moiety of the tRNA wobble base queuine. Biochemistry 32:7811-7817. [DOI] [PubMed] [Google Scholar]
- 37.Slany, R. K., and H. Kersten. 1994. Genes, enzymes and coenzymes of queuosine biosynthesis in procaryotes. Biochimie 76:1178-1182. [DOI] [PubMed] [Google Scholar]
- 38.Terleckyj, B., N. P. Willett, and G. D. Shockman. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vander Wauven, C., A. Pierard, M. Kley-Raymann, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Lanen, S. G., and D. Iwata-Reuyl. 2003. Kinetic mechanism of the tRNA-modifying enzyme S-adenosylmethionine:tRNA ribosyltransferase-isomerase (QueA). Biochemistry 42:5312-5320. [DOI] [PubMed] [Google Scholar]
- 41.Van Lanen, S. G., S. D. Kinzie, S. Matthieu, T. Link, J. Culp, and D. Iwata-Reuyl. 2003. tRNA modification by S-adenosylmethionine:tRNA ribosyltransferase-isomerase. Assay development and characterization of the recombinant enzyme. J. Biol. Chem. 278:10491-10499. [DOI] [PubMed] [Google Scholar]
- 42.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wexler, D. L., M. C. Hudson, and R. A. Burne. 1993. Streptococcus mutans fructosyltransferase (ftf) and glucosyltransferase (gtfBC) operon fusion strains in continuous culture. Infect. Immun. 61:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winterhoff, N., R. Goethe, P. Gruening, M. Rohde, H. Kalisz, H. E. Smith, and P. Valentin-Weigand. 2002. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the arginine deiminase system of Streptococcus pyogenes. J. Bacteriol. 184:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuniga, M., M. Champomier-Verges, M. Zagorec, and G. Perez-Martinez. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuniga, M., G. Perez, and F. Gonzalez-Candelas. 2002. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 25:429-444. [DOI] [PubMed] [Google Scholar]