Bacteria belonging to the genus Lactobacillus are members of the lactic acid bacteria (LAB), a broadly defined group characterized by the formation of lactic acid as the sole or main end product of carbohydrate metabolism. They can be found in plants or material of plant origin, silage, fermented food (yogurt, cheese, olives, pickles, salami, etc.), as well as in the oral cavities, gastrointestinal tracts (GIT), and vaginas of humans and animals (31). In particular, the Lactobacillus species found in the GIT have received tremendous attention due to their health-promoting properties. They are commonly used as probiotics, which are defined by the FAO/WHO as live microorganisms that when administered in adequate amounts confer a health benefit on the host.
The economic success and exciting prospects of probiotic products have accelerated research on intestinal lactobacilli. Genomics of Lactobacillus species is booming, and the genomes of five strains that belong to species commonly found in human fecal samples have recently been sequenced (50). Several comparative and functional genomic investigations have been conducted to gain information about the functionality of lactobacilli in the GIT (69). Unfortunately, a major misconception regarding the ecological role of lactobacilli in the intestinal tract has been embraced by many scientists working in the field. Specifically, there has been a general and persistent assumption that a large number of Lactobacillus species form stable and numerically significant populations in the human intestinal tract, especially in the small intestine, where they are presumed to form epithelial associations (101). Considering how widespread and accepted this perception is, there is surprisingly little experimental evidence that supports it. Ecological observations for the prevalence and dynamics of fecal Lactobacillus populations and the findings obtained with comparative genomics do indicate now that the ecological role of most types of intestinal lactobacilli, and their relationship with the human host, should be reconsidered.
In this review, evidence is summarized that suggests that only a small number of Lactobacillus species are true inhabitants of the mammalian intestinal tract and that most lactobacilli present are allochthonous members derived from fermented food, the oral cavity, or more proximal parts of the GIT. It is further explained why this knowledge provides information valuable for selecting strains for fundamental research of the ecological role of lactobacilli in the GIT, for their use as probiotics in foods and supplements, and for pharmaceutical applications.
THE GASTROINTESTINAL MICROBIOTA
The vertebrate GIT, including that of humans, is home to a vast collection of microbial, mostly bacterial, species, which is referred to as the gut microbiota. Comparisons of the characteristics of germ-free animals and those of conventional animals have clearly demonstrated that the gut microbiota has considerable influence on host biochemistry, physiology, immunology, and low-level resistance to gut infections (7, 30). Because of the variations in physical and chemical properties in the different compartments of the GIT, specific microbial communities exist in the stomach, small intestine, and large intestine (93). In monogastric animals, the largest numbers of bacteria reside in the distal gut (colon), reaching densities of around 1011 microbes per gram of luminal contents (90). The carbon and energy requirements of the enormous numbers of microbes residing in the colon are met by two sources: by complex carbohydrates, proteins, and fats that have escaped digestion in the small bowel and by the components of host secretions (mucins) and sloughed epithelial cells. Although nutrient availability is highest proximal to sites of absorption (e.g., the stomach and the first two-thirds of the small bowel), these sites contain relatively small numbers of microbes in humans. Microbial numbers are restricted in these areas because of the pH of the stomach contents (as low as pH 2), the toxicity of bile salts, and the relatively swift flow of the digesta (93). The population density and diversity increase from the proximal small intestine (103 microbes per ml luminal contents in the duodenum) to the ileum (up to 108) to the colon (24). In contrast to humans, however, some animal species have relatively large numbers of bacteria (mainly lactobacilli) in the proximal gut (e.g., the forestomachs of rodents, the crops of chickens, and the pars oesophageas of pigs) (92, 93). The reason for this special foregut association is likely due to the adherence of lactobacilli to the surface of the nonsecretory epithelium lining of these sites, which enables the bacteria to form a biofilm-like structure that provides a bacterial inoculum of the digesta (92).
Traditionally, gut microbiota research relied on techniques that required cultivation of the microbes (91). In the last decade, however, culture-independent molecular approaches have been intensively applied to the study of the microbial diversity in the gut ecosystem. The most comprehensive and probably least biased investigation of microbial diversity within the mammalian gut has come from direct sequencing of the 16S rRNA genes (48). The sequences are obtained from DNA extracted from gut samples, using PCR in combination with primers that are conserved for large groups of microbes (4, 22, 26). These molecular techniques have revealed that the diversity of the gut microbiota has been greatly underestimated (25). Although a complete catalogue of the members of the collective human gut microbiome is not yet available, more then 10,000 different species are estimated to be present (25), among which a large majority of these microbes are resilient to cultivation by currently available methodologies (90).
WHO'S WHO IN THE GUT
The astounding degree of microbial diversity in the GIT indicates a multitude of ecological niches. Many niches are likely to be determined by anatomical, immunological, and physiological characteristics of the host species. However, many niches are also generated through the development of complex food webs (niche construction) where the product of one microbe becomes the substrate for another (18, 48). Evolutionary theory predicts that in a spatially heterogeneous environment, vacant niches become occupied by organisms, and natural selection favors the emergence of ecological specialists that are highly adapted to the available niches (40). During the gradual colonization of the human GIT in early life, all niches in the GIT are likely to become occupied by well-adapted microbes, many of which are probably maternally acquired (48). Since every ecological niche can support the existence of only one type (according to the niche exclusion theory), it is extremely difficult for an organism that is accidentally or intentionally introduced into the gut to gain access (32). These ecological principles explain why the population levels and species compositions of the gastrointestinal microbiota remain remarkably constant over time in adult humans, and the phenomenon is referred to as colonization resistance or competitive exclusion (7, 82, 112). The bacteria that occupy a niche in the GIT are true residents or autochthonous (i.e., found where they are formed) components, as defined by Savage more than 30 years ago (80). Other bacteria are just “hitchhiking” through the gut and are allochthonous (i.e., formed in another place). An allochthonous organism in one section of the gut, however, may represent an autochthonous member of a more proximal niche that has been dislodged (shed), or it can be derived from ingested food and water (7, 111). Autochthonous strains have a long-term association with a particular host, and they form stable populations of a characteristic size in a particular region of the gut (80). It is often difficult to determine whether or not a particular microorganism is truly autochthonous to a particular host (7). However, following the succession and population dynamics of a bacterial group within the gut microbiota does permit the identification of some allochthonous bacteria: they do not persist within the ecosystem and are detectable only for a limited time. As shown below, the identification of the exact ecological status of individual Lactobacillus species in the human GIT remains a major challenge.
THE GOOD, THE BAD, AND THE UGLY
At the beginning of the last century, Elie Metchnikoff (1845 to 1916), a Nobel Prize winner for work on phagocytosis, proposed that the gut microbiota produces small amounts of toxic substances that damage the nervous and vascular systems and ultimately lead to aging (59). Metchnikoff suggested that the administration of bacteria present in fermented milk products would “implant” these beneficial, lactic acid-producing bacteria in the intestinal tract and would “arrest intestinal putrefaction and must at the same time postpone and ameliorate old age.” Metchnikoff's theories were based on two observations. First, Bulgarian peasants, assumed to have a long life expectancy, consumed large amounts of fermented milk products (97). Second, the natural fermentation of food by lactic acid-producing microbes prevented the growth of putrefactive organisms. Metchnikoff concluded “as lactic fermentation serves so well to arrest putrefaction in general, why should it not be used for the same purpose within the digestive tube?” Taken as the proof of its efficacy, milk fermented with the “Bulgarian bacillus” of Metchnikoff subsequently enjoyed considerable popularity in western Europe (94). Overall, Metchnikoff's theories remain very influential today and have contributed to the conviction that lactobacilli exert important functional attributes that promote health in the human GIT.
Although Metchnikoff's theories focused on LAB that were introduced into the digestive tract through the consumption of fermented food, he argued that each bacterium was “able to take its place in the intestinal flora of man” (59). Accordingly, in the era following Metchnikoff, lactobacilli were identified as one of the dominant organisms in the human gut (91). Anaerobic bacteriology was not yet invented, and most gut microbes escaped cultivation due to their strict anaerobic nature. In contrast, lactobacilli (together with clostridia, enterococci, and Escherichia coli) could be cultured with relative ease due to their higher oxygen tolerances. Consequently, lactobacilli gained a reputation as numerically dominant intestinal inhabitants, and even the advent of anaerobic culture techniques did little to correct this situation. Lactobacilli are still listed as numerically dominant organisms of the human gut in current microbiology text books (52, 70, 76), and even researchers working on functional and applied aspects of intestinal lactobacilli have continued to adhere to this dogma (11, 42, 57, 69, 71, 97).
FALL FROM GLORY
It is somehow intriguing how lactobacilli could maintain a reputation as numerically important intestinal inhabitants, given that the vast majority of experimental studies conducted after 1960 clearly showed that they form marginal populations in the human gut. When total anaerobic culturing techniques are used, lactobacilli form a very small proportion of the cultivable human fecal microbiota and can rarely be cultured at population levels exceeding 108 CFU per gram. Most studies report averages of around 106 CFU per gram (16, 17, 23, 62, 96, 104). This accounts for only about 0.01% of the total cultivable counts. Subject-to-subject variation is significant, and lactobacilli are not detectable in around 25% of human fecal samples (24, 96). The findings obtained by culture are in good agreement with culture-independent molecular approaches. In one study, fecal samples from 11 subjects were analyzed by fluorescent in situ hybridization (FISH) in combination with fluorescence microscopy, using a a LAB158 system Lactobacillus-Enterococcus targeted probe. Results revealed an average of 4.1 × 106 cells per gram of wet feces, which is around 0.01% of the total bacterial count (33). Quantification of lactobacilli in fecal samples from three human subjects, with a Lactobacillus-specific quantitative real-time PCR, revealed levels between 107 and 108 target cells per of gram of feces (74). In contrast to the studies described above, it was reported that the Lactobacillus-Enterococcus group constitutes 6.6% of the human fecal microbiota, on average, when assessed by dot plot hybridization using the LAB158 probe (57). Provided that the rRNA abundance measured with dot plot hybridizations correlates with cell numbers, this finding indicates an average presence of 1010 lactobacilli and enterococci per gram of human feces. Such a high value is not supported by any finding using alternative methods, and it represents 100-fold the proportion of bacteria found by FISH using the same probe (33). It is also 10-fold higher than the values obtained using dot plot hybridization with the Lacto722 probe, although this probe also detects streptococci (86). In this respect, it is important to point out that as the probes used for the quantification of lactobacilli by FISH are not specific for lactobacilli, the real numbers of lactobacilli could be even less.
High-throughput analysis of 16S rRNA sequences retrieved directly by PCR now allows a comprehensive view of the microbial diversity of the human GIT (25). A quantitative assessment of the results obtained from these studies, with a focus on the prevalence and diversity of Lactobacillus operational taxonomic units (OTUs), is shown in Table 1. Eckburg and coworkers (22) studied 11,831 bacterial near-full-length 16S rRNA sequences retrieved from cecal, colonial, and fecal samples (including those from biopsy samples) of three human subjects and, remarkably, found not one single Lactobacillus sequence. Lactobacilli were also absent from the libraries generated from several studies of a smaller scale (34, 35, 37, 90). Ley and coworkers studied fecal samples from 12 human subjects and found only 6 sequences to account for lactobacilli in a total of 18,348 sequences (49). To date, significant proportions of lactobacilli could be found only in two 16S rRNA libraries obtained from human samples (26, 36). In a study of impressive scale, Frank and colleagues (26) presented a comprehensive molecular-based analysis of the bacterial diversity of gut tissue samples obtained from patients suffering from inflammatory bowel disease (IBD), as well as from non-IBD controls. Around 5% of the sequences obtained from the colons of non-IBD patients accounted for lactobacilli (Table 1). Hayashi and coworkers (36) found that 12.9% of the sequences in libraries generated from jejunal, ileal, cecal, and rectosigmoidal (luminal) samples of elderly subjects accounted for lactobacilli. However, in both studies, the vast majority of the Lactobacillus sequences did represent species that are not considered real inhabitants of the GIT (e.g., L. delbrueckii and L. mali), suggesting that these bacteria were introduced through food. Overall, the comprehensive molecular-phylogenetic analysis of the human gut microbiota now provides clear evidence for the numerically minor proportion of lactobacilli.
TABLE 1.
Representation of Lactobacillus sequences in molecular-phylogenetic analysis of human gastrointestinal microbiota
| Reported sample site(s) or material (reference[s]) | No. of subjects | Total no. of sequences | No. of Lactobacillus sequences | % of Lactobacillus sequences |
|---|---|---|---|---|
| Stomach tissue (10) | 23 | 1,833 | 4 | 0.22 |
| Small intestine tissue, non-IBD (26) | 20 | 1,638 | 5 | 0.31 |
| Jejunum, ileum tissue (107) | 1 | 173 | 0 | <0.6 |
| Jejunum and ileal lumen (36) | 3 | 545 | 87a | 16 |
| Ileal and colon tissue (109) | 2 | 361 | 0 | <0.3 |
| Colon and rectal tissue (107) | 1 | 174 | 0 | <0.6 |
| Colon and rectal lumen (36) | 3 | 545 | 54b | 9.9 |
| Cecal, colon, and rectal tissue and feces (22) | 3 | 11,831 | 0 | <0.01 |
| Colon tissue, non-IBD (26) | 40 | 3,214 | 157c | 4.9 |
| Colon tissue (22) | 3 | 110 | 0 | <1 |
| Feces (34, 35) | 4 | 927 | 0 | <0.11 |
| Feces (90) | 1 | 284 | 0 | <0.4 |
| Feces (49) | 12 | 18,348 | 6d | 0.03 |
The species detected were L. mali (85 sequences) and L. reuteri (2 sequences).
The species detected were L. reuteri (27 sequences), L. mali (20 sequences), and L. delbrueckii (7 sequences).
The main species detected were L. delbrueckii (108 sequences), L. rhamnosus (38 sequences), L. reuteri, and L. animalis (each 5 sequences).
Sequence identification was performed using the Classifier tool of the Ribosomal Database Project II (108) with a confidence threshold of 80%; the complete sequence data set was kindly provided by Ruth Ley (Washington University, St. Louis, MO).
One could now speculate that lactobacilli are underrepresented in 16S rRNA libraries due to a PCR bias that discriminates against Lactobacillus sequences. However, this objection is unfounded since Lactobacillus sequences are actually overrepresented (compared to results obtained by culture) in libraries of intestinal samples of mice, rats, pigs, and chicken (Table 2). Furthermore, it is often argued that the study of fecal samples does not provide accurate information concerning the intestinal microbiota and that the small numbers of lactobacilli in human fecal samples might in fact represent remnants of larger populations colonizing a more proximal part of the GIT or mucosal sites. In fact, lactobacilli are among the most common bacteria in the stomach, duodenum, and jejunum of humans, as found by cultivation approaches (62, 72). However, as shown in Table 1, molecular investigations of the bacterial populations present in the stomach, small intestine, and mucosal biopsies have shown that Lactobacillus sequences are present only in small proportions (<1%) in most of these samples. In this respect, it should be considered that Lactobacillus populations that can be cultured from the stomach and small intestine are generally rather small (<104 bacteria per ml) and that most bacteria present are likely to be transients from the oral cavity or from food (7). Taken together, the molecular-phylogenetic characterization of samples taken from throughout the human GIT does not support the hypothesis that more proximal or mucosal sites harbor greater populations of lactobacilli, and it appears that lactobacilli are greatly outnumbered by organisms yet to be cultured.
TABLE 2.
Representation of Lactobacillus sequences in the molecular-phylogenetic analysis of the gastrointestinal microbiota of animals
| Reported animal site(s) or sample (reference[s]) | No. of animals | Total no. of sequences | No. of Lactobacillus sequences | % of Lactobacillus sequences |
|---|---|---|---|---|
| Pig ileum, cecum, and colon lumen (46) | 24 | 4,270 | 674 | 15.8 |
| Mouse small and large intestine lumens and tissue and feces (78) | 70 | 8 | 11.4 | |
| Mouse cecum lumen (47) | 5,089 | 205a | 4 | |
| Rat feces (12) | 109 | 25 | 22.9 | |
| Chicken ileum and cecum lumens (44, 51) | 1,393 | 490 | 35.2 |
Sequence identification was performed using the Classifier tool of the Ribosomal Database Project II (108) with a confidence threshold of 80%; the complete sequence data set was kindly provided by Ruth Ley (Washington University, St. Louis, MO).
UPS AND DOWNS
Stability is a general characteristic for microbial ecosystems (2). Intestinal ecosystems are no exception, and although they are dynamic, they remain remarkably resistant and resilient to chaotic blooms of subpopulations and pathogens (48). Functional redundancy in the microbiota confers stability, and if it is perturbed, homeostatic reactions come into place and restore a reasonably stable equilibrium. Molecular fingerprinting of 16S rRNA genes by denaturing and temperature gradient gel electrophoresis (DGGE and TGGE, respectively) is a simple way to show stability of the gut microbiota in healthy adult humans. Several studies revealed that the total bacterial population, as well as bacterial groups such as bifidobacteria, Bacteroides spp., and clostridia, show a high degree of temporal stability down to the species level (82, 96, 100, 112). However, the situation is very different for lactobacilli. DGGE in combination with primers for LAB showed that the Lactobacillus populations in fecal samples from most human subjects show temporal dynamics that are characterized by fluctuations and a lack of stability (82, 100, 104). The temporal fluctuations of Lactobacillus populations are also evident when the succession of isolates (strains) in human fecal samples is studied. Early pioneering studies, conducted between 1960 and 1980 by Gerhard Reuter and Tomotari Mitsuoka, showed both persistent and transient Lactobacillus strains in human feces (45, 61, 63, 73). Based on current taxonomic criteria, the persistent strains identified in these studies belonged to the L. gasseri, L. crispatus, L. reuteri, L. salivarius, and L. ruminis species (62, 72). These early findings were confirmed more recently by Tannock and coworkers (96), who followed the temporal succession of Lactobacillus strains by molecular strain typing (41, 96). Human subjects that had a stable and large (>106 CFU per gram) fecal population of lactobacilli maintained single strains that predominated throughout the period of investigation (up to 15 months). These strains belonged to the L. ruminis and L. salivarius species. Although lactobacilli could be cultured from all subjects in these studies, several of the subjects also had periods when no lactobacilli were detectable. Most strains were detected only in one or two fecal samples from the majority of subjects and then went missing. These sporadic strains belonged to the L. acidophilus, L. crispatus, L. gasseri, and L. plantarum species and the L. casei group (L. casei, L. paracasei, and L. rhamnosus) (96).
There are 17 Lactobacillus species that are associated with the human GIT, some of which were only recently detected by molecular techniques using PCR primers specific for LAB (Table 3). However, the studies cited above show that caution is advised when particular Lactobacillus species are described as real (autochthonous) inhabitants. Species such as L. acidophilus, L. casei, L. paracasei, L. rhamnosus, L. delbrueckii, L. brevis, L. johnsonii, L. plantarum, and L. fermentum have, so far, not been reported to form stable populations in the gut and are likely to be allochthonous. Most of these species are regularly present in fermented foods, and they are common inhabitants of the oral cavity (Table 3). The results from feeding studies of lactobacilli indicate that the survival of lactobacilli that originate from food during gastrointestinal passage is comparable to that of probiotic strains. They can be cultured from fecal samples in numbers comparable to that of resident lactobacilli when they are consumed in cell numbers not uncommon for fermented foods (Table 4). Lactobacilli are present in human saliva in various numbers but often attain populations exceeding 105 CFU per ml (1, 16, 43, 56). The average output of saliva is 1,000 to 1,500 ml per day, which, when swallowed, potentially introduces doses of oral lactobacilli into the GIT that are comparable to those used in probiotic feeding trials. Interestingly, the species that predominate in the oral cavity, such as L. acidophilus, L. gasseri, L. crispatus, L. plantarum, L. salivarius, L. brevis, L. rhamnosus, L. paracasei, and L. vaginalis, are also frequently isolated from human feces, and the species composition present in the oral cavity and in fecal samples coincides in some humans (16, 60). Dal Bello and Hertel showed that several fecal and oral isolates from three subjects isolated at the same time point were of the same randomly amplified polymorphic DNA type, suggesting that these fecal isolates originated from the oral cavity (16). Several Lactobacillus species, such as L. salivarius and L. gasseri, might therefore be allochthonous to the human intestinal tract but autochthonous to the oral cavity (72).
TABLE 3.
Lactobacillus species commonly detected in human feces, saliva, and food
| Species | Fecesa | Oral cavity | Food |
|---|---|---|---|
| L. acidophilus | + | + | |
| L. crispatus | + (P) | + | |
| L. gasseri | + (P) | + | |
| L. johnsonii | + | + | |
| L. salivarius | + (P) | + | |
| L. ruminis | + (P) | ||
| L. casei | + | + | + |
| L. paracasei | + | + | + |
| L. rhamnosus | + | + | + |
| L. plantarum | + | + | + |
| L. reuteri | + (P) | (+)b | |
| L. fermentum | + | + | + |
| L. brevis | + | + | + |
| L. delbrueckii | + | + | |
| L. sakei | + | + | |
| L. vaginalis | + | + | |
| L. curvatus | + | + |
TABLE 4.
Dose and recovery of allochthonous lactobacilli in human feces
| Bacteria | Daily dose (cells/ml) | Reisolation (CFU/g feces) | Reference(s) |
|---|---|---|---|
| Probiotics | |||
| L. rhamnosus GG | 1010 | 105-108 | 38 |
| L. casei strain Shirota | 1011 | Around 107 | 88 |
| L. rhamnosus DR20 | 109 | 105-106 | 96 |
| Food lactobacilli | |||
| L. paracasei | 109 | 107-108 | 13 |
| L. delbrueckii | 1010 | 105-108 | 38 |
| L. casei | 1010 | 105-108 | 38 |
| Oral lactobacilli | |||
| Ca. 20% of subjects >106 CFU/ml saliva | >109a | 1, 16, 43 | |
| Ca. 40% of subjects >105 CFU/ml saliva | >108a |
Values are based on a daily saliva output of >1,000 ml.
HOW DO AUTOCHTHONOUS LACTOBACILLI PERSIST IN THE GUT?
Most lactobacilli present in the GIT of mice, rats, pigs, and chickens are clearly autochthonous, since they form stable populations throughout the life of the animal host, they can be cultured in large numbers, and they are present in almost all animals (62, 92). As shown in Table 2, clones derived from lactobacilli are common representatives in 16S rRNA gene libraries derived from intestinal samples of these animals. Unlike the human stomach, which is lined with a glandular mucosa, the stomachs of pigs, mice, and rats and the crops of birds are lined, at least partly, with a nonglandular, squamous stratified epithelium (92). These regions are densely colonized by lactobacilli which adhere directly to the epithelium and form a layer of bacterial cells. The epithelial associations formed by lactobacilli show characteristics of bacterial biofilms because the bacteria are firmly attached to a surface (epithelium) and are embedded in a matrix of extracellular polymeric substances (27, 81).
Strains closely related to L. reuteri and L. johnsonii are clearly autochthonous to the rodent and porcine gut because they have been detected there in several studies in almost all animals (12, 46, 78). These species have recently been used to identify bacterial factors that allow lactobacilli to persist in the guts of mice. These studies have begun to provide mechanistic explanations for the ecological success of lactobacilli as a result of the applications of in vivo expression technology, microarray transcriptome analysis, and the investigation of the ecological performance of isogenic mutants (19, 95, 102, 103, 105, 106). The bacterial factors identified in these studies are summarized in Table 5. Although knowledge about their exact ecological function is still rudimentary, the findings suggest that bacterial adherence to the squamous stratified epithelium of the forestomach of mice is a key feature. Proteins such as the large surface protein (Lsp) seem to initiate adherence, while extracellular polysaccharides appear to contribute to the matrix and facilitate cell aggregation. Furthermore, several bacterial factors (e.g., MsrB, Dlt, and immunoglobulin A [IgA] protease) were identified as important, allowing the bacteria to overcome adverse environmental conditions generated through high acidity or innate and adaptive host defenses (nitric oxide and IgA). Bacterial factors with similar function have recently been shown to be important for Bacteroides thetaiotaomicron in the murine gut (68). Peterson and coworkers (68) showed that a capsular polysaccharide (CPS4) with unknown ecological function was essential for the competitiveness of the organism. B. thetaiotaomicron avoided CPS4-specific IgA recognition by downregulating epitope expression in vivo. In the absence of IgA (in Rag1−/− mice, which lack T and B cells), B. thetaiotaomicron triggered expression of inducible nitric oxide synthase in the small intestine, and the organism responded to the oxidative challenge by elevating expression of operons involved in nitric oxide metabolism. Accordingly, MsrB and the IgA protease of lactobacilli might contribute to ecological performance by counteracting nitric oxide and IgA exposure in the gut. It is striking that all of the factors identified as contributors to the persistence of lactobacilli in the murine gut (d-alanylation of TA, epithelial adhesion, repair of oxidative damage of proteins, luxS-dependent production of AI-2, extracellular polysaccharide formation, and proteolytic degradation of immunoglobulins) are also important contributors to bacterial virulence, thus emphasizing that commensal lactobacilli and bacterial pathogens apply similar strategies to occupy niches within the mammalian host.
TABLE 5.
Genetic factors shown to contribute toward ecological performance of lactobacilli in the gut of mice
| Loci | Protein encoded | Strain | Why studied? | Putative function in the GIT | Reference |
|---|---|---|---|---|---|
| lsp | Large surface protein | L. reuteri 100-23Ca | Dominant surface protein | Adherence | 102 |
| msrB | Methionine sulfoxide reductase B | L. reuteri 100-23C | Gene expression specifically induced in vivo | Reduction of oxidized methionine residues, resistance to nitric oxide produced by epithelial cells | 102 |
| luxS | LuxS | L. reuteri 100-23C | Importance of AI-2 on the formation of biofilms by gram-positive bacteria | Quorum sensing (AI-2) production/metabolic importance as part of activated methyl cycle | 95 |
| dltA | d-Alanine-d-alanyl carrier protein ligase (Dcl) | L. reuteri 100-23 | Importance of the dlt operon for biofilm formation and adhesion of gram-positive bacteria | Resistance against low pH values and defensins | 105 |
| gtfA | Glycosyltransferase A | L. reuteri TMW1.106 | Importance of EPS for bacterial biofilm formationb | Cell aggregation, biofilm formation | 106 |
| inu | Inulosucrase | L. reuteri TMW1.106 | Importance of EPS for bacterial biofilm formationb | Cell aggregation, biofilm formation | 106 |
| LJ1680 | IgA protease | L. johnsonii NCC533 | In vivo expressed and associated with a long gut persistence phenotype | Degradation of IgA | 19 |
| LJ1654 to LJ1656 | PTS transporter | L. johnsonii NCC533 | In vivo expressed and associated with a long gut persistence phenotype | Sugar utilization | 19 |
Plasmid-free variant of Lactobacillus reuteri 100-23.
EPS, extracellular polysaccharide.
It is important to recognize that the ecological cohesions discovered in mice do not necessarily account for the corresponding persistence in the human gut, due to significant anatomical differences. Most importantly, a stratified, squamous epithelium is not present in the human stomach. Still, the adherence of lactobacilli to epithelia or mucus is often considered to contribute to the persistence of lactobacilli in the human GIT (69, 101). It has been shown that some lactobacilli have the ability to bind to intestinal mucus and polymers associated with the surface of enterocytes (64, 75), and putative adherence factors of lactobacilli have been identified (101). The ecological relevance of these factors in the human GIT remains to be determined in vivo. In this respect, it is important to emphasize that colonization of mucus associated with tissue surfaces by members of the gastrointestinal microbiota is very limited in humans, and the numbers of bacteria obtained from washed tissue surfaces are considerably lower than those observed in studies of rodents (93). Evidence for significant in vivo association of lactobacilli with the columnar epithelium in the intestinal tract of humans is still inconclusive, and more work is needed to determine if the association with the epithelium contributes to the persistence of lactobacilli in the human gut. Although stratified squamous epithelia are not present in the human gut, they seem to be key factors to Lactobacillus colonization, as habitats with high numbers of lactobacilli contain such epithelia (e.g., the human mouth and vagina and the proximal GIT of rodents, pigs, horses, and birds). Adherence to these epithelia appears to be more relevant than adherence to columnar epithelia or mucus present in the intestinal tract, and the identification of adherence mechanisms to squamous cells would therefore teach us a lot about how lactobacilli manage to colonize their mammalian hosts.
In contrast to that of rodents and pigs, significant epithelial associations of gut bacteria or biofilms have not been described in the human gut. Commensal bacteria appear to live in suspension with limited contact with epithelial cells (99). Rapid generation times are therefore vital for the bacteria to avoid washout. Numerically dominant human gut organisms such as Bacteroides thetaiotaomicron and Bifidobacterium longum have highly evolved “glycobiomes” which consist of an elaborate apparatus for acquiring and hydrolyzing dietary and host-derived polysaccharides associated with a large repertoire of environmentally regulated expression systems (83, 110). Complete pathways for the synthesis of amino acids, nucleotides, and some key vitamins were identified. It appears that Bacteroides spp. and bifidobacteria base their ecological competitiveness on the utilization of complex nutrients, using well-regulated pathways to save energy and assure high proliferation rates in the lumen of the gut. How lactobacilli facilitate rapid growth in the human intestinal tract remains dubious, as they are fastidious organisms with nutritional requirements one would consider disadvantageous in regions distal to host nutrient absorption. Lactobacilli require amino acids, peptides, nucleic acid derivatives, vitamins, salts, fatty acid esters, and fermentable carbohydrates for growth, and they have very limited abilities to utilize complex carbohydrates (39). The analysis of genome sequences for several intestinal Lactobacillus species (L. acidophilus, L. salivarius, L. plantarum, L. gasseri, and L. johnsonii) did not reflect an adaptation to the intestinal tract, as the physiology based on genome annotations is in striking contrast to that of the dominant gut inhabitants Bacteroides thetaiotaomicron and Bifidobacterium longum (3, 15, 42, 55, 71). It is of course possible that lactobacilli occupy specific niches in the human GIT and have evolved to become ecological specialists, in contrast to Bacteroides thetaiotaomicron and Bifidobacterium longum, which appear to be generalists with large genomes (40). Lactobacilli could utilize simple carbohydrates that result from the degradation of complex carbohydrates by other microbes. Alternatively, some species such as L. acidophilus, L. plantarum, and L. paracasei are able to metabolize complex prebiotic carbohydrates that remain untouched by human enzymes and which could serve as nutrients in the intestinal tract (5, 6, 29, 79). However, these species still lack pathways for the synthesis of most amino acids, nucleotides, and vitamins. The significant auxotrophy revealed by genome characterizations has led researchers to speculate that lactobacilli may inhabit the nutrient-rich upper GIT of humans in higher numbers (3, 71). However, as shown in Table 1, this view is not supported by recent molecular characterizations of the microbiota present at these sites. Overall, the findings obtained with the analysis of the currently available Lactobacillus genomes provide further support for their allochthony in the human intestinal tract.
ARE THE MAJORITY OF LACTOBACILLI IN THE INTESTINAL TRACT OF RODENTS, PIGS, AND CHICKENS ALLOCHTHONOUS?
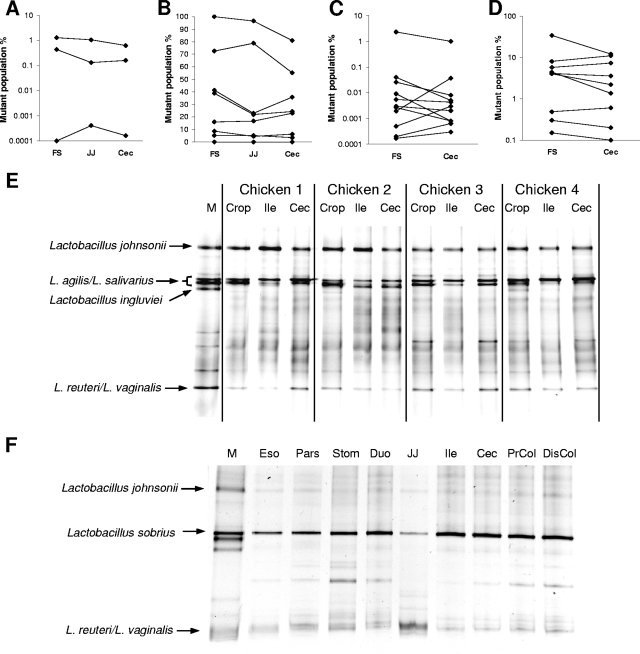
As noted above, most Lactobacillus species found in the human intestinal tract do not appear to be true inhabitants, and it remains unclear how autochthonous species satisfy their fastidious nutritional requirements in regions distal to host nutrient absorption. Nevertheless, lactobacilli are present throughout the GIT of mice, rats, pigs, and chickens in high numbers, including the large intestine (around 109 cells per gram). How do lactobacilli maintain such high cell numbers in the distal GIT of these animals? Like lactobacilli in conventional animals, Lactobacillus reuteri colonizes Lactobacillus-free mice throughout the gut and stably maintains cell numbers of around 109 cells per gram in the forestomach, around 107 cells per gram in the jejunum, and around 108 cells per gram in the cecum (102, 103, 105). These significant numbers certainly do imply that L. reuteri does inhabit all these different sites. One could also assume that the different anatomical and physiological conditions present throughout the gut would account for distinct bacterial traits to be required for colonization. Hence, genes that contribute to ecological performance in one compartment would not necessarily affect fitness throughout the gut. However, an unexpected finding in experiments with isogenic L. reuteri mutants was that gene inactivation always affected the mutant populations in the entire GIT of mice, independent of gene function (102, 105, 106). This was especially surprising for bacterial factors involved in adherence and biofilm formation, as significant adhesion of lactobacilli to the columnar epithelial lining of the intestinal tract has not been described in mice. So, it is unlikely that inactivation of Lsp, a protein involved in adherence to the forestomach epithelium, would result in reduced population levels in the distal intestinal tract (102). An even more puzzling finding was that the proportion of the mutants in the cecum always mirrored that in the forestomach in individual animals (Fig. 1A to D). As a conclusion, these findings suggest that the cecal L. reuteri population is composed of remnants of the forestomach population and point to the forestomach as the real habitat of L. reuteri. L. reuteri is therefore likely to be allochthonous to the murine intestinal tract.
FIG. 1.
(A to D) Competition experiments performed between the wild-type L. reuteri strains 100-23C (A and B), 100-23 (C), and TMW 1.106 (D) and the isogenic mutants with insertional inactivations of the lsp (A), msrB (B), dltA (C), and inu (D) genes (102, 105, 106). Mixtures of mutants and wild type (1:1) were used to inoculate Lactobacillus-free mice, and the percentages of mutants in the total Lactobacillus population were determined at 7 days in the forestomach (FS), jejunum (JJ), and cecum (Cec). Data points of individual animals are connected by lines. (E) DGGE analysis of PCR-amplified 16S rRNA gene fragments obtained with the primers Lac1 and Lac2GC and DNA isolated from the crop, ileum (Ile), and cecum (Cec) of four chickens (age, 42 days) that were floor reared at the University of Nebraska (I. Martínez, S. Scheideler, and J. Walter, unpublished data). M, marker representing species isolated from the chickens. DGGE was performed as described by Walter et al. (104). (F) DGGE analysis of PCR-amplified 16S rRNA gene fragments obtained with the primers Lac1 and Lac2GC and DNA isolated from the esophagus close to the stomach (Eso), the pars esophagus (Pars), the stomach contents (Stom), the duodenum (Duo), jejunum (JJ), ileum (Ile), cecum (Cec), proximal colon (PrCol), and distal colon (DisCol) of a male, castrated pig (age, 10 weeks) that was reared at the University of Nebraska (I. Martínez, T. Burkey, and J. Walter, unpublished data). M, marker representing species commonly present in pigs. DGGE was performed as described by Walter et al. (104).
It remains to be determined whether this also accounts for other Lactobacillus species present in the intestinal tract of rodents, pigs, and birds. Comparison of the population composition of the forestomach and cecum of BALB/c mice by DGGE and sequencing of bands revealed that all lactobacilli detectable in the cecum (three OTUs) were also present in the forestomach (58). Similarly, in chickens, DGGE analysis with LAB-specific primers revealed that the molecular fingerprint detected in the cecum was virtually identical to that of the crop (Fig. 1E). In addition, the Lactobacillus succession that has been observed in the crop of chicks is remarkably similar to that in the ileum (94), suggesting that the Lactobacillus microbiota in the intestinal tract of these animals consists of bacteria originating from the crop. In pigs, DGGE analysis with LAB-specific primers revealed that the same molecular fingerprint could be detected throughout the entire GIT, from the distal esophagus to the distal colon (Fig. 1F). These findings suggest that numerically dominant Lactobacillus populations present in the rodent, pig, and chicken intestinal tract are allochthonous and that they originate from the forestomach, pars esophagus, and crop, respectively. The identification and characterization of Lactobacillus strains autochthonous to the distal intestinal tract of such animals would be of great interest, since traits that enable the strains' colonization might be similar to traits of lactobacilli autochthonous to the human large bowel. These bacteria, together with their animal hosts, would provide a good model system to study ecological interactions that are likely to be equivalent in humans.
IMPLICATIONS FOR FUNCTIONAL AND BIOMEDICAL RESEARCH
Lactobacilli offer exciting research opportunities, both in terms of biomedical applications and in acquiring fundamental knowledge about the functionality of gut microbes (94). The tools for genetic modification, identification, detection, and functional analysis of lactobacilli have improved tremendously over the last 2 decades. More and more Lactobacillus genomes are becoming available, allowing systematic comparative and functional genomic studies to investigate ecological and probiotic functionality. There is no doubt that the means necessary to carry out detailed and informative studies of gastrointestinal lactobacilli now exist. However, it is important to consider the ecological characteristics of individual species and their relationship with their host in such studies. Unfortunately, the ecological status of Lactobacillus species in the human gut has generally not been taken into consideration by researchers working in the field, despite its important implications. Comparative genomic investigations to identify colonization determinants require exact knowledge about the origin of strains in order to link genome features to ecological function. The ecological status of most intestinal isolates, including the strains for which genome sequences are available, is at best uncertain. Furthermore, most of the Lactobacillus strains currently used as probiotics are not adequate model organisms with which to study ecological aspects of gut colonization, as they belong to species that have never been shown to form stable populations in this ecosystem. It would be of great value to include Lactobacillus strains, having strong evidence as autochthonous organisms, in comparative and functional genomic investigations.
The bacteria residing in the mammalian gut and their hosts are likely to have coevolved over a long conjoined history and, by doing so, have developed an intimate and complex symbiotic relationship. The mechanisms underlying these interactions are likely to be specific for a particular microbe and its host and are probably influenced by other partners of the gut microbiota. Therefore, investigations of the host/microbe interplay in gut ecosystems should be conducted within an ecological context. Most importantly, this research requires the examination of bacterial species proven to be autochthonous in a particular host. This is particularly important when the organism's response and behavior in the GIT is studied by global transcriptome analysis using microarrays. The physiology and expression of phenotypic traits of an autochthonous gut organism colonizing the GIT is a dynamic entity that reflects the microbe's adaptation to the ecosystem and its specific host. In contrast, the response of an allochthonous organism to the gut environment is likely to be based on signals that are generic (e.g., stress response, basic metabolism) and, hence, will reveal neither much about the environment from which the organism originates nor how autochthonous lactobacilli manage to live in the gut.
It has been clearly shown that gut microbes benefit their host in many aspects (4). Gut bacteria can enhance host immune functions and the mucosal barrier, and they provide protection against incoming microbes (97). These interactions comprise modulation of signal transduction pathways and gene expression in epithelial and immune cells, and their high level of complexity makes it unlikely that they have emerged by coincidence. In contrast, one would predict that mutually beneficial microbial activities have been shaped by natural selection during coevolution, as they promote host fitness (4, 48). As a consequence, gut inhabitants that share long evolutionary histories with their host species are likely to possess adaptive health attributes that can be explored when these organisms are used as probiotics. It is therefore reasonable to consider that autochthonous strains constitute better probiotic strains for some applications. Indeed, many researchers consider human origin as an important criterion for the selection of probiotics (21, 66, 77). However, although most probiotic strains originate from human gut or fecal samples, they show a poor persistence after administration is stopped (66). This is generally believed to be due to competitive exclusion conferred by the resident gut bacteria and to individual differences between human subjects. In addition, human subjects are different, and a strain isolated from one individual would not necessarily be compatible with the intestinal ecosystem of another individual. Although these are legitimate claims, most strains currently used as probiotics do belong to species which are likely to be allochthonous to the human intestinal tract, and their failure to persist might reflect a lack of competitiveness in the gut ecosystem. It would be fascinating to investigate the probiotic characteristics of strains proven to be autochthonous, both in relation to persistence and health benefits. Is the strain autochthonous for one person a better “universal colonizer”? Of course, even autochthonous Lactobacillus strains would not be compatible with the intestinal environment and immune system of most individuals. Still, an autochthonous strain is adapted to the GIT, and its ecological fitness, metabolic activity, physiology, and ability to persist and produce microbial products that define its probiotic functionality in the gut should be higher than those of allochthonous strains. It has been shown that lactobacilli and other LAB could be genetically modified so that their cells produced bioactive substances of therapeutic value and delivered them upon ingestion to the gut mucosa (85, 89). For this purpose, it appears that the utilization of autochthonous strains makes it more likely that the recombinant organisms will persist, metabolize, and produce sufficient amounts of the therapeutic compound at a desired location in the gut.
It is now generally recognized that the health benefits of probiotics are conferred mainly though a stimulation or modulation of the immune system (66). Several animal and human studies have provided unequivocal evidence that specific strains of probiotics are able to stimulate as well as regulate several aspects of natural and acquired immune responses, which opens opportunities to treat or prevent specific diseases that have an immunological etiology (28). When host immune functions are targeted, it is again likely that the evolutionary history of the probiotic strain is of paramount importance. The autochthonous microbe-immune system relationship in healthy animals is characterized by tolerance, while the exposure to allochthonous bacteria results in a stronger immune response (8, 9). Duchmann and coworkers showed that tolerance selectively exists to intestinal biota from autologous but not heterologous intestinal samples and that the latter resulted in strong responses from blood and mucosal lymphocytes (20). It appears that gut bacteria have evolved properties for avoiding an immune response from their host. Indeed, gut bacteria possess factors that induce antigen-specific regulatory T cells which actively contribute to tolerance development (87, 98). As a consequence, autochthonous bacteria might be more promising candidates for probiotics aimed at suppressing an inappropriate immune response, desirable in the treatment of inflammatory bowel diseases (IBD). L. reuteri, which is autochthonous to rodents and humans, has been shown to modulate macrophage and dendritic cell functions in a way one would expect to favor immunological tolerance (14, 67, 87). Accordingly, strains of L. reuteri are especially successful in the prevention of colitis in several animal models (54, 67). On the other hand, the activation of the immune system (such as enhanced phagocytosis and adjuvant effects) observed after the administration of some probiotic strains may reflect the allochthonous nature of the bacteria, and these bacteria might be more effective for the treatment or prevention of infectious and rotavirus-caused diarrhea (53, 84). One would assume that allochthonous organisms are also more successful in the prevention of atopic diseases in early life because the immune system will experience novel antigenic complexes with the encounter of the bacterial strains. It has been shown that virtually all health benefits and effects on host cells reported for probiotics are strain dependent (53). Mechanistic explanations for this strain specificity are so far lacking, but it is likely that the distinct evolutionary histories of currently used probiotic strains are at least partly responsible for their different effects.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
The scientific data presented in this review indicate that most Lactobacillus species found in the mammalian intestinal tract are in fact not true intestinal inhabitants. They probably originate from more proximal or exogenous sources where the nutrient requirements of these fastidious organisms are satisfied. Future research is needed to identify the autochthonous Lactobacillus microbiota of the mammalian intestine. Strains that form stable populations (over several months) in the intestinal tract without having significant upstream populations would show clear characteristics of autochthonous intestinal inhabitants. In humans, fecal isolates of subjects fed a diet devoid of lactobacilli could be compared to oral isolates by using discriminative strain typing methods to identify strains autochthonous to the GIT. Strains whose ecological status is clearly identified are good candidates with which to elucidate ecological cohesions that take place within the gut environment and should be included in functional and comparative genomic investigations to reveal how lactobacilli make a living in the intestinal tract. A better understanding of the ecology of lactobacilli will help us to more systematically develop probiotic applications.
It has become more and more evident that shifts in gut commensal populations and an aberrant immune reaction toward these microbes are associated with several disease conditions such as allergies, IBD, obesity, and colon cancer. Redress of these ecological and immunological imbalances, for instance by probiotics, has the potential to ameliorate and prevent disease (25). For lactobacilli to become successful in this respect, ecological and functional aspects of the strains should already be considered when candidates are screened. As noted by Morelli, there is considerable doubt about the real value of the current selection criteria for probiotics, such as their tolerance to the hostile conditions of the stomach and the small intestine and their ability to adhere to intestinal surfaces of epithelial cell lines (65). The ecological origin of the probiotic strain remains important, but this requires much more than just picking a fecal isolate. In the future, strain selection could be based on criteria such as ecological performance, persistence, and evolutionary history. Autochthonous strains that naturally persist in human subjects over long periods are tested by nature for their functionality in the gut, and they are likely to possess adaptive traits to benefit their human host. Strain selection should also be targeted directly at the alleviation or prevention of specific medical conditions. This is more difficult, as it requires a mechanistic understanding of the effect one wants to achieve, but ex vivo experiments with immune cells isolated from humans are likely to become very valuable in this respect.
It is important to note that the majority of traditional probiotic strains are probably allochthonous to the intestinal tract, and they show very little ability to persist in the human gut. These strains might nonetheless be excellent probiotics with respect to activation of the immune system. As there is no indication that colonization is required for the health benefits of these strains, research of traditional probiotic strains should focus less on the investigation of ecological fitness and the identification of putative colonization determinants and more on the provision of mechanistic explanations for the health benefits that have been achieved in clinical trials.
Acknowledgments
I thank Daniel Frank (Norman Pace laboratory, University of Colorado, Boulder) and Ruth Ley (Jeffrey Gordon Laboratory, Washington University, St. Louis, MO) for providing comprehensive sequence data sets of their recent molecular characterization of the gut microbiota (26, 47, 49). I acknowledge various inspiring discussions with Gerald Tannock (University of Otago, Dunedin, New Zealand). I am indebted to Robert Hutkins (University of Nebraska, Lincoln) for many lively discussions and critical reading of the manuscript and to Andrew Benson (University of Nebraska, Lincoln) for many helpful comments. I thank Sheila Scheideler and Thomas Burkey (University of Nebraska, Lincoln) for providing gut samples from chicken and swine and Inés Martínez for performing the DGGE analysis.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Ahola, A. J., H. Yli-Knuuttila, T. Suomalainen, T. Poussa, A. Ahlstrom, J. H. Meurman, and R. Korpela. 2002. Short-term consumption of probiotic-containing cheese and its effect on dental caries risk factors. Arch. Oral Biol. 47:799-804. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, M. 1971. Microbial ecology. John Wiley and Sons, New York, NY.
- 3.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 5.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 100:8957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrangou, R., M. A. Azcarate-Peril, T. Duong, S. B. Conners, R. M. Kelly, and T. R. Klaenhammer. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. USA 103:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg, R. D. 1996. The indigenous gastrointestinal microflora. Trends Microbiol. 4:430-435. [DOI] [PubMed] [Google Scholar]
- 8.Berg, R. D., and D. C. Savage. 1975. Immune responses of specific pathogen-free and gnotobiotic mice to antigens of indigenous and nonindigenous microorganisms. Infect. Immun. 11:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg, R. D., and D. C. Savage. 1972. Immunological responses and microorganisms indigenous to the gastrointestinal tract. Am. J. Clin. Nutr. 25:1364-1371. [DOI] [PubMed] [Google Scholar]
- 10.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 103:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booijink, C. C., E. G. Zoetendal, M. Kleerebezem, and W. M. de Vos. 2007. Microbial communities in the human small intestine: coupling diversity to metagenomics. Future Microbiol. 2:285-295. [DOI] [PubMed] [Google Scholar]
- 12.Brooks, S. P., M. McAllister, M. Sandoz, and M. L. Kalmokoff. 2003. Culture-independent phylogenetic analysis of the faecal flora of the rat. Can. J. Microbiol. 49:589-601. [DOI] [PubMed] [Google Scholar]
- 13.Bunte, C., C. Hertel, and W. P. Hammes. 2000. Monitoring and survival of Lactobacillus paracasei LTH 2579 in food and the human intestinal tract. Syst. Appl. Microbiol. 23:260-266. [DOI] [PubMed] [Google Scholar]
- 14.Christensen, H. R., H. Frokiaer, and J. J. Pestka. 2002. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J. Immunol. 168:171-178. [DOI] [PubMed] [Google Scholar]
- 15.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeno-Tarraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 103:6718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dal Bello, F., and C. Hertel. 2006. Oral cavity as natural reservoir for intestinal lactobacilli. Syst. Appl. Microbiol. 29:69-76. [DOI] [PubMed] [Google Scholar]
- 17.Dal Bello, F., J. Walter, W. P. Hammes, and C. Hertel. 2003. Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition. Microb. Ecol. 45:455-463. [DOI] [PubMed] [Google Scholar]
- 18.Day, R. L., K. N. Laland, and F. J. Odling-Smee. 2003. Rethinking adaptation: the niche-construction perspective. Perspect. Biol. Med. 46:80-95. [DOI] [PubMed] [Google Scholar]
- 19.Denou, E., R. D. Pridmore, B. Berger, J. M. Panoff, F. Arigoni, and H. Brussow. 2008. Identification of genes associated with the long gut persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190:3161-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duchmann, R., I. Kaiser, E. Hermann, W. Mayet, K. Ewe, and K. H. Meyer zum Buschenfelde. 1995. Tolerance exists towards resident intestinal flora but is broken in active inflammatory bowel disease (IBD). Clin. Exp. Immunol. 102:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunne, C., L. Murphy, S. Flynn, L. O'Mahony, S. O'Halloran, M. Feeney, D. Morrissey, G. Thornton, G. Fitzgerald, C. Daly, B. Kiely, E. M. Quigley, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 1999. Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie van Leeuwenhoek 76:279-292. [PubMed] [Google Scholar]
- 22.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finegold, S. M., H. R. Attebery, and V. L. Sutter. 1974. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am. J. Clin. Nutr. 27:1456-1469. [DOI] [PubMed] [Google Scholar]
- 24.Finegold, S. M., V. L. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal flora, p. 3-31. In D. J. Hentges (ed.), Human intestinal microbiota in health and disease. Academic Press, New York, NY.
- 25.Frank, D. N., and N. R. Pace. 2008. Gastrointestinal microbiology enters the metagenomics era. Curr. Opin. Gastroenterol. 24:4-10. [DOI] [PubMed] [Google Scholar]
- 26.Frank, D. N., A. L. St. Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz, and N. R. Pace. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 104:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuller, R., and B. E. Brooker. 1974. Lactobacilli which attach to the crop epithelium of the fowl. Am. J. Clin. Nutr. 27:1305-1312. [DOI] [PubMed] [Google Scholar]
- 28.Gill, H., and J. Prasad. 2008. Probiotics, immunomodulation, and health benefits. Adv. Exp. Med. Biol. 606:423-454. [DOI] [PubMed] [Google Scholar]
- 29.Goh, Y. J., J. H. Lee, and R. W. Hutkins. 2007. Functional analysis of the fructooligosaccharide utilization operon in Lactobacillus paracasei 1195. Appl. Environ. Microbiol. 73:5716-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon, H. A., and L. Pesti. 1971. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 35:390-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammes, W. P., and R. F. Vogel. 1995. The genus Lactobacillus, p. 19-54. In B. J.-B. Wood and W. H. Holzapfel (ed.), The genera of lactic acid bacteria, vol. 2. Blackie Academic and Professional, London, United Kingdom. [Google Scholar]
- 32.Hardin, G. 1960. The competitive exclusion principle. Science 131:1292-1297. [DOI] [PubMed] [Google Scholar]
- 33.Harmsen, H. J., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Fecal microbial diversity in a strict vegetarian as determined by molecular analysis and cultivation. Microbiol. Immunol. 46:819-831. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46:535-548. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi, H., R. Takahashi, T. Nishi, M. Sakamoto, and Y. Benno. 2005. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J. Med. Microbiol. 54:1093-1101. [DOI] [PubMed] [Google Scholar]
- 37.Hold, G. L., S. E. Pryde, V. J. Russell, V. J. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 38.Jacobsen, C. N., V. Rosenfeldt Nielsen, A. E. Hayford, P. L. Moller, K. F. Michaelsen, A. Paerregaard, B. Sandstrom, M. Tvede, and M. Jakobsen. 1999. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kandler, O., and N. Weiss. 1986. Regular, nonsporing gram-positive rods, p. 1208-1234. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, MD. [Google Scholar]
- 40.Kassen, R., and P. B. Rainey. 2004. The ecology and genetics of microbial diversity. Annu. Rev. Microbiol. 58:207-231. [DOI] [PubMed] [Google Scholar]
- 41.Kimura, K., A. L. McCartney, M. A. McConnell, and G. W. Tannock. 1997. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl. Environ. Microbiol. 63:3394-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klock, B., M. Svanberg, and L. G. Petersson. 1990. Dental caries, mutans streptococci, lactobacilli, and saliva secretion rate in adults. Community Dent. Oral Epidemiol. 18:249-252. [DOI] [PubMed] [Google Scholar]
- 44.Lan, P. T., H. Hayashi, M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of cecal microbiota in chicken by the use of 16S rDNA clone libraries. Microbiol. Immunol. 46:371-382. [DOI] [PubMed] [Google Scholar]
- 45.Lerche, M., and G. Reuter. 1961. Isolierung und Differenzierung anaerober Lactobacillaceae aus dem Darm erwachsener Menschen (Beitrag zum Lactobacillus Bifidus-Problem). Zentralbl. Bakteriol. 180:324-356. [Google Scholar]
- 46.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ley, R. E., D. A. Peterson, and J. I. Gordon. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837-848. [DOI] [PubMed] [Google Scholar]
- 49.Ley, R. E., P. J. Turnbaugh, S. Klein, and J. I. Gordon. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022-1023. [DOI] [PubMed] [Google Scholar]
- 50.Liu, M., F. H. van Enckevort, and R. J. Siezen. 2005. Genome update: lactic acid bacteria genome sequencing is booming. Microbiology 151:3811-3814. [DOI] [PubMed] [Google Scholar]
- 51.Lu, J., U. Idris, B. Harmon, C. Hofacre, J. J. Maurer, and M. D. Lee. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madigan, M. T., and J. M. Martinko. 2006. Biology of microorganisms, 11th ed. Pearson Prentice Hall, Upper Saddle River, NJ.
- 53.Madsen, K. 2006. Probiotics and the immune response. J. Clin. Gastroenterol. 40:232-234. [DOI] [PubMed] [Google Scholar]
- 54.Madsen, K. L., J. S. Doyle, L. D. Jewell, M. M. Tavernini, and R. N. Fedorak. 1999. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology 116:1107-1114. [DOI] [PubMed] [Google Scholar]
- 55.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marsh, P., and M. V. Martin. 1999. Oral microbiology, 4th ed. Butterworth-Heinemann, Oxford, United Kingdom.
- 57.Marteau, P., P. Pochart, J. Dore, C. Bera-Maillet, A. Bernalier, and G. Corthier. 2001. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol. 67:4939-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McBurney, W. T. 2003. Analysis of the gut microbiota of rodents. Ph.D. thesis. University of Otago, Dunedin, New Zealand.
- 59.Metchnikoff, E. 1907. The prolongation of life. Optimistic studies. William Heinemann, London, United Kingdom.
- 60.Mikelsaar, M., R. M. Mändar, and E. Sepp. 1998. Lactic acid microbiota in the human microbial ecosystem and its development, p. 279-342. In S. Salminen and A. von Wright (ed.), Lactic acid bacteria: microbiology and functional aspects. Marcel Dekker, Inc., New York, NY.
- 61.Mitsuoka, T. 1969. Comparative studies on lactobacilli from the faeces of man, swine and chickens. Zentralbl. Bakteriol. 210:32-51. (In German.) [PubMed] [Google Scholar]
- 62.Mitsuoka, T. 1992. The human gastrointestinal tract, p. 69-114. In B. J. B. Wood (ed.), The lactic acid bacteria in health and disease, vol. 1. Elsevier Applied Science, London, United Kingdom. [Google Scholar]
- 63.Mitsuoka, T., K. Hayakawa, and N. Kimura. 1975. The fecal flora of man. III. Communication: the composition of Lactobacillus flora of different age groups. Zentralbl. Bakteriol. 232:499-511. (In German.) [PubMed] [Google Scholar]
- 64.Miyoshi, Y., S. Okada, T. Uchimura, and E. Satoh. 2006. A mucus adhesion promoting protein, MapA, mediates the adhesion of Lactobacillus reuteri to Caco-2 human intestinal epithelial cells. Biosci. Biotechnol. Biochem. 70:1622-1628. [DOI] [PubMed] [Google Scholar]
- 65.Morelli, L. 2000. In vitro selection of probiotic lactobacilli: a critical appraisal. Curr. Issues Intest. Microbiol. 1:59-67. [PubMed] [Google Scholar]
- 66.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie van Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 67.Peña, J. A., A. B. Rogers, Z. Ge, V. Ng, S. Y. Li, J. G. Fox, and J. Versalovic. 2005. Probiotic Lactobacillus spp. diminish Helicobacter hepaticus-induced inflammatory bowel disease in interleukin-10-deficient mice. Infect. Immun. 73:912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peterson, D. A., N. P. McNulty, J. L. Guruge, and J. I. Gordon. 2007. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2:328-339. [DOI] [PubMed] [Google Scholar]
- 69.Pfeiler, E. A., and T. R. Klaenhammer. 2007. The genomics of lactic acid bacteria. Trends Microbiol. 15:546-553. [DOI] [PubMed] [Google Scholar]
- 70.Prescott, L. M., J. P. Harley, and D. A. Klein. 2005. Microbiology, 6th ed. McGraw-Hill, Boston, MA.
- 71.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 73.Reuter, G. 1965. Untersuchungen über die Zusammensetzung und die Beeinflussbarkeit der menschlichen Magen- und Darmflora unter besonderer Berücksichtigung der Laktobazillen. Ernährungsforschung 10:429-435. [Google Scholar]
- 74.Rinttila, T., A. Kassinen, E. Malinen, L. Krogius, and A. Palva. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97:1166-1177. [DOI] [PubMed] [Google Scholar]
- 75.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433-442. [DOI] [PubMed] [Google Scholar]
- 76.Ryan, K. J. 2004. Normal microbial flora. In K. J. Ryan and C. G. Ray (ed.), Medical microbiology. An introduction to infectious diseases, 4th ed. McGraw-Hill, New York, NY.
- 77.Saarela, M., G. Mogensen, R. Fonden, J. Matto, and T. Mattila-Sandholm. 2000. Probiotic bacteria: safety, functional and technological properties. J. Biotechnol. 84:197-215. [DOI] [PubMed] [Google Scholar]
- 78.Salzman, N. H., H. de Jong, Y. Paterson, H. J. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148:3651-3660. [DOI] [PubMed] [Google Scholar]
- 79.Saulnier, D. M., D. Molenaar, W. M. de Vos, G. R. Gibson, and S. Kolida. 2007. Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Appl. Environ. Microbiol. 73:1753-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 81.Savage, D. C., R. Dubos, and R. W. Schaedler. 1968. The gastrointestinal epithelium and its autochthonous bacterial flora. J. Exp. Med. 127:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scanlan, P. D., F. Shanahan, C. O'Mahony, and J. R. Marchesi. 2006. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J. Clin. Microbiol. 44:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schiffrin, E. J., F. Rochat, H. Link-Amster, J. M. Aeschlimann, and A. Donnet-Hughes. 1995. Immunomodulation of human blood cells following the ingestion of lactic acid bacteria. J. Dairy Sci. 78:491-497. [DOI] [PubMed] [Google Scholar]
- 85.Seegers, J. F. 2002. Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 20:508-515. [DOI] [PubMed] [Google Scholar]
- 86.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smits, H. H., A. Engering, D. van der Kleij, E. C. de Jong, K. Schipper, T. M. van Capel, B. A. Zaat, M. Yazdanbakhsh, E. A. Wierenga, Y. van Kooyk, and M. L. Kapsenberg. 2005. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 115:1260-1267. [DOI] [PubMed] [Google Scholar]
- 88.Spanhaak, S., R. Havenaar, and G. Schaafsma. 1998. The effect of consumption of milk fermented by Lactobacillus casei strain Shirota on the intestinal microflora and immune parameters in humans. Eur. J. Clin. Nutr. 52:899-907. [DOI] [PubMed] [Google Scholar]
- 89.Steidler, L. 2003. Genetically engineered probiotics. Best Pract. Res. Clin. Gastroenterol. 17:861-876. [DOI] [PubMed] [Google Scholar]
- 90.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tannock, G. W. 1999. Analysis of the intestinal microflora: a renaissance. Antonie van Leeuwenhoek 76:265-278. [PubMed] [Google Scholar]
- 92.Tannock, G. W. 1992. Lactic microbiota of pigs, mice and rats, p. 21-48. In B. J. B. Wood (ed.), The lactic acid bacteria in health and disease, vol. 1. Elsevier Applied Science, London, United Kingdom. [Google Scholar]
- 93.Tannock, G. W. 1995. Normal microflora: an introduction to microbes inhabiting the human body. Chapman and Hall, London, United Kingdom.
- 94.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 70:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tannock, G. W., S. Ghazally, J. Walter, D. Loach, H. Brooks, G. Cook, M. Surette, C. Simmers, P. Bremer, F. Dal Bello, and C. Hertel. 2005. Ecological behavior of Lactobacillus reuteri 100-23 is affected by mutation of the luxS gene. Appl. Environ. Microbiol. 71:8419-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tannock, G. W., K. Munro, H. J. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tappenden, K. A., and A. S. Deutsch. 2007. The physiological relevance of the intestinal microbiota: contributions to human health. J. Am. Coll. Nutr. 26:679S-683S. [DOI] [PubMed] [Google Scholar]
- 98.Tsuji, N. M. 2006. Antigen-specific CD4(+) regulatory T cells in the intestine. Inflamm. Allergy Drug Targets 5:191-201. [DOI] [PubMed] [Google Scholar]
- 99.van der Waaij, L. A., H. J. Harmsen, M. Madjipour, F. G. Kroese, M. Zwiers, H. M. van Dullemen, N. K. de Boer, G. W. Welling, and P. L. Jansen. 2005. Bacterial population analysis of human colon and terminal ileum biopsies with 16S rRNA-based fluorescent probes: commensal bacteria live in suspension and have no direct contact with epithelial cells. Inflamm. Bowel Dis. 11:865-871. [DOI] [PubMed] [Google Scholar]
- 100.Vanhoutte, T., G. Huys, E. De Brandt, and J. Swings. 2004. Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol. Ecol. 48:437-446. [DOI] [PubMed] [Google Scholar]
- 101.Velez, M. P., S. C. De Keersmaecker, and J. Vanderleyden. 2007. Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol. Lett. 276:140-148. [DOI] [PubMed] [Google Scholar]
- 102.Walter, J., P. Chagnaud, G. W. Tannock, D. M. Loach, F. Dal Bello, H. F. Jenkinson, W. P. Hammes, and C. Hertel. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71:979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Walter, J., N. C. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69:2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Walter, J., D. M. Loach, M. Alqumber, C. Rockel, C. Hermann, M. Pfitzenmaier, and G. W. Tannock. 2007. D-Alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100-23 results in impaired colonization of the mouse gastrointestinal tract. Environ. Microbiol. 9:1750-1760. [DOI] [PubMed] [Google Scholar]
- 106.Walter, J., C. Schwab, D. M. Loach, M. G. Ganzle, and G. W. Tannock. 2008. Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiology 154:72-80. [DOI] [PubMed] [Google Scholar]
- 107.Wang, M., S. Ahrne, B. Jeppsson, and G. Molin. 2005. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 54:219-231. [DOI] [PubMed] [Google Scholar]
- 108.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang, X., S. P. Heazlewood, D. O. Krause, and T. H. Florin. 2003. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J. Appl. Microbiol. 95:508-520. [DOI] [PubMed] [Google Scholar]
- 110.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 111.Xu, J., and J. I. Gordon. 2003. Inaugural article: honor thy symbionts. Proc. Natl. Acad. Sci. USA 100:10452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zoetendal, E. G., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]