Abstract
The microbial communities of three different sulfidic and acidic mine waste tailing dumps located in Botswana, Germany, and Sweden were quantitatively analyzed using quantitative real-time PCR (Q-PCR), fluorescence in situ hybridization (FISH), catalyzed reporter deposition-FISH (CARD-FISH), Sybr green II direct counting, and the most probable number (MPN) cultivation technique. Depth profiles of cell numbers showed that the compositions of the microbial communities are greatly different at the three sites and also strongly varied between zones of oxidized and unoxidized tailings. Maximum cell numbers of up to 109 cells g−1 dry weight were determined in the pyrite or pyrrhotite oxidation zones, whereas cell numbers in unoxidized tailings were significantly lower. Bacteria dominated over Archaea and Eukarya at all tailing sites. The acidophilic Fe(II)- and/or sulfur-oxidizing Acidithiobacillus spp. dominated over the acidophilic Fe(II)-oxidizing Leptospirillum spp. among the Bacteria at two sites. The two genera were equally abundant at the third site. The acidophilic Fe(II)- and sulfur-oxidizing Sulfobacillus spp. were generally less abundant. The acidophilic Fe(III)-reducing Acidiphilium spp. could be found at only one site. The neutrophilic Fe(III)-reducing Geobacteraceae as well as the dsrA gene of sulfate reducers were quantifiable at all three sites. FISH analysis provided reliable data only for tailing zones with high microbial activity, whereas CARD-FISH, Q-PCR, Sybr green II staining, and MPN were suitable methods for a quantitative microbial community analysis of tailings in general.
Mining residues from mining activities are dumped as waste rock or as tailings, which are metal-degraded materials from ore processing. Both kinds of dumps often contain sulfide minerals such as pyrite (FeS2) or pyrrhotite (Fe1−xS, x = 0 to 0.125) and release acidic metal-rich waters known as acid mine drainage (AMD)/acid rock drainage (ARD) because of chemical and microbial sulfide oxidation processes, e.g.,
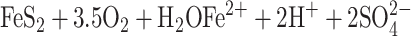 |
Over a period of several years, an oxidized zone with depleted sulfide content, low pH, and enrichment of secondary minerals develops above an unoxidized zone with unaltered material in the waste dump (e.g., references 4, 14, 21, 40, and 49).
Several geomicrobiological investigations of sulfidic mine waste dumps located in different climate zones have been undertaken to gather information about microbial processes and diversity in these extreme environments. At such sites, aerobic, acidophilic, chemolithotrophic Bacteria or Archaea dissolve metal sulfides by oxidizing Fe(II) and sulfur compounds and generate AMD/ARD. Products resulting from these oxidation processes can be used by Fe(III)- and sulfate-reducing prokaryotes. When Fe(III) (hydr)oxides are dissolved, adsorbed or precipitated metals are released. Sulfate-reducing Bacteria or Archaea may also precipitate metals as metal sulfides (22, 49, 50).
Primarily cultivation techniques have been used to enumerate prokaryotes involved in oxidation and reduction processes in sulfidic mine waste dumps (3, 17, 52, 57, 64). Cultivation techniques yield cell numbers merely according to physiological properties; therefore, only a subset of the whole microbial community is detected. Up to this point, only qualitative molecular biological tests were applied to sulfidic mine dumps such as cloning and subsequent sequencing (7), denaturing gradient gel electrophoresis (12, 38), and terminal restriction fragment length polymorphism (7, 13). In addition, protein and lipid analysis (37, 58) were performed with tailing samples. These investigations provided valuable information about the microbial diversity in mine dumps but little information about the quantities of the different microbial groups or species. The molecular biological quantification technique fluorescence in situ hybridization (FISH) and metagenomic and proteomic techniques have been previously applied to quantify acidophiles in water samples from AMD sites (2, 6, 15, 20, 23, 46, 56, 61) but not solid samples from mine dumps.
In this study, the microbial communities in three mine tailing dumps, each with a different mineralogy and located in different climate zones, were quantified by cultivation and molecular biological methods. Depth profiles of cell numbers of different microorganisms were created using Sybr green II direct counting, quantitative real-time PCR (Q-PCR), FISH, catalyzed reporter deposition-FISH (CARD-FISH), and the most probable number (MPN) cultivation technique. Some of the data from MPN and total number determination of Bacteria and Archaea were previously published in connection with geochemical and mineralogical studies, as were data from determinations of metal sulfide oxidation rates determined by calorimetry (21, 25, 30, 31, 55). Here, a detailed Q-PCR and FISH analysis of microorganisms relevant for Fe and S oxidation and reduction processes is presented for the first time.
MATERIALS AND METHODS
Site description and sampling. (i) Uncovered pyrrhotite-containing tailing dump, Selebi-Phikwe, Botswana.
The tailing dump in Botswana exists in a semiarid climate with an average annual temperature of 21°C. At the time of sampling in 2003, it was still in operation and uncovered. It was approximately 40 m high and spread over an area of approximately 1 km2. It contained Ni, Cu, Zn, and Co sulfidic ore processing waste accumulated over a period of about 32 years. The solid material of the original tailings consisted of 11% pyrrhotite, about 1.5% other metal sulfides, and hornblende and feldspar as gangue minerals. A set of 24 drilling samples was taken to a depth of 26 m. The content of total reduced inorganic sulfur was 4.1% on average in the investigated samples, and total organic carbon (TOC) was below 0.1%. The geochemistry of the tailing dump and the sampling procedure have been previously described (55).
(ii) Covered pyrite-containing tailings in impoundment 1, Kristineberg, northern Sweden.
The climate in Kristineberg, northern Sweden, is humid with an annual precipitation between 400 and 800 mm per year and an annual mean temperature of 0.7°C. The tailing dump had been in operation for about 10 years and was left unremediated for approximately 40 years before it was covered in 1996 with a soil cover consisting of 0.3 m compacted till and 1.5 m unspecific till. Before covering, sulfide oxidation occurred in distinct depth layers (oxidized tailings). The tailing dump consists of waste from Zn and Cu sulfidic ore processing, covers an area of 0.1 km2, and is 6 to 7 m high. In the unoxidized tailings, the content of sulfide minerals ranges from 10 to 30% and is completely dominated by pyrite, while in the oxidized tailings the sulfide content is generally around 1 to 2%. In 2003, nine samples were taken by drilling to a depth of 6.5 m. The TOC was below 0.8%. The geochemistry and mineralogy of the tailing dump and the sampling procedure have been described by others in detail (8, 9, 25-27, 30, 39, 41).
(iii) Uncovered pyrite- and arsenopyrite-containing tailing dump, Freiberg, Germany.
The tailing dump in Freiberg, Germany, is located in a temperate zone with an annual mean precipitation of 763 mm per year and an annual mean temperature of 7.7°C. Its height is 30 m, and it is spread over an area of 0.06 km2. It is unremediated and partly covered by a layer of soil less than 0.2 m in thickness. The tailing dump consists of waste from Pb and Zn sulfidic ore processing with the main metal sulfides being pyrite, arsenopyrite, sphalerite, and galena. Three thin, cemented layers occur in a depth range of 0.6 to 0.63 m. They separate an upper zone (metal sulfide content of ≤0.1%), which underwent oxidation, from a zone of active oxidation (metal sulfide content of ≤1%) beneath it. An unoxidized zone with a metal sulfide content of 1% occurs at a depth below 0.85 m. In 2005, 21 samples were taken down to a depth of 1.2 m in an outcrop profile from the center of the tailing dump. TOC was below 0.1%. The geochemistry and mineralogy of the tailing dump and the sampling procedure have been previously described (21).
Quantification of microorganisms. (i) Sybr green II direct counting and MPN.
Total cell numbers were determined in formaldehyde-fixed samples (36) by staining with Sybr green II as described previously (63). The MPN technique was used to enumerate acidophilic chemolithoautotrophic Fe(II) oxidizers and sulfur oxidizers. For quantifying Fe(II) oxidizers, the medium described by Leathen et al. (34) at pH 3.5 was used: (NH4)2SO4, 0.15 g; KCl, 0.05 g; MgSO4·7H2O, 0.50 g; K2HPO4, 0.05 g; Ca(NO3)2, 0.01 g; FeSO4·7H2O, 1.00 g in 1,000 ml of distilled water. The following medium described by Starkey (60) at pH 3.5 was used for quantifying sulfur oxidizers: (NH4)2SO4, 0.30 g; KH2PO4, 3.50 g; CaCl2, 0.25 g; MgSO4, 0.50 g; Fe(SO4)3, 0.01 g; elemental sulfur, 10 g in 1,000 ml of distilled water.
(ii) FISH and CARD-FISH.
Living Bacteria (probes EUB338 I to EUB338 III) and Archaea (ARCH915) were quantified by CARD-FISH and by FISH in formaldehyde-fixed samples on filters as previously published (11, 43, 44, 54, 59). FISH was also applied for quantifying specific Fe- and S-oxidizing and/or -reducing prokaryotes by using the following probes: LF655 (specific for Leptospirillum species of groups I, II, and III), ACM732 (specific for the genus Acidimicrobium and relatives), SUL228 (specific for the genus Sulfobacillus) (5), Acdp821 (specific for the genus Acidiphilium), Thio820 (specific for Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans) (42), TF539 (specific for A. ferrooxidans) (56), and LGC0355 (specific for gram-positive bacteria with low G+C content [23]). The detection limit for the microscopic methods Sybr green II, FISH, and CARD-FISH was 105 cells g−1 dry weight each. The specificity for selected probes was tested with different pure cultures.
(iii) Real-time PCR.
The DNA was extracted from 1 to 5 g of a frozen tailing sample following a modified FastDNA Spin Kit for Soil (Bio 101) protocol (62). Q-PCR was used to quantify Bacteria, Archaea, Geobacteraceae, Leptospirillum spp., Acidithiobacillus caldus, and Sulfobacillus spp. by targeting their 16S rRNA genes; Eukarya by targeting their 18S rRNA genes; and sulfate-reducing prokaryotes by targeting their specific functional gene dsrA as described previously (35, 51). Furthermore, the 16S rRNA gene of Acidithiobacillus spp. was quantified using a modified protocol published by Zammit et al. (65). Mastermix (Eurogentec) with Sybr green I (12.5 μl) was used with 1 μl of each primer (250 nM; 27F [33] and At.f384R [5′-CATTGCTTCGTCAGGGTTG-3′] [65]) and 1.25 μl bovine serum albumin in a total reaction volume of 25 μl. The cycling parameters were as follows: one cycle of 94°C for 10 min and 35 cycles of 94°C for 45 s, 61°C for 1 min, and 72°C for 45 s.
Each DNA extract was measured in triplicate and in at least two dilutions to check for PCR inhibition. After each Q-PCR run, melting curves were measured for Sybr green I assays. The primer specificity for the specific Q-PCR assays was confirmed by sequence alignment in databases (Blast, Ribosomal Database Project) and tested using DNA of A. ferrooxidansT, A. caldusT, A. thiooxidansT, Leptospirillum ferrooxidansT, Leptospirillum ferriphilumT, and Sulfobacillus acidophilusT. To convert DNA copy numbers to cell numbers, the following conversion factors were used: 4.1 for Bacteria and Geobacteraceae, 1.5 for Archaea (29), 1 for the dsrA gene (51), 12 for Acidithiobacillus spp. and A. caldus, 6 for Sulfobacillus spp., and 10 for Leptospirillum spp. (65). The detection limits for Q-PCR analyses were 103 DNA copies g−1 dry weight for the assays specific for Bacteria and Acidithiobacillus spp.; 102 DNA copies g−1 dry weight for the assays specific for Geobacteraceae, Eukarya, Leptospirillum spp., and the dsrA gene; and 101 DNA copies g−1 dry weight for the assays specific for Archaea, Sulfobacillus spp., and A. caldus.
RESULTS
Oxidized and unoxidized tailing zones.
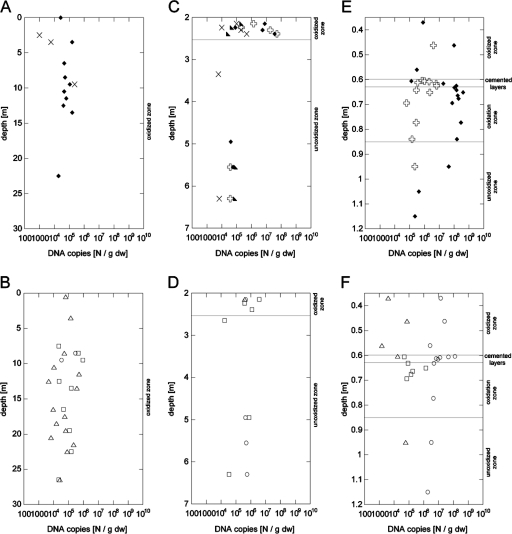
We observed significant differences in the location and thickness of oxidized zones with reduced sulfide content due to sulfide weathering and unoxidized zones in the three sulfidic mine tailing dumps. In the active tailing dump in Selebi-Phikwe, Botswana, the entire investigated depth of 26 m comprised an oxidized zone defined by the occurrence of brownish Fe(III) (hydr)oxides and a low pH of 3 to 4 measured in the paste of a sample (Fig. 1B). In contrast, the remediated tailing dump in Kristineberg, Sweden, was divided into an approximately half-meter-thick oxidized zone with an average pH of 3.5 measured in the paste of a sample below a 2-m-thick cover of soil and above the unoxidized tailings (pH 4.9) (Fig. 1D). A special situation was found in the inactive tailing dump in Freiberg, Germany, where a zone of cemented layers in 0.6- to 0.63-m depth (pH 3 to 4) divided the previously oxidized tailings above (pH 4 to 5) from the zone of active oxidation below (pH ∼3). The zone of unoxidized tailings (pH 5 to 7) was located below an 0.85-m depth (Fig. 1F). The TOC was generally low in all tailing dumps; thus, metal sulfides must provide the main energy source for microbial activity.
FIG. 1.
The pH values (×) and cell numbers of samples from mine tailing dumps located in Selebi-Phikwe, Botswana (A and B); Kristineberg, Sweden (C and D); and Freiberg, Germany (E and F). The cell numbers were detected by Sybr green II direct counting (•), CARD-FISH for Bacteria ( ), the MPN technique for detection of Fe(II) oxidizers (○), Q-PCR for Bacteria (▵), and Q-PCR for Archaea (□). The number of Archaea detected by CARD-FISH was below the detection limit of 105 cells g−1 dry weight (dw) at all three sites. Note different zones and depth scales. (Panels A and B are both adapted from references 30 and 55 with permission from Elsevier. Panel C is adapted from reference 25 with permission of the publisher [copyright 2008 American Chemical Society] and reference 30 with permission from Elsevier. Panel D is adapted from reference 30 with permission from Elsevier. Panels E and F are both adapted from reference 31 with permission and reference 21 with permission from Elsevier.)
Quantification of total cells, Bacteria, Archaea, and cultivable Fe(II) oxidizers.
The quantitative microbial community analysis showed significantly different depth profiles of cell numbers for the various microbial groups (Fig. 1 and 2). Sybr green II total cell counts comprised the highest cell numbers in all three tailings over the whole depth profiles (106 to 109 cells g−1 dry weight) (Fig. 1A, C, and E). Bacterial cell numbers determined via Q-PCR were somewhat lower but of the same order of magnitude as the total cell counts at all three sites. Furthermore, Archaea were detected via Q-PCR in the oxidized zone of the tailing dump in Sweden and above the zone of cemented layers in the tailing dump in Germany but not in Botswana. By the use of CARD-FISH, Bacteria were detected in different proportions of total cell numbers at the different sites, while Archaea remained below the detection limit of 105 cells g−1 dry weight at all three sites.
FIG. 2.
DNA copy numbers of the 16S rRNA genes of Acidithiobacillus spp. (⧫), Acidithiobacillus caldus (◣), Leptospirillum spp. ( ), Sulfobacillus spp. (×), Geobacteraceae (▵), the dsrA gene of sulfate reducers (□), and the 18S rRNA gene of Eukarya (○) in samples from mine waste tailings located in Selebi-Phikwe, Botswana (A and B); Kristineberg, Sweden (C and D); and Freiberg, Germany (E and F). Note different zones and depth scales. dw, dry weight.
The highest cell numbers (108 to 109 cells g−1 dry weight) were detected with all methods in the active oxidation zone (0.63- to 0.85-m depth) in the tailing dump in Germany. Above and below this zone, the cell numbers of living and cultivable cells detected by CARD-FISH and MPN, respectively, were significantly lower than cell numbers detected by Sybr green and Q-PCR (Fig. 1E and F). In the tailing dump in Botswana, cell numbers generally did not show a depth trend except for the MPN numbers of Fe(II) oxidizers, which decreased toward the tailing surface correlating with the water content (data not shown). CARD-FISH and MPN detectable cell numbers generally exhibited a high proportion of Sybr green and Q-PCR detectable cell numbers (Fig. 1A and B). A pronounced depth trend was observed in Sweden, where the cell numbers detected in the oxidized zone of the tailing dump (107 to 108 cells g−1 dry weight) were approximately 1 order of magnitude higher than those in the unoxidized zone (Fig. 1C and D).
Quantification of microorganisms of specific groups detected via Q-PCR analysis.
Since the Q-PCR data showed high bacterial cell numbers, this method was chosen for a detailed microbial community analysis by the quantification of genes of particular microbial phylogenetic groups, genera, or species relevant for mine tailings (Fig. 2). In the tailing dump in Botswana, the DNA copy numbers of all specific genes were between 104 and 106 copies g−1 dry weight (Fig. 2A and B). In many samples of the investigated depth profile, the acidophilic Fe(II) and/or sulfur oxidizers Acidithiobacillus spp., the Fe(III) reducers Geobacteraceae, and sulfate reducers (dsr) were detected. The 16S rRNA genes specific for the acidophilic Fe(II) oxidizers Leptospirillum spp. and the acidophilic sulfur oxidizer A. caldus were below the detection limit of 103 copies g−1 dry weight. The acidophilic Fe(II) and sulfur oxidizer Sulfobacillus spp. and eukaryotic 18S rRNA genes were detected in only a few samples.
In the tailing dump in Sweden, Leptospirillum spp. and Acidithiobacillus spp. were most abundant in the oxidized zone with the highest DNA copy numbers of up to 107 copies g−1 dry weight (Fig. 2C and D). DNA gene copy numbers specific for Eukarya, Geobacteraceae, sulfate reducers, Sulfobacillus spp., and A. caldus were between 104 and 106 copies g−1 dry weight in both zones.
In the tailing dump in Germany, the microbial community was dominated by Acidithiobacillus spp., which were most abundant in the oxidation zone below the cemented layers with the maximum DNA gene copy number of 108 copies g−1 dry weight and exceeded the abundance of Leptospirillum spp. by 3 orders of magnitude in higher DNA gene copy numbers (Fig. 2E and F). While the DNA gene copy numbers of Acidithiobacillus spp. were most abundant, A. caldus was not detected by Q-PCR. Also, Sulfobacillus spp. were below the detection limit of 103 copies g−1 dry weight. DNA gene copy numbers between 103 and 106 copies g−1 dry weight were obtained for the 16S rRNA gene of Geobacteraceae and for dsr. The latter gene was detected only in the zone of active oxidation. Eukaryotic 18S rRNA genes could be found throughout the entire investigated depth in high amounts (106 to 108 copies g−1 dry weight).
Quantification of microorganisms of specific groups detected via FISH analysis.
In addition to the detailed quantitative microbial community analysis using Q-PCR, FISH analyses of five selected samples from each tailing dump were performed with different specific probes. For comparison, the cell numbers obtained by FISH as well as CARD-FISH, Q-PCR, and MPN for the selected samples are shown in Table 1. By the use of CARD-FISH and Q-PCR, Bacteria were detected in all samples in similar numbers and usually slightly below the total cell counts. By the use of FISH, Bacteria could not be quantified for all samples and the bacterial cell numbers were significantly lower than those determined by CARD-FISH in samples from Botswana and Sweden. In contrast, for the five samples from the zone of active oxidation processes in the tailing dump in Germany, similar bacterial numbers were obtained for FISH and CARD-FISH, even though the brightness of the cell signals was considerably different for the two methods (Fig. 3). Archaea were quantified via Q-PCR in samples from Sweden but not by CARD-FISH or FISH. No other specific groups were detected via FISH in the latter tailing dump.
TABLE 1.
Cell numbers of five selected samples from each mine tailing dump located in Botswana, Sweden, and Germany determined by different quantification methodsa
| Organism type | Method | Cell no. (log n/g dry wt) for location, zone, and depth (m) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Selebi-Phikwe, Botswana (oxidized zone) |
Kristineberg, Sweden |
Freiberg, Germany (oxidation zone) |
||||||||||||||
| Oxidized zone |
Unoxidized zone (2.65 m) | |||||||||||||||
| 6.5 | 8.5 | 9.5 | 11.5 | 17.5 | 2.15 | 2.24 | 2.3 | 2.39 | 0.63 | 0.64 | 0.65 | 0.68 | 0.77 | |||
| Total | Sybr green | 7.6 | 8.1 | 7.9 | 8.7 | 8.2 | 8.9 | 8.6 | 9.0 | 8.5 | 6.5 | 9.4 | 9.3 | 9.4 | 9.6 | 8.3 |
| Bacteria | Q-PCR | 7.1 | 7.5 | 8.1 | 8.1 | 6.7 | 7.7 | 7.1 | 7.8 | 3.8 | 3.9 | 8.1 | 8.4 | 8.8 | 8.4 | 8.9 |
| CARD-FISH | 6.9 | 7.5 | 7.6 | 7.7 | 6.5 | 7.7 | 6.8 | 7.8 | 7.1 | 6.0 | 7.9 | 8.8 | 8.4 | 8.5 | 8.2 | |
| FISH | 5.8 | 6.4 | ND | 6.3 | 5.1 | ND | 6.3 | ND | ND | ND | 8.1 | 8.9 | 8.4 | 8.6 | 8.4 | |
| Archaea | Q-PCR | ND | ND | ND | ND | ND | 5.9 | 5.4 | 6.8 | 7.9 | ND | ND | ND | ND | ND | ND |
| Fe(II) oxidizers | MPN | 5.5 | 7.1 | 7.1 | 7.1 | 5.4 | 5.5 | 3.8 | 2.8 | 6.8 | ND | 8.8 | 8.8 | 9.2 | 8.8 | 8.8 |
| S oxidizers | MPN | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | 6.8 | 6.2 | 6.5 | 7.8 | 7.8 |
| Acidithiobacillus spp. | Q-PCR | 3.6 | 3.6 | 4.0 | 3.7 | ND | 5.8 | 3.8 | 5.7 | 6.5 | ND | 7.1 | 7.1 | 7.5 | 7.2 | 7.4 |
| FISH | ND | 5.8 | ND | 5.8 | ND | ND | ND | ND | ND | ND | 8.0 | 8.8 | 8.1 | 8.6 | 7.7 | |
| A. ferrooxidans | FISH | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 7.7 | 8.8 | 8.4 | 8.5 | 7.4 |
| A. caldus | Q-PCR | ND | ND | ND | ND | ND | 4.2 | 3.6 | ND | 3.3 | ND | ND | ND | ND | ND | ND |
| Leptospirillum spp. | Q-PCR | ND | ND | ND | ND | ND | 5.1 | 4.3 | 6.3 | 6.7 | ND | 5.8 | 4.5 | 5.4 | ND | 4.4 |
| L. ferrooxidans | FISH | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 5.9 | ND | 5.8 | ND | ND |
| Sulfobacillus spp. | Q-PCR | ND | ND | 4.6 | ND | ND | 4.2 | 3.2 | 4.5 | 4.9 | ND | ND | ND | ND | ND | ND |
| Acidiphilium spp. | FISH | ND | 5.3 | ND | 5.6 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Geobacteraceae | Q-PCR | ND | 4.1 | ND | 5.1 | 4.1 | 5.0 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Sulfate reducers (dsr) | Q-PCR | ND | 5.7 | 6.0 | ND | ND | 6.6 | 5.6 | ND | 6.1 | 4.2 | ND | ND | 6.1 | 5.1 | ND |
Cell numbers were below detection limits for FISH and CARD-FISH for Archaea, FISH for Acidimicrobium and relatives, FISH for gram-positive Bacteria with low G+C contents, and FISH for Sulfobacillus spp. in the investigated samples of all three sites. Conversion factors for calculating cell numbers from DNA copy numbers and detection limits of the methods are given in Materials and Methods. NM, not measured; ND, not detected.
FIG. 3.
FISH, CARD-FISH, and DAPI (4′,6′-diamidino-2-phenylindole) fluorescence images for a selected sample from the oxidation zone of the sulfidic mine waste dump in Freiberg, Germany. (A and C) Weak FISH (A) and bright CARD-FISH (C) signals of cells detected with probes EUB338 I to III specific for Bacteria. (B and D) Same microscopic fields as panels A and C, respectively, with UV excitation (DAPI staining).
In the tailing dump in Botswana, Acidithiobacillus spp. were detected via FISH only in two of five of the investigated samples and in significantly lower cell numbers than those detected via MPN for Fe(II) oxidizers. The cell numbers were somewhat higher than those detected with Q-PCR for Acidithiobacillus spp. FISH probes specific for the acidophilic Fe(II) oxidizers A. ferrooxidans and L. ferrooxidans did not show quantifiable positive signals. Acidiphilium species-specific FISH probes showed cell numbers in the same samples and of the same order of magnitude as those for the Acidithiobacillus species-specific FISH probes.
In contrast to samples from the tailing dumps in Botswana and Sweden, the five samples from the zone of active oxidation in the tailing dump in Germany revealed cell numbers detected by FISH for Acidithiobacillus spp. and A. ferrooxidans that were of the same order of magnitude as those of the cell numbers revealed by FISH, CARD-FISH, and Q-PCR for Bacteria as well as the cell numbers obtained by MPN for Fe(II) oxidizers. L. ferrooxidans was the only other microorganism which was detected by FISH, and it was significantly less abundant, in agreement with the Q-PCR data for this site.
DISCUSSION
Sulfide weathering in tailings.
The microbial communities in sulfidic mine waste tailings were comprehensively quantified by molecular biological methods for the first time in this study. All three investigated tailing dumps showed an intense colonization by microorganisms. Depth profiles of cell numbers showed that the compositions of the microbial communities are significantly different at the three sites and also varied greatly between zones of oxidized and unoxidized tailings.
Physical, geochemical, and mineralogical parameters of the tailings, e.g., temperature, pH, oxygen diffusion, or mineral reactivity, determine the composition of the microbial communities. Metal sulfide oxidation is the main energy-delivering process for microorganisms in sulfidic mine tailings (4, 49); thus, cell numbers are generally higher in the oxidized than in the unoxidized tailings in this study. Previous studies of sulfidic mine waste dumps showed that the microbial metal sulfide oxidation activity correlates with cell numbers of acidophilic Fe(II)-oxidizing microorganisms (16, 52, 53, 55). Microcalorimetric measurements of the tailing samples used in this study exhibited distinct differences in the potential pyrrhotite and pyrite oxidation rates between the oxidized zones in Botswana (17.5 μg pyrrhotite kg−1 tailings s−1) and in Sweden (0.9 μg pyrite kg−1 tailings s−1). The distinctly lower oxidation rates in Sweden were presumably caused by a lower temperature and, based on the till cover, by a lower oxygen diffusion rate and less water infiltration combined with a 20- to 100-times-less-reactive main metal sulfide (pyrite) than that in Botswana (pyrrhotite) (26, 30, 55).
In the tailing dump in Germany, microbial growth and the formation of cemented layers were probably favored by the temperate climate, the unremediated state of the dump, and the relatively high content in the tailings of arsenopyrite, sphalerite and galena, which are more reactive than pyrite (21, 31).
Comparison of results for cultivation and different molecular biological methods.
It was proven that the quantification methods used here are applicable to mine tailings. The fluorescent dye Sybr green II binds to all DNA of living and dead cells and therefore provided the highest cell numbers (total cell counts). The Q-PCR method quantifies amplifiable DNA presumably of living, and to some extent dead, cells and mainly yielded somewhat lower cell numbers than the total cell counts. Variation of Q-PCR cell numbers may be explained either by DNA preservation and/or degradation or by variable 16S rRNA operon numbers and/or variable genome copy numbers of distinct species (51). CARD-FISH and FISH probes target quickly degradable rRNA and, therefore, detect only living cells (44, 54). The bacterial numbers determined by Q-PCR and CARD-FISH were of the same order of magnitude for most of the samples, which means that a very high proportion of the total cell numbers in the tailings were alive.
In agreement with this interpretation, high MPNs of living acidophilic, chemolithoautotrophic Fe(II) oxidizers were found. The C-free medium used here at pH 3.5 provides cell numbers comparable with those of the molecular methods, which also detected the chemolithoautotrophic Fe(II) oxidizers Acidithiobacillus spp. and Leptospirillum spp. In the active oxidation zone (0.63- to 0.85-m depth) in the tailing dump in Germany, the MPNs were as high as the bacterial numbers determined by Q-PCR, CARD-FISH, and FISH, which argues for a high microbial activity and growing cells with a high ribosome content. In the other two tailings, the bacterial FISH numbers were lower (or below the detection limit) than the bacterial CARD-FISH numbers; thus, the microbial activity and the ribosome content of the Bacteria were lower, and only the more-sensitive method CARD-FISH enabled quantification of most cells. As a consequence, reliable FISH data for tailings can be obtained only for zones of high microbial activity, whereas CARD-FISH, Q-PCR, and MPN are suitable for tailings in general.
Quantification of microbial communities.
At all the sites investigated in this study, Bacteria dominated the microbial community. Archaea and Eukarya were less abundant. The autotrophic (or facultatively autotrophic) and acidophilic Fe(II)- and S-oxidizing bacterial genera Acidithiobacillus, Leptospirillum, and Sulfobacillus were detected by Q-PCR and FISH analysis in different numbers in the three tailings. Acidithiobacillus was detected at all three sites and overall dominated over Leptospirillum and Sulfobacillus. Leptospirillum spp. occurred in numbers similar to those of Acidithiobacillus spp. only in Sweden, occurred in lower numbers in Germany, and occurred not at all in Botswana. Sulfobacillus occurred in even lower numbers than Leptospirillum in Sweden, occurred in only a few samples in Botswana, and occurred not at all in Germany. In contrast to these findings, Diaby et al. (13) reported that Leptospirillum and Sulfobacillus relatives dominated over Acidithiobacillus in a pyrite-containing porphyry copper tailing impoundment in Chile based on terminal restriction fragment length polymorphism analysis. This technique allows calculation of a relative abundance of species in a clone library but cannot provide cell numbers of particular species, as was possible in this study. However, the results of this study and those of Diaby et al. (13) clearly demonstrate that the composition of the Fe(II)- and S-oxidizing bacterial community varies greatly at the different tailing sites. It is not yet understood which physical, chemical, and mineralogical parameters favor particular genera and species based on their physiological properties in mine tailings.
Previous bioleaching and AMD studies revealed that pH, redox potential, and Fe concentration control the composition of the acidophilic Fe(II)- and S-oxidizing community. In bioleaching processes which operate at a pH of <2, a high redox potential, and a high Fe(III) concentration, the Fe(II) oxidizers Leptospirillum spp. but not A. ferrooxidans has been shown to be dominant (47, 48). At the Iron Mountain AMD site, the dominance of Leptospirillum spp. over Acidithiobacillus spp. was shown by FISH analysis of extremely acidic water of pH ∼0.5 to 1.5 (6, 15, 56). Both genera occurred on average in similar numbers in the Tinto River at a pH of ∼2.0 to 2.5 (20). In less-acidic AMD with a pH of 2.7 to 3.7 from a mine in Norway, A. ferrooxidans dominated over Leptospirillum spp. (28), in agreement with this study of oxidized tailings with a similar pH range of ∼3 to 4. As shown by culture studies, the pH optimum for Leptospirillum spp. (1.3 to 3.0) is below that of Acidithiobacillus spp. (2.0 to 4.0) (50), supporting the findings of the ecological studies. Besides pH, redox potential, and Fe concentration, the availability of oxygen and inorganic sulfur compounds should be relevant. While Leptospirillum spp. are obligate aerobic Fe(II) oxidizers, A. ferrooxidans and Sulfobacillus spp. are more versatile and are able to oxidize Fe(II) and sulfur compounds aerobically as well as to reduce Fe(III) anaerobically (22, 50).
Quantifiable heterotrophic, facultative anaerobic Fe(III)-reducing Bacteria belonging to the acidophilic genus Acidiphilium and the neutrophilic Fe(III)-reducing family Geobacteraceae were detected in this study. Acidiphilium spp. were detected in the tailing dump in Botswana and previously in other sulfidic mine waste dumps (13, 52). The heterotrophic genus is widely distributed in bioleaching and AMD environments and in the case of the species Acidiphilium acidophilum is also able to oxidize inorganic sulfur compounds (22, 28, 50). Different studies implicate synergistic interactions between autotrophic and heterotrophic microorganisms by removing metabolic by-products of the autotrophs within the community (1, 24, 45, 52). Geobacteraceae were present at all three sites of this study. In other studies of sulfidic mine waste Geobacteraceae were detected by 16S rRNA gene sequence analysis (10) and neutrophilic Fe(III)-reducing microorganisms were found by cultivation techniques (52, 64). Microenvironments may protect the anaerobic and presumably neutrophilic Bacteria from acidity and oxygen (17, 52).
Sulfate-reducing Bacteria were quantified via their functional gene dsr coding for dissimilatory sulfite reductase. So far, other attempts to amplify the dsr gene in AMD sites with extremely low pH and high iron concentrations have been unsuccessful (1). Sulfate-reducing Bacteria, e.g., Desulfosarcina spp. and Desulfotomaculum spp., were regularly detected in sulfidic mine tailings (3, 7, 13, 17, 18, 64). These anaerobic and presumably neutrophilic Bacteria might be protected by microenvironments from acidity and oxygen, or they are able to remove oxygen by active respiration (32).
The high abundance of Fe(II)- and S-oxidizing, as well as of Fe(III)- and sulfate-reducing, Bacteria in the tailing dumps studied here and elsewhere shows that biogeochemical Fe and S cycling are predominant processes mediated by the activity of Bacteria.
In contrast to the dominant Bacteria, Archaea were detected only in low numbers in the oxidized zones of two tailings in this study. Archaea were not detected at all in the tailing deposit in Chile (13). The reason that Archaea do not predominate in mine tailings is most likely that the described species relevant for Fe and S cycling are extremophiles. Ferroplasma spp. are often found in AMD and bioleaching operations at very acidic conditions (pH of <2) and very high iron and sulfate concentrations (1, 12, 20, 50), conditions which usually do not prevail in mine tailings. Extremely thermophilic Archaea (e.g., Sulfolobus and Metallosphaera) have been found only at sites of high temperatures in sulfidic waste rock dumps because of high rates of pyrite oxidation (19, 52), but not yet in mine tailings.
For the first time, it is reported that the eukaryotic 18S rRNA gene was quantified in sulfidic tailing dumps in this study. The 18S rRNA gene occurred at all sites in lower copy numbers than the 16S rRNA gene of Acidithiobacillus spp., except for the unoxidized tailings in Sweden. Up to this point, only AMD sites such as the Rio Tinto and Iron Mountain sites were investigated in this regard and Eukarya such as algae, ciliates, flagellates, amoebae, and fungi were detected (1, 2, 66). To determine the impact of Eukarya on the microbial community in sulfidic mine waste, further studies will be necessary.
In conclusion, microorganisms occur in significantly different numbers at different sulfidic mine waste tailing zones and sites. Bacteria which are relevant for biogeochemical Fe and S cycling dominate the microbial community. The molecular methods applied here have proven to be useful for the quantification of microbial communities in sulfidic tailing dumps. These methods should also be useful for the monitoring of bioleaching processes such as heap or tank leaching.
Acknowledgments
We thank T. Graupner, R. B. Herbert, Jr., and H. Vogel for organizing the sampling and the drilling team of DGS Botswana for their sampling efforts. The English was thankfully improved by T. Wright and two anonymous reviewers.
The sampling was supported by the DGS Botswana and the BCL mine in Selebi-Phikwe, the German BMBF (grant 0330523), and the Swedish MISTRA research program MiMi.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. [DOI] [PubMed] [Google Scholar]
- 2.Baker, B. J., M. A. Lutz, S. C. Dawson, P. L. Bond, and J. F. Banfield. 2004. Metabolically active eukaryotic communities in extremely acidic mine drainage. Appl. Environ. Microbiol. 70:6264-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blowes, D. W., T. Al, L. Lortie, W. D. Gould, and J. L. Jambor. 1995. Microbiological, chemical, and mineralogical characterization of the Kidd Creak mine tailings impoundment, Timmins area, Ontario. Geomicrobiol. J. 13:13-31. [Google Scholar]
- 4.Blowes, D. W., C. J. Ptacek, J. L. Jambor, and C. G. Weisener. 2003. The geochemistry of acid mine drainage, p. 149-204. In B. S. Lollar (ed.), Environmental geochemistry, vol. 9. Treatise on geochemistry. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 5.Bond, P. L., and J. F. Banfield. 2001. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol. 41:149-161. [DOI] [PubMed] [Google Scholar]
- 6.Bond, P. L., G. K. Druschel, and J. F. Banfield. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruneel, O., R. Duran, K. Koffi, C. Casiot, A. Fourçans, F. Elbaz-Poulichet, and J.-C. Personné. 2005. Microbial diversity in a pyrite-rich tailings impoundment (Carnoulès, France). Geomicrobiol. J. 22:249-257. [Google Scholar]
- 8.Carlsson, E., J. Thunberg, B. Öhlander, and H. Holmström. 2002. Sequential extraction of sulfide-rich tailings remediated by the application of till cover, Kristineberg mine, northern Sweden. Sci. Total Environ. 299:207-226. [DOI] [PubMed] [Google Scholar]
- 9.Carlsson, E., B. Öhlander, and H. Holmström. 2003. Geochemistry of the infiltrating water in the vadose zone of a remediated tailings impoundment, Kristineberg mine, northern Sweden. Appl. Geochem. 18:659-674. [Google Scholar]
- 10.Cummings, D. E., A. W. March, B. Bostick, S. Spring, Jr., F. Caccavo, S. Fendorf, and F. Rosenzweig. 2000. Evidence for microbial Fe(III) reduction in anoxic, mining-impacted lake sediments (Lake Coeur d'Alene, Idaho). Appl. Environ. Microbiol. 66:154-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 12.Demergasso, C. S., P. A. Galleguillos, L. V. Escudero, V. J. Zepeda, D. Castillo, and E. O. Casamayor. 2005. Molecular characterization of microbial populations in a low-grade copper ore bioleaching test heap. Hydrometallurgy 80:241-253. [Google Scholar]
- 13.Diaby, N., B. Dold, H.-R. Pfeifer, C. Holliger, D. B. Johnson, and K. Hallberg. 2007. Microbial communities in a porphyry copper tailings impoundment and their impact on the geochemical dynamics of the mine waste. Environ. Microbiol. 9:298-307. [DOI] [PubMed] [Google Scholar]
- 14.Dold, B. 2006. Element flows associated with marine shore mine tailings deposits. Environ. Sci. Technol. 40:752-758. [DOI] [PubMed] [Google Scholar]
- 15.Edwards, K. J., T. M. Gihring, and J. F. Banfield. 1999. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl. Environ. Microbiol. 65:3627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elberling, B., A. Schippers, and W. Sand. 2000. Bacterial and chemical oxidation of pyritic mine tailings at low temperatures. J. Contam. Hydrol. 41:225-238. [Google Scholar]
- 17.Fortin, D., B. Davis, and T. J. Beveridge. 1996. Role of Thiobacillus and sulfate-reducing bacteria in iron biocycling in oxic and acidic mine tailings. Microbiol. Ecol. 21:11-24. [Google Scholar]
- 18.Fortin, D., M. Roy, J.-P. Rioux, and P.-J. Thibault. 2000. Occurrence of sulfate-reducing bacteria under a wide range of physico-chemical conditions in Au and Cu-Zn mine tailings. Microbiol. Ecol. 33:197-208. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs, T., H. Huber, K. Teiner, S. Burggraf, and K. O. Stetter. 1995. Metallosphaera prunae, sp. nov., a novel metal-mobilizing, thermoacidophilic archaeum, isolated from a uranium mine in Germany. Syst. Appl. Microbiol. 18:560-566. [Google Scholar]
- 20.González-Toril, E., E. Llobet-Brossa, E. O. Casamayor, R. Amann, and R. Amils. 2003. Microbial ecology of an extreme acidic environment, the Tinto River. Appl. Environ. Microbiol. 69:4853-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graupner, T., A. Kassahun, D. Rammlmair, J. Meima, D. Kock, M. Furche, A. Fiege, A. Schippers, and F. Melcher. 2007. Formation of sequences of cemented layers and hardpans within sulfide-bearing mine tailings (mine district Freiberg, Germany). Appl. Geochem. 22:2486-2508. [Google Scholar]
- 22.Hallberg, K. B., and D. B. Johnson. 2001. Biodiversity of acidophilic prokaryotes. Adv. Appl. Microbiol. 49:37-84. [DOI] [PubMed] [Google Scholar]
- 23.Hallberg, K. B., K. Coupland, S. Kimura, and D. B. Johnson. 2006. Macroscopic streamer growths in acidic, metal-rich mine waters in north Wales consist of novel and remarkably simple bacterial communities. Appl. Environ. Microbiol. 72:2022-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallmann, R., A. Friedrich, H.-P. Koops, A. Pommerening-Röser, K. Rohde, C. Zenneck, and W. Sand. 1993. Physiological characteristics of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans and physicochemical factors influence microbial metal leaching. Geomicrobiol. J. 10:193-206. [Google Scholar]
- 25.Herbert, R. B., Jr., and A. Schippers. 2008. Iron isotope fractionation by biogeochemical processes in mine tailings. Environ. Sci. Technol. 42:1117-1122. [DOI] [PubMed] [Google Scholar]
- 26.Höglund, L. O., and R. B. Herbert, Jr. (ed.). 2004. MiMi—performance assessment main report, MiMi 2003:3. The MISTRA-programme MiMi, mitigation of the environmental impact from mining waste. MiMi Print, Luleå, Sweden. http://www.mistra-research.se/program/mimi/guide/home/documentation/mimiperformanceassessmentmainreport.4.c791f4103209a06ec80008117.html.
- 27.Holmström, H., U. J. Salmon, E. Carlsson, P. Petrov, and B. Öhlander. 2001. Geochemical investigations of sulfide-bearing tailings at Kristineberg, northern Sweden, a few years after remediation. Sci. Total Environ. 273:111-133. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, D. B., S. Rolfe, K. B. Hallberg, and E. Iversen. 2001. Isolation and phylogenetic characterization of acidophilic microorganisms indigenous to acidic drainage waters at an abandoned Norwegian copper mine. Environ. Microbiol. 3:630-637. [DOI] [PubMed] [Google Scholar]
- 29.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kock, D., and A. Schippers. 2006. Geomicrobiological investigation of two different mine waste tailings generating acid mine drainage. Hydrometallurgy 83:167-175. [Google Scholar]
- 31.Kock, D., T. Graupner, D. Rammlmair, and A. Schippers. 2007. Quantification of microorganisms involved in cemented layer formation in sulfidic mine waste tailings (Freiberg, Saxony, Germany). Adv. Mater. Res. 20/21:481-484. [Google Scholar]
- 32.Krekeler, D., A. Teske, and H. Cypionka. 1998. Strategies of sulphate-reducing bacteria to escape oxygen stress in a cyanobacterial mat. Microb. Ecol. 25:89-96. [Google Scholar]
- 33.Lane, D. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics, John Wiley & Sons, Inc., New York, NY.
- 34.Leathen, W. W., L. D. MacIntyre, and S. A. Braley. 1951. A medium for the study of the bacterial oxidation of ferrous iron. Science 114:280-281. [DOI] [PubMed] [Google Scholar]
- 35.Liu, C.-Q., J. Plumb, and P. Hendry. 2006. Rapid specific detection and quantification of bacteria and archaea involved in mineral sulfide bioleaching using real-time PCR. Biotechnol. Bioeng. 94:330-336. [DOI] [PubMed] [Google Scholar]
- 36.Llobet-Brossa, E., R. Rosselló-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Londry, K. L., and B. L. Sherriff. 2005. Comparison of microbial biomass, biodiversity, and biogeochemistry in three contrasting gold mine tailings deposits. Geomicrobiol. J. 22:237-247. [Google Scholar]
- 38.Macur, R. E., J. T. Wheeler, T. R. McDermott, and W. P. Inskeep. 2001. Microbial populations associated with the reduction and enhanced mobilization of arsenic in mine tailings. Environ. Sci. Technol. 35:3676-3682. [DOI] [PubMed] [Google Scholar]
- 39.Müller, B., M. D. Axelsson, and B. Öhlander. 2003. Analyses of trace elements on quartz surfaces in sulfidic mine tailings from Kristineberg (Sweden) a few years after remediation. Environ. Geol. 45:98-105. [DOI] [PubMed] [Google Scholar]
- 40.Nordstrom, D. K., and C. N. Alpers. 1999. Geochemistry of acid mine waters, p. 133-160. In G. S. Plumlee and M. J. Logsdon (ed.), The environmental geochemistry of mineral deposits. Part A: processes, techniques, and health issues. The Society of Economic Geologists, Littleton, CO.
- 41.Öhlander, B., E. Carlsson, H. Holmström, and P. Elander (ed.). 2004. MiMi—characterization of the tailings and the till cover in impoundments 1 and 1B, Kristineberg mine, northern Sweden. Report MiMi 2001:2. The MISTRA-programme MiMi, mitigation of the environmental impact from mining waste. MiMi Print, Luleå, Sweden. http://www.mistra-research.se/download/18.c791f4103209a06ec80005691/2001_2.pdf.
- 42.Peccia, J., E. A. Marchand, J. Silverstein, and M. Hernandez. 2000. Development and application of small-subunit rRNA probes for assessment of selected Thiobacillus species and members of the genus Acidiphilium. Appl. Environ. Microbiol. 66:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pernthaler, J., F. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. Mar. Microbiol. 30:207-226. [Google Scholar]
- 45.Pronk, J. T., and D. B. Johnson. 1991. Oxidation and reduction of iron by acidophilic bacteria. Geomicrobiol. J. 10:153-171. [Google Scholar]
- 46.Ram, R. J., N. C. VerBerkmoes, M. P. Thelen, G. W. Tyson, B. J. Baker, R. C. Blake II, M. Shah, R. L. Hettich, and J. F. Banfield. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915-1920. [PubMed] [Google Scholar]
- 47.Rawlings, D. E., and D. B. Johnson. 2007. The microbiology of biomining: development and optimization of mineral-oxidizing microbial consortia. Microbiology 153:315-324. [DOI] [PubMed] [Google Scholar]
- 48.Rawlings, D. E., H. Tributsch, and G. S. Hansford. 1999. Reasons why ‘Leptospirillum’-like species rather than Thiobacillus ferrooxidans are the dominant iron-oxidizing bacteria in many commercial processes for the biooxidation of pyrite and related ores. Microbiology 145:5-13. [DOI] [PubMed] [Google Scholar]
- 49.Schippers, A. 2004. Biogeochemistry of metal sulfide oxidation in mining environments, sediments and soils, p. 49-62. In J. P. Amend, K. J. Edwards, and T. W. Lyons (ed.), Sulfur biogeochemistry—past and present. Special paper 379. Geological Society of America, Boulder, CO.
- 50.Schippers, A. 2007. Microorganisms involved in bioleaching and nucleic acid-based molecular methods for their identification and quantification, p. 3-33. In E. R. Donati and W. Sand (ed.), Microbial processing of metal sulfides. Springer, New York, NY.
- 51.Schippers, A., and L. Neretin. 2006. Quantification of microbial communities in near-surface and deeply buried marine sediments on the Peru continental margin using real-time PCR. Environ. Microbiol. 8:1251-1260. [DOI] [PubMed] [Google Scholar]
- 52.Schippers, A., R. Hallmann, S. Wentzien, and W. Sand. 1995. Microbial diversity in uranium mine waste heaps. Appl. Environ. Microbiol. 61:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schippers, A., P.-G. Jozsa, W. Sand, Z. M. Kovacs, and M. Jelea. 2000. Microbiological pyrite oxidation in a mine tailings heap and its relevance to the death of vegetation. Geomicrobiol. J. 17:151-162. [Google Scholar]
- 54.Schippers, A., L. N. Neretin, J. Kallmeyer, T. G. Ferdelman, B. A. Cragg, R. J. Parkes, and B. B. Jørgensen. 2005. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433:861-864. [DOI] [PubMed] [Google Scholar]
- 55.Schippers, A., D. Kock, M. O. Schwartz, M. E. Böttcher, H. Vogel, and M. Hagger. 2007. Geomicrobiological and geochemical investigation of a pyrrhotite-containing mine waste tailings dam near Selebi-Phikwe in Botswana. J. Geochem. Explor. 92:151-158. [Google Scholar]
- 56.Schrenk, M. O., K. J. Edwards, R. M. Goodman, R. J. Hamers, and J. F. Banfield. 1998. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans—implications for generation of acid mine drainage. Science 279:1519-1522. [DOI] [PubMed] [Google Scholar]
- 57.Southam, G., and T. J. Beveridge. 1992. Enumeration of thiobacilli within pH-neutral and acidic mine tailings and their role in the development of secondary mineral soil. Appl. Environ. Microbiol. 58:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Southam, G., and T. J. Beveridge. 1993. Examination of lipopolysaccharide (O-antigen) populations of Thiobacillus ferrooxidans from two mine tailings. Appl. Environ. Microbiol. 59:1283-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 60.Starkey, R. L. 1925. Concerning the physiology of Thiobacillus thiooxidans, an autotrophic bacterium oxidizing sulfur under acid conditions. J. Bacteriol. 10:135-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 62.Webster, G., C. J. Newberry, J. Fry, and A. J. Weightman. 2003. Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: a cautionary tale. J. Microbiol. Methods 55:155-164. [DOI] [PubMed] [Google Scholar]
- 63.Weinbauer, M. G., C. Beckmann, and M. G. Höfle. 1998. Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl. Environ. Microbiol. 64:5000-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wielinga, B., J. K. Lucy, J. N. Moore, O. F. Seastone, and J. E. Gannon. 1999. Microbiological and geochemical characterization of fluvially deposited sulfidic mine tailings. Appl. Environ. Microbiol. 65:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zammit, C. M., L. A. Mutch, H. R. Watling, and E. L. J. Watkin. Evaluation of quantitative real time polymerase chain reaction for enumeration of biomining microorganisms in culture. Hydrometallurgy, in press.
- 66.Zettler, L. A. A., F. Gómez, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Eukaryotic diversity in Spain's river of fire. Nature 417:137. [DOI] [PubMed] [Google Scholar]