Abstract
Giardiasis is a notifiable disease of high prevalence in New Zealand, but there is limited knowledge about the sources of Giardia duodenalis genotypes that can potentially cause human infections. Dairy calves are one environmental source of Giardia isolates, but it is unknown whether they harbor genotypes that are potentially capable of causing infections in humans. To address these questions, 40 Giardia isolates from calves and 30 from humans, living in the same region and collected over a similar period, were genotyped using the β-giardin gene. The G. duodenalis genetic assemblages A and B were identified from both calves and humans, and genotype comparisons revealed a substantial overlap of identical genotypes from the two hosts for both assemblages. Significantly, no assemblage E (the genotype commonly found in cattle elsewhere in the world) has been detected in New Zealand livestock to date. Given recent and rapid land use conversions to dairy farming in many South Island regions of New Zealand, an increasingly large concentration of domestic cattle harboring genotypes potentially capable of causing infections in humans is particularly concerning.
Giardia duodenalis (synonyms are G. intestinalis and G. lamblia) is a common protozoan parasite that infects a wide range of mammalian hosts. In humans, it is one of the most frequently identified protozoan parasites causing gastrointestinal disease worldwide (8, 19). Originally thought to show host specificity, substantial evidence suggests G. duodenalis is better considered a species complex, as multiple and genetically distinct assemblages have been characterized. The following seven assemblages are currently recognized for G. duodenalis: A and B, the only assemblages capable of infecting humans but also found in a wide range of other mammalian hosts; C and D, isolated from dogs; E, isolated from livestock (cattle, sheep, and pigs); F, isolated from felines; and G, isolated from rats (18).
A significant prevalence of Giardia isolates in dairy cattle was first reported during the early 1980s, since which time an increasing focus has been on the identification of potentially zoonotic assemblages in such hosts (10). High levels of prevalence of G. duodenalis have been reported in newborn calves worldwide, with the majority of molecular genotyping studies identifying livestock assemblage E (6, 23, 24). However, of particular concern recently has been the frequent identification of assemblage A and the intermittent identification of assemblage B from cattle, the same Giardia assemblages found in human infections (15, 16). Despite this concern, only two studies to date have compared G. duodenalis genotypes from humans and calves living in the same geographical location (15, 16).
In New Zealand, the intensification of existing dairy farms and the conversion to dairy farming of land from lower-density sheep and beef farming are causing concern, with reports of declines in water quality coincident with such land use changes (4, 20). In addition to physical alterations to the landscape, concerns for public health have arisen, as environmental sources of pathogens capable of causing human infections could potentially be transmitted by contact with and consumption of contaminated water (5). While the characterization of identical Giardia assemblages from dairy cattle and humans does not confirm an active transmission pathway between them, the potential for zoonotic transmission may exist and should be considered when managing public health risks (15), particularly in areas with rapidly increasing cattle density (26).
Giardiasis is a notifiable disease with an already high incidence in the New Zealand population, yet few studies have assessed Giardia genotypes from humans. Similarly, it is unknown which genotypes cattle harbor, even though recent studies of newborn dairy calves found an average Giardia prevalence of 31%, with such animals capable of excreting up to 105 cysts per gram of feces per day for up to 2 weeks (5, 26, 28). This lack of knowledge becomes particularly important in areas currently undergoing rapid intensification of dairying, as it is unclear whether Giardia found in dairy cattle actually poses a threat to the public's health (2). Given the disparities in Giardia prevalence and genotypes in cattle reported from different regions of the world, it would be unsafe to rely on results from studies in other countries to estimate the potential risk of zoonotic transmission in New Zealand (13, 23) (Table 1). Therefore, in the present study, we genotyped Giardia isolates from dairy calves and infected humans located in the same region and spanning the same time period. Our objectives were to determine the Giardia genotypes present in calves and humans in a previously uncharacterized New Zealand region and to assess the potential for zoonotic transmission between these two hosts in a region undergoing rapid land use change to intensive dairy farming.
TABLE 1.
Comparison of the prevalence rates of Giardia isolates in cattle and the assemblages identified from humans and cattle worldwide between 2000 and 2007
| Publication | Location | Host species (no.) | Prevalence in calves (%) | Assemblage in:
|
|
|---|---|---|---|---|---|
| Humans (%) | Calves (%) | ||||
| Caccio et al. (7) | Italy | Human (30) | A (80), B (20) | ||
| Becher et al. (6) | Australia | Calf (31) | 89 | E (100) | |
| Lalle et al. (15) | Italy | Calf (37), human (24) | Not reported | A (45), B (41), A and B (14) | A (50), B (21), E (13), A and B (8), A and E (8) |
| Trout et al. (24) | United States, East Coast | Calf (237) | 52 | A (13), E (87) | |
| Trout et al. (23) | United States, East Coast | Calf (204) | 36 | A (9), E (91) | |
| Gelanew et al. (12) | Ethiopia | Human (59) | A (52), B (22), A and F (12), A and B (14) | ||
| Mendonça et al. (17) | Portugal | Calf (14) | 14 | A (14), B (7), E (79) | |
| Hunt et al. (14) | New Zealand, North Island | Calf (15) | 41 | A (73), B (27) | |
| Learmonth et al. (16) | New Zealand, North Island | Calf (26), human (2) | 10 | A (50), B (50) | A (55), B (45) |
| Winkworth et al. (this study) | New Zealand, South Island | Calf (40), human (30) | 31 | A (77), B (23) | A (88), B (12) |
MATERIALS AND METHODS
Study area.
At the time of this study, 322 dairy farms were operating in the Otago region of the South Island of New Zealand, milking approximately 153,111 head of cattle (3). The geographical locations of the dairy farms used in the current research spanned an area dominated by year-round dairying with spring and winter milking. The chosen sites were representative of the recent intensification of dairy farming in the Otago region.
Sample collection.
Giardia samples were genotyped from dairy calves born in the Otago region during the spring calving seasons (August to October) of 2005 and 2006. Cysts were purified from calf feces collected in a parallel study investigating the prevalence of Giardia isolates in the Otago region (26). In the prevalence study, fecal samples from mobs of calves were collected from 10 individual farms during the first 7 weeks of the animals' lives. Samples for the current genotype analysis were chosen to represent multiple farms and different weeks during each calving season, providing an appropriate overview of the Giardia assemblages present in newborn calves from the region (Table 2). During the same two spring calving seasons and the year in between, Giardia isolates from humans were collected from individuals screened at medical laboratories in the Otago region who showed active infections of giardiasis.
TABLE 2.
Spatial and temporal distribution of Giardia isolates sequenced from calf isolates collected in the Otago region of New Zealand and the assemblage(s) identified
| Collection time, yr and wk | No. of individual farms | Total no. of isolates | Assemblage(s) identified |
|---|---|---|---|
| 2005 | |||
| 1 | 3 | 4 | A |
| 2 | 2 | 1 | A |
| 3 | 2 | 2 | A |
| 4 | 2 | 2 | A |
| 5 | 5 | 5 | A |
| 6 | 3 | 3 | A |
| 7 | 2 | 3 | A |
| 2006 | |||
| 1 | 2 | 3 | A and B |
| 2 | 2 | 3 | A |
| 3 | 4 | 7 | A and B |
| 4 | 1 | 1 | B |
| 5 | 1 | 1 | A |
| 6 | 2 | 4 | A |
| 7 | 1 | 1 | A |
Cyst isolation.
Giardia isolates were purified from fecal samples using a modified diethyl ether procedure to remove large fecal debris and fats, followed by either sucrose flotation or immunomagnetic bead separation to remove smaller particulate matter (25). Concentrating cysts prior to nucleic acid extraction ensured that higher yields of nucleic acid concentrations were obtained for subsequent PCR amplification.
Approximately 1 g of feces was homogenized with 7 ml Milli-Q water in a 15-ml tube (BD Biosciences, San Jose, CA) and poured through a standard tea strainer into a 15-ml conical centrifuge tube (BD Biosciences, San Jose, CA). After straining of the samples, 3 ml of diethyl ether was added, and the suspension was mixed vigorously for 1 min and then immediately centrifuged at 750 × g for 1 min in a swing bucket rotor. Taking care not to disturb the pelleted cysts, the resulting plug of fat was detached from the tube's edge by encircling it with a pipette tip, then poured off along with the supernatant.
Samples containing more than 105 Giardia cysts per ml were concentrated using a sucrose flotation method following the diethyl ether purification. Cysts were resuspended in Milli-Q water and transferred to a clean 50-ml conical centrifuge tube (BD Biosciences, San Jose, CA), with a final volume of 20 ml. Taking care not to disrupt or mix the two layers, 30 ml 1 M sucrose was layered underneath this suspension. Samples were immediately centrifuged at 1,050 × g for 10 min in a swing bucket rotor using slow acceleration and no brake. Of the 50-ml total volume, the top 20 ml of water, the water-sucrose interface, and the first 5 ml of sucrose were transferred to a second clean 50-ml conical centrifuge tube, resuspended in Milli-Q water to a final volume of 50 ml, and centrifuged again. All except the last 5 ml and pelleted cysts were discarded, with the purified cysts resuspended in the remaining 5 ml.
Fecal samples containing a low number of cysts were purified using immunomagnetic bead separation following the diethyl ether concentration step. Pelleted cysts were resuspended in 2 ml Milli-Q water and 1 ml was transferred to a clean 1.5-ml centrifuge tube (Axygen, Union City, CA) containing 100 μl each of 10× SL buffers A and B and 20 μl anti-Giardia Dynabeads (Dynal Biotech Pty Ltd., Australia). Tubes were incubated on a rotary mixer at 60 rpm for an hour at room temperature to promote the formation of bead-Giardia complexes. Following incubation, the samples were transferred to a magnetic rack and rocked gently back and forth for a minute to capture the bead-Giardia complexes. Taking care not to disturb the bead-Giardia complex, and without removing the tubes from the magnet, the supernatant was removed, the complex washed once with 100 μl 10× SL buffer A premixed in 900 μl Milli-Q water, and the supernatant discarded once more. To dislodge Giardia cells from the bead complex, the tubes were removed from the magnet, mixed with 50 μl 0.1 M HCl (VWR International Ltd., Poole, United Kingdom), and incubated for 10 min at room temperature. The dislodged beads were captured by returning the tube to the magnet, and the cysts, now contained in the supernatant, were transferred to a new 1.5-ml tube. To neutralize the HCl, 5 μl 5 M sodium hydroxide (VWR International Ltd., Poole, United Kingdom) was added to the purified cysts and the samples were maintained at 4°C until use.
Nucleic acid extraction.
Nucleic acid was extracted from purified cysts using a QIAamp DNA stool mini kit (Qiagen, Valencia, CA) by following the manufacturer's instructions, with three exceptions. First, as samples contained concentrated cysts instead of fecal matter, 50 μl of each sample was added directly to 1.2 ml of buffer ASL, mixed by vortexing for 1 min, and then incubated at 95°C (instead of 70°C) for 10 min. Second, following the removal of inhibitors with the InhibitEX steps, volumes were doubled for the remaining steps, bar that of elution. Finally, when the extracted DNA was eluted, half the recommended volume of buffer AE was used due to the low DNA concentrations recovered using this procedure. The concentration of purified DNA was quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and stored at −20°C until required.
PCR amplification.
In order to discriminate between the highly conserved sequences typically observed for the G. duodenalis assemblages, the β-giardin gene assay was used (7). The β-giardin protein is considered unique to Giardia spp., effectively eliminating the chance of cross-amplification of host, nontarget protozoan, fungal, algal, or bacterial templates. This assay also avoids the technical problems associated with the high GC content of small-subunit rRNA amplification (7). Nested-PCR amplifications were performed on all extracted DNA using primers described by Cacciò et al. (7) and Lalle et al. (15). The PCR mix for both primer sets comprised the Qiagen components of 1× PCR buffer, 2× Q-Solution, 1.25 mM MgCl2, 1 unit of Taq DNA polymerase (Qiagen, Valencia, CA), 250 μM of each deoxynucleoside triphosphate (Bioline, Randolph, MA), and 200 to 300 nM of each primer (Invitrogen, Auckland, New Zealand). Between 1 and 10 ng of purified DNA was added as template to the initial PCR amplification (primers G7 and G759) and Milli-Q water to a total volume of 50 μl. For the nested PCR, 2 μl from the completed initial PCR amplification was added as template to the reaction mix, and the volume of Milli-Q water was adjusted to give a final total volume of 50 μl. For both primer pairs, PCR consisted of an initial denaturing step of 98°C for 5 min, a set of 45 cycles comprising 98°C for 30 s, 55°C for 30 s, and 72°C for 45 s, and a final extension at 72°C for 7 min. PCR products from the nested-PCR amplification, but not the initial amplification, were visualized on Tris-buffered 1% agarose gels (VWR International Ltd., Poole, United Kingdom; Pure Science Ltd., Wellington, New Zealand) containing 3% SybrSafe DNA gel stain (Invitrogen, Auckland, New Zealand).
All products amplified with the nested primers were purified to a ready-for-sequencing state by using a PureLink PCR purification kit (Invitrogen, Auckland, New Zealand) by following the manufacturer's instructions and eluted in 30 μl. Purified nested-PCR product concentrations were quantified with a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE).
DNA sequencing.
All nested-PCR products were sequenced using the ABI Prism BigDye Terminator version 3.1 ready reaction cycle sequencing kit (Applied BioSystems, Foster City, CA). Sequencing reactions comprised 3.2 pmol of the nested primer, the manufacturer's components of 3.5 μl sequencing buffer and 1 μl Big Dye Terminator version 3.1, 10 to 20 ng template, and Milli-Q water to a total volume of 20 μl. Every nested-PCR product was sequenced in its entirety in two overlapping bidirectional reactions by following the manufacturer's recommended cycle protocol, with one exception: all denaturating temperature steps were increased from 96°C to 98°C. Sequencing reactions were cleaned using a Qiagen DyeEX 2.0 spin kit (Qiagen, Valencia, CA) by following the manufacturer's directions and air dried at 50°C. Capillary separation sequencing was performed at the Allan Wilson Centre for Molecular Ecology and Evolution, Massey University, within 48 h.
Sequence analysis.
Bidirectional sequences for all amplified Giardia isolates were assembled using Sequencher version 4.7 (Gene Codes Corporation, Ann Arbor, MI). Individual isolates were compared to identify nucleotide differences, followed by comparison with previously published Giardia sequences from Lalle et al. (15). A genotype network was reconstructed using the algorithm of statistical parsimony described by Templeton et al. (21) and implemented in TCS version 1.21 (9).
Nucleotide sequence accession numbers.
Representatives for each of the assemblages identified in this study have been submitted to GenBank under the following accession numbers: EU274389 to EU274415 and EU275983 (Table 3).
TABLE 3.
Details of the 26 Giardia genotypes identified in this study, assemblage A and B consensus sequences, and the 24 novel genotypesa
| Assemblage (total no. of isolates) | Isolate name | No. of isolates | GenBank accession no. | Base change(s) from the consensus sequence |
|---|---|---|---|---|
| A (58) | ||||
| Calf consensus | 24 | EU274404 | ||
| Human consensus | 13 | EU274390 | ||
| Calf 4 | 1 | EU274409 | G-368-A | |
| Calf 5 | 1 | EU274412 | T-162-A | |
| Calf 6 | 1 | EU274413 | C-288-T; A-39-G | |
| Calf 9 | 1 | EU274414 | T-423-C; C-161-T | |
| Calf 10 | 1 | EU274403 | A-380-G; A-245-T | |
| Calf 11 | 1 | EU274415 | A-286-G; T-199-C | |
| Calf 15 | 1 | EU274406 | T-243-C; A-154-G | |
| Calf 17 | 1 | EU274407 | C-230-T; A-116-G | |
| Calf 48 | 1 | EU274408 | G-216-A; A-121-G | |
| Calf 53 | 1 | EU274410 | A-431-G | |
| Calf 57 | 1 | EU274411 | A-175-G; −20-T; C-161-T | |
| Human 10 | 1 | EU275983 | A-31-G; G-474-A; A-195-G | |
| Human 17 | 1 | EU274392 | G-98-A | |
| Human 26 | 1 | EU274394 | C-233-T | |
| Human 27 | 1 | EU274395 | T-78-C | |
| Human 28 | 1 | EU274396 | A-305-G; A-170-G | |
| Human 30 | 1 | EU274398 | T-387-C | |
| Human 33 | 1 | EU274399 | T-16-C | |
| Human 35 | 1 | EU274400 | A-172-G; C-77-T | |
| Human 36 | 1 | EU274401 | A-31-G; A-361-C | |
| Human 39 | 1 | EU274402 | G-314-A | |
| B (12) | ||||
| Calf consensus | 5 | EU274405 | ||
| Human consensus | 4 | EU274397 | ||
| Human 1 | 1 | EU274389 | A-63-G; T-228-C; C-375-T | |
| Human 16 | 1 | EU274391 | A-63-G; T-228-C; C-375-T; T-297-C; C-211-T; C-67-T | |
| Human 25 | 1 | EU274393 | A-63-G; T-228-C; C-375-T; T- 297-C |
Nucleotide substitutions from the respective consensus sequences are shown (original base-nucleotide position-substituted base), as are the number of isolates found representing each genotype.
RESULTS
Human isolates.
Human fecal samples containing Giardia parasites were collected between 10 August 2005 and 30 October 2006. This collection period was chosen to encompass both calving seasons as well as the period in between when dairy animals are on farm fields. Thirty isolates spanning the entire collection period were sequenced.
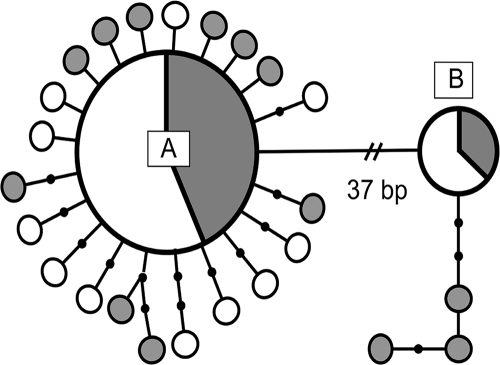
A total of 498 bp were sequenced for all 30 isolates and covered the same region as that sequenced for the calf isolates (Table 3). Two main Giardia genotypes were identified from the human isolates, corresponding to Giardia assemblages A and B, and differed from each other by 37 nucleotide substitutions (Fig. 1). Twenty-three isolates were identified as assemblage A and showed 99% sequence similarity to assemblage A isolate AY545642 of Lalle et al. (15), while the remaining seven isolates showed 99% sequence similarity to assemblage B isolate AY647265 of Lalle et al. (15).
FIG. 1.
Network showing the 26 individual Giardia duodenalis genotypes in the Otago region of New Zealand. The assemblage A and B consensus genotypes were separated by 37 bp. The sizes of the circles are proportional to the genotype frequency, and the proportion of shading represents the frequency of genotype from humans (gray) and calves (white). The smallest gray and white circles represent single genotypes. Lines connecting genotypes represent one mutation, and small black circles represent inferred mutations (or genotypes) not observed in the data.
First focusing on the 23 assemblage A isolates, 13 of the isolates were identical to each other and only differed by one base pair from AY545642; thus, it was designated the human assemblage A consensus sequence (GenBank accession no. EU274390) (Table 3). In comparison to this consensus sequence, the remaining 10 isolates displayed a variety of nucleotide substitutions; 6 isolates each differed by one unique base pair (GenBank accession no. EU274392, EU274394-5, EU274398-9, and EU274402), 2 isolates differed by two unique base pairs (GenBank accession no. EU274396 and EU274400), and lastly, 2 isolates shared one nucleotide substitution but differed from each other by a further one (GenBank accession no. EU274401) and two (GenBank accession no. EU275983) nucleotide substitutions, respectively (Table 3).
Turning now to the seven assemblage B isolates, four isolates were identical to the human assemblage B consensus sequence (GenBank accession no. EU274397), while the remaining three isolates differed by three (GenBank accession no. EU274389), four (GenBank accession no. EU274393), and six (GenBank accession no. EU274391) unique nucleotide substitutions, respectively (Table 3).
In general, the human isolates did not share identical nucleotide substitutions; of the 13 assemblage A and B isolates displaying substitutions, 8 had novel substitutions. Of the five isolates that did share identical nucleotide substitutions, two assemblage A isolates shared a single nucleotide substitution but differed by an additional two base pairs. The remaining three isolates, all assemblage B, shared the same initial substitution of three base pairs, but an additional nucleotide substitution differentiated two of the isolates from each other, while a further two base pairs differentiated the third.
Calf isolates.
Calf fecal samples were collected from 12 August to 30 September 2005 and 18 August to 6 October 2006. Of the 372 calf fecal samples collected from 10 Otago farms in 2005 and 2006 in which Giardia was detected, 40 (11%) were chosen to span all farms, collection dates (1- to 7-week-old calves), and seasons. The number of isolates sequenced from each week and the number of farms represented by those samples are shown in Table 2.
A total of 498 bp were sequenced for all 40 calf isolates. Two main Giardia genotypes, corresponding to Giardia assemblages A and B, were identified from calves born during the 2005 and 2006 spring calving seasons (Fig. 1). The two assemblages differed by 37 nucleotide substitutions. Assemblage A was detected in 35 of the 40 calf isolates and over the 498 bp sequenced differed by one base pair from the previously published assemblage A isolate AY545642 of Lalle et al. (15). The assemblage A isolates came from all 10 farms and were identified in both 2005 and 2006. Twenty-four of the 35 assemblage A isolates were identical to each other and were thus designated the calf assemblage A consensus sequence (GenBank accession no. EU274404) (Table 3). In comparison to the assemblage A consensus, of the remaining 11 isolates, 3 differed by one base pair each (GenBank accession no. EU274409-10 and EU274412) and 7 differed by two base pairs each (GenBank accession no. EU274403, EU274413-15, and EU274406-8), while, lastly, 1 isolate differed by three base pairs (GenBank accession no. EU274411) (Table 3).
Turning now to the five isolates identified as assemblage B, these displayed 99% sequence similarity to assemblage B of Lalle et al. (15). The five assemblage B isolates were identical (calf assemblage B consensus sequence, GenBank accession no. EU274405) (Table 3), originated from 2 of the 10 farms, and were only detected in 2006.
Eleven of the 40 calf isolates sequenced contained nucleotide substitutions; however, all of the nucleotide substitutions detected were unique to those individual isolates, with no identical substitutions shared by isolates.
Several different genotype patterns were observed from the farms during the two seasons. For example, both assemblage A and B sequences were detected on the same sampling date on two farms, identical assemblage sequences were detected on different sampling dates on two farms, and nonidentical sequences of the same assemblage were identified on different sampling dates on two farms, while, lastly, one nonidentical and two identical sequences from the same assemblage were found on one farm.
Combined analysis.
An analysis combining isolates originating from calves and humans identified similar patterns to those seen for each group individually (Fig. 1). Thus, assemblage A accounted for approximately five times more isolates than assemblage B (58 isolates as opposed to 12). As with the separate calf and human analyses, assemblages A and B differed by 37 nucleotide substitutions, with assemblage A displaying fewer nucleotide substitutions than assemblage B, in comparison to the published sequences from Lalle et al. (15). Within each assemblage, both identical and nonidentical sequences were detected throughout the collection period, and of the 70 isolates, nucleotide substitutions were observed from a total of 24 (13 from humans and 11 from calves). Identical nucleotide substitutions were not observed for both human and calf isolates; rather, matching substitutions were only detected within each host species and not between them.
Overall, 58 (35 from calves and 23 from humans) of the 70 isolates were identified as showing sequence identity to assemblage A. Twenty-four calf isolates and 13 human isolates had identical sequences over the 498 bp analyzed, while 21 further isolates (11 from calves and 10 from humans) showed 99% similarity to Lalle et al. (15) assemblage A isolate AY545642. Both single and multiple nucleotide substitutions were observed in assemblage A isolates.
Twelve of the 70 isolates were characterized as assemblage B (five from calves and seven from humans). Four human and five calf isolates displayed identical nucleotide sequences across the 498 bp analyzed and 99% sequence similarity to Lalle et al. (15) assemblage B isolate AY647265. Three isolates (25%) had nucleotide substitutions from the assemblage B consensus sequence, compared with 21 isolates (36%) displaying substitutions from the assemblage A consensus sequence. Further, no assemblage B samples displayed single nucleotide substitutions from the assemblage B consensus sequence; rather, multiple substitutions of three, four, and six nucleotides were observed.
DISCUSSION
Human-isolated Giardia genotypes.
Human rates of giardiasis in New Zealand are high compared with rates from economically and developmentally similar countries like Canada and the United States (42 cases per 100,000 people in the population compared to 15 and 8 per 100,000, respectively) (11), and an increase in cases over recent years is concerning (2). Surprisingly, the current study, which detected both genetic assemblages A and B, is the first comprehensive analysis in either the North or South Islands of New Zealand of G. duodenalis genotypes from humans.
Globally, a higher proportion of human isolates have been genotyped as assemblage A than B (7, 12, 15, 17), with genotypic variation also being disproportional; assemblage A displays fewer multiple nucleotide substitutions than assemblage B (7, 12). Assemblage B genotypes were characterized by multiple substitutions of at least three base pairs in the current study, while assemblage A genotypes displayed a larger proportion of single and double substitutions from the consensus. For example, all 4 assemblage B genotypes that differed from the consensus sequence did so by three or more substitutions, while only 2 of 21 assemblage A genotypes that differed from the consensus sequence had three substitutions. Though the genetic variability detected in the current study corresponded with rates reported from other studies, any significance of the differences is unclear (12).
Calf-isolated Giardia genotypes.
Significant increases have been observed in dairy cattle numbers during the previous decade, with that in the Otago region in the South Island increasing by over 295% (1), yet the current study is the first to genotype Giardia isolates from dairy calves in the South Island. G. duodenalis assemblages A and B were detected in dairy calves during the current study, echoing studies of dairy animals in Italy and Portugal (15, 17). Detailed genotypic comparisons could not be made with a previous North Island study because different molecular locations were analyzed; however, both studies identified the same main two assemblages (16, 26). The occurrence of assemblage B in 2006 but not 2005, and detection of assemblage A from calves born in both years, would be consistent with a shift in genotype composition in South Island calves, but further study is needed to confirm this.
In contrast to studies conducted elsewhere (6, 15, 17, 22-24), the livestock Giardia assemblage E has so far not been detected in either New Zealand's North Island (14, 16) or South Island (this study). This is a remarkable difference, given the ubiquitous presence of assemblage E elsewhere and the fact that now 100 Giardia isolates from cattle have been genotyped in New Zealand (14, 16, 17, 19, 24). Whether assemblage E is truly absent from New Zealand's national dairy herd remains to be seen.
Zoonotic transmission.
The current study provides the first definitive genotypic assessment of Giardia spp. isolated from cattle and human hosts in New Zealand and surprisingly demonstrates the possibility of the dairy herd as a potential reservoir for human infection. It is not clear whether zoonotic transmission of Giardia isolates has occurred in New Zealand; however, links between cattle and human disease have previously been found for Cryptosporidium, a similar waterborne parasite also of high prevalence in human infections. For example, Learmonth et al. (16) found the national dairy herd to be a potential reservoir of genotypes capable of causing Cryptosporidium infections in humans in the North Island.
Comparing β-giardin gene sequences from the two hosts, the current study identified both identical and nonidentical genotypes for G. duodenalis assemblages A and B. The significant overlap of identical β-giardin gene sequences observed between humans and calves for assemblage A and B implies zoonotic transmission may have occurred between the two hosts; however, this cannot be established solely from the presence of identical genotypes in different hosts. Nevertheless, these results justify the previously unsubstantiated cause for concern for public health in regions like those in the South Island of New Zealand, where dairy farming has intensified rapidly. Multiple transmission pathways are known for Giardia spp., but transmission via water may be of particular significance in areas with large numbers of domestic cattle. This is because dairy cattle are typically maintained at high densities and produce large quantities of fecal waste. Further, the cysts excreted in their feces are environmentally resistant, immediately infectious to susceptible hosts (3, 5), and are prone to moving passively across landscapes and into waterways via runoff (27).
Echoing results from similar investigations, the current study found that zoonotic transmission may not necessarily be solely confined to assemblage A, as had earlier been suggested (15, 16, 19). Olson et al. (19) proposed that cattle were not a significant Giardia reservoir for human infection, as they were most commonly infected with assemblage E. However, increasing evidence of both assemblages A and B in calves suggests they may be a potential, and noteworthy, reservoir for human infection by Giardia spp., especially in New Zealand where assemblage E has yet to be detected in cattle. With this in mind, the significance of New Zealand's dairy herd as a potential reservoir of zoonotic Giardia organisms is probably greater than that of dairy cattle in other countries.
Acknowledgments
This work was funded by the Miss E. L. Hellaby Indigenous Grasslands Research Trust and the New Zealand Tertiary Education Commission Bright Futures Scheme.
We thank the farmers located across the Lower Taieri Plain who enabled our sample collection and Southern Community Laboratories for the Giardia-positive human specimens.
Footnotes
Published ahead of print on 20 June 2008.
REFERENCES
- 1.Anonymous. 2004. Agricultural production statistics: June 2004. Statistics New Zealand, Wellington.
- 2.Anonymous. 2006. Annual summary of outbreaks in New Zealand. Institute of Environmental Science and Research Limited, Wellington, New Zealand.
- 3.Anonymous. 2007. Dairy statistics 2005-2006. Livestock Improvement Corporation Limited, Hamilton, New Zealand.
- 4.Anonymous. 2003. Otago Regional Council report on the surface water quality of the upper Taieri River catchment, November 2001-December 2002. Otago Regional Council, Dunedin, New Zealand.
- 5.Appelbee, A., L. Frederick, T. Heitman, and M. Olson. 2003. Prevalence and genotyping of Giardia duodenalis from beef calves in Alberta, Canada. Vet. Parasitol. 112:289-294. [DOI] [PubMed] [Google Scholar]
- 6.Becher, K., I. Robertson, D. Fraser, D. Palmer, and R. Thompson. 2004. Molecular epidemiology of Giardia and Cryptosporidium infections in dairy calves originating from three sources in Western Australia. Vet. Parasitol. 123:1-9. [DOI] [PubMed] [Google Scholar]
- 7.Cacciò, S., M. De Giacomo, and E. Pozio. 2002. Sequence analysis of the β-giardin gene and development of a polymerase chain reaction-restriction fragment length polymorphism assay to genotype Giardia duodenalis cysts from human faecal samples. Int. J. Parasitol. 32:1023-1030. [DOI] [PubMed] [Google Scholar]
- 8.Cacciò, S., R. Thompson, J. McLauchlin, and H. Smith. 2005. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol. 21:430-437. [DOI] [PubMed] [Google Scholar]
- 9.Clement, M., D. Posada, and K. Crandall. 2000. TCS: a computer model to estimate gene genealogies. Mol. Ecol. 9:1657-1659. [DOI] [PubMed] [Google Scholar]
- 10.Deshpande, P., and U. Shastri. 1981. Incidence of Giardia infection in calves in Maharashtra state, India. Trop. Anim. Health Prod. 13:34. [DOI] [PubMed] [Google Scholar]
- 11.ESR. 2004. Annual summary of outbreaks in New Zealand 2003. Institute of Environmental Science and Research Limited, Wellington, New Zealand.
- 12.Gelanew, T., M. Lalle, A. Hailu, E. Pozio, and S. Cacciò. 2007. Molecular characterization of human isolates of Giardia duodenalis from Ethiopia. Acta Trop. 102:92-99. [DOI] [PubMed] [Google Scholar]
- 13.Hamnes, I., B. Gjerde, and L. Roberston. 2006. Prevalence of Giardia and Cryptosporidium in dairy calves in three areas of Norway. Vet. Parasitol. 140:204-216. [DOI] [PubMed] [Google Scholar]
- 14.Hunt, C., G. Ionas, and T. Brown. 2000. Prevalence and strain differentiation of Giardia intestinalis in calves in the Manawatu and Waikato regions of North Island, New Zealand. Vet. Parasitol. 91:7-13. [DOI] [PubMed] [Google Scholar]
- 15.Lalle, M., E. Pozio, G. Capelli, F. Bruschi, D. Crotti, and S. Cacciò. 2005. Genetic heterogeneity at the β-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int. J. Parasitol. 35:207-213. [DOI] [PubMed] [Google Scholar]
- 16.Learmonth, J., G. Ionas, A. Pita, and R. Cowie. 2003. Identification and genetic characterisation of Giardia and Cryptosporidium strains in humans and dairy cattle in the Waikato region of New Zealand. Water Sci. Technol. 47(3):21-26. [PubMed] [Google Scholar]
- 17.Mendonça, C., A. Almeida, A. Castro, M. Delgado, S. Soares, J. da Costa, and N. Canada. 2007. Molecular characterisation of Cryptosporidium and Giardia isolates from cattle in Portugal. Vet. Parasitol. 147:47-50. [DOI] [PubMed] [Google Scholar]
- 18.Monis, P., R. Andrews, G. Mayrhofer, and P. Ey. 2003. Genetic diversity within the morphological species Giardia intestinalis and its relationship to host origin. Infect. Genet. Evol. 3:29-38. [DOI] [PubMed] [Google Scholar]
- 19.Olson, M., R. O'Handley, B. Ralston, T. McAllister, and R. Thompson. 2004. Update on Cryptosporidium and Giardia infections in cattle. Trends Parasitol. 20:185-191. [DOI] [PubMed] [Google Scholar]
- 20.PCE. 2004. Growing for good: intensive farming, sustainability and New Zealand's environment. Parliamentary Commissioner for the Environment, Wellington, New Zealand.
- 21.Templeton, A., K. Crandall, and C. Sing. 1992. A cladistic-analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA-sequence data. III. Cladogram estimation. Genetics 132:619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trout, J., M. Santín, and R. Fayer. 2007. Prevalence of Giardia duodenalis genotypes in adult dairy cows. Vet. Parasitol. 147:205-209. [DOI] [PubMed] [Google Scholar]
- 23.Trout, J., M. Santín, E. Greiner, and R. Fayer. 2006. Prevalence and genotypes of Giardia duodenalis in 1-2 year old dairy cattle. Vet. Parasitol. 140:217-222. [DOI] [PubMed] [Google Scholar]
- 24.Trout, J., M. Santín, E. Greiner, and R. Fayer. 2005. Prevalence and genotypes of Giardia duodenalis in post-weaned dairy calves. Vet. Parasitol. 130:177-183. [DOI] [PubMed] [Google Scholar]
- 25.Webster, K., H. Smith, M. Giles, L. Dawson, and L. Roberston. 1996. Detection of Cryptosporidium parvum oocysts in faeces: comparison of conventional coproscopical methods and the polymerase chain reaction. Vet. Parasitol. 61:5-13. [DOI] [PubMed] [Google Scholar]
- 26.Winkworth, C., C. Matthaei, and C. Townsend. 2008. Prevalence of Giardia and Cryptosporidium spp. in calves from a region in New Zealand experiencing intensification of dairying. N. Z. Vet. J. 56:15-20. [DOI] [PubMed] [Google Scholar]
- 27.Winkworth, C., C. Matthaei, and C. Townsend. Recently planted vegetation strips reduce Giardia runoff reaching waterways. J. Environ. Qual., in press. [DOI] [PubMed]
- 28.Xiao, L. 1994. Giardia infection in farm animals. Parasitol. Today 10:436-438. [DOI] [PubMed] [Google Scholar]