Abstract
Bacterial communities associated with sediment particles were examined using PCR-denaturing gradient gel electrophoresis and 16S rRNA gene sequencing. Particle size influenced community structure, with attached bacterial assemblages separating into 63- to 125-, 125- to 1,000-, and 1,000- to 2,000-μm fractions. Differences were particularly pronounced for the Verrucomicrobia-Planctomycetes, whose numbers were significantly reduced on coarser particles.
Bacterial communities associated with particles in aquatic systems clearly differ from free-living communities (1, 2, 3, 4). However, few studies have examined differences in bacterial assemblages associated with different sizes of particles. DNA-DNA hybridizations have shown that microbial communities associated with different sizes of particulate organic matter are similar (18). The same approach suggested that there are differences in the communities associated with different sizes of detrital particles, with diversity potentially increasing as particle size decreased (21). A similar phenomenon has been reported for soils, where particle size may be more important in structuring bacterial communities than nutrient status (17). However, whether particle size influences bacterial community structure in aquatic systems is still unclear. To address this issue, we investigated the compositions of bacterial communities associated with different sizes of particles in stream bed sediments.
Sediment was collected on two dates (12 February 2007 and 21 March 2007) from Cypress Creek, a third-order stream with a sandy bottom in northern Mississippi. The sediment was wet sieved through a series of sieves (pore diameters, 2,000, 1,000, 500, 250, 125, and 63 μm) to separate particles into five fractions: 63 to 125, 125 to 250, 250 to 500, 500 to 1,000, and 1,000 to 2,000 μm. DNA was extracted from each fraction (0.5 g) using a bead-beating procedure (Power Soil DNA; Mo Bio, Carlsbad, CA), and 323 bp of the 16S rRNA gene was amplified for denaturing gradient gel electrophoresis (DGGE) analysis using primers Bac1070f and Univ1392GC (7). DGGE was performed using a 40 to 70% denaturing gradient, and community banding patterns were converted to binary data (presence or absence of bands). Binary data were analyzed using Gingko/VegAna software (Department of Vegetal Biology, University of Barcelona).
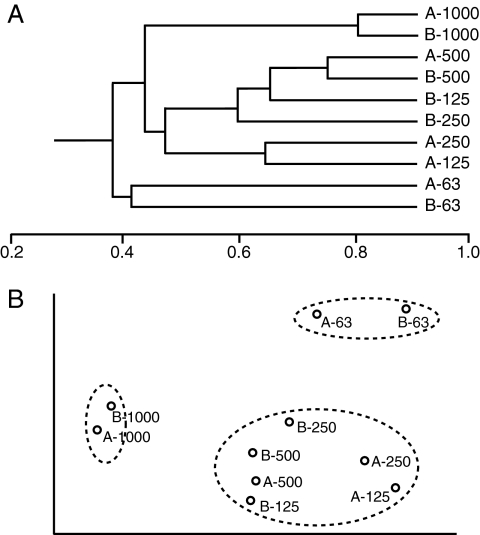
Bacterial community profiles for particles that were different sizes and for different sampling dates were compared using the Jaccard similarity index and cluster analysis based on the unweighted pair group method with average means (13). Communities were separated mostly by particle size, as opposed to sampling date, into three main groups: the communities on the 1,000- to 2,000-μm particles (which were very similar), the communities on the 63- to 125-μm particles, and the communities on the remaining intermediate-size particles (Fig. 1A). Nonmetric multidimensional scaling showed the same pattern, with clear separation between these three groups of particles (Fig. 1B).
FIG. 1.
Multivariate plots based on Jaccard similarities of bacterial communities on different sizes of stream sediment particles as determined from DGGE profiles. Samples were taken on two dates (indicated by the prefixes A [12 February 2007] and B [21 March 2007]) and from five size ranges of particles: 63 to 125 μm (63), 125 to 250 μm (125), 250 to 500 μm (250), 500 to 1,000 μm (500), and 1,000 to 2,000 μm (1000). (A) Cluster analysis based on the unweighted pair group method with average means. The scale indicates the Jaccard similarity scores. (B) Nonmetric multidimensional scaling plot (three dimensions, stress 0.05; the first two dimensions are shown) with circles indicating groups of samples.
Based on these patterns, we selected three samples (samples A-63, A-250, and A-1000) for more detailed analysis. A larger fragment of the 16S rRNA gene was amplified (with primers Bac8f and Univ1492 [7]), and clone libraries were generated for each sample. Approximately 600 bp of the insert was sequenced for the first 96 clones in each library. Sequences were aligned with the greengenes 16S rRNA gene database using NAST (5, 6), resulting in elimination of 12 to 17 erroneous sequences from each sample. A total of 244 aligned sequences were imported into ARB (12), and alignments were checked manually. Sequences were incorporated into an existing phylogenetic tree of 8,600 16S rRNA gene sequences using the “Quick add by parsimony” function, a procedure that should minimize distortions that can arise from analysis of short, divergent sequences (8).
Across all particles, 15 recognized lineages of Bacteria were identified, along with two candidate lineages. The majority of the phylotypes recovered from each clone library were affiliated with the Proteobacteria, Acidobacteria, Verrucomicrobia, and Bacteroidetes (Table 1), lineages that are typically well represented in 16S rRNA gene clone libraries generated from soils and freshwater sediments (2, 9, 14). Of these groups, the proportions of ribotypes affiliated with the Verrucomicrobia and, to a lesser extent, the Bacteroidetes were reduced on the larger particles (1,000 to 2,000 μm), whereas the proportion of Betaproteobacteria appeared to be elevated (Table 1).
TABLE 1.
16S rRNA gene sequences affiliated with different phylogenetic groups of Bacteria in clone libraries generated from DNA recovered from three different size ranges of stream bed sediment particles
| Phylogenetic group | No. of 16S rRNA clones identified for the following particle sizes:
|
||
|---|---|---|---|
| 63 to 125 μm | 250 to 500 μm | 1,000 to 2,000 μm | |
| Alphaproteobacteria | 8 | 2 | 8 |
| Betaproteobacteria | 14 | 13 | 25 |
| Gammaproteobacteria | 1 | 0 | 0 |
| Deltaproteobacteria | 7 | 3 | 8 |
| Bacteroidetes | 8 | 8 | 4 |
| Nitrospira | 0 | 3 | 3 |
| Verrucomicrobia | 15 | 18 | 2 |
| Planctomycetes | 2 | 6 | 1 |
| Acidobacteria | 22 | 16 | 16 |
| Actinobacteria | 0 | 3 | 2 |
| Chlorobi | 1 | 4 | 2 |
| Thermus-Deinococcus | 0 | 1 | 0 |
| Spirochetes | 1 | 0 | 0 |
| Cyanobacteria | 1 | 0 | 0 |
| Chloroflexi | 4 | 1 | 6 |
| Uncultured (TM7, OP11) | 0 | 1 | 4 |
| Total | 84 | 79 | 81 |
The phylogenetic tree was imported into the online UniFrac interface (10, 11) to specifically test for differences among the three clone libraries based on phylogenetic relationships. That is, did bacterial assemblages associated with different sizes of particles show evidence of different evolutionary adaptation to the particles? The pairwise UniFrac significance test probabilities for different communities were 1.00 (A-63 and A-250), 0.24 (A-63 and A-1000), and <0.03 (A-250 and A-1000), suggesting that there were significant differences between the two finer fractions and the coarsest particles. Lineage-specific analysis revealed that the differences were primarily due to the Verrucomicrobia-Planctomycetes, whose numbers were significantly reduced in the clone library generated from 1,000- to 2,000-μm particles (P = 0.028, G-test).
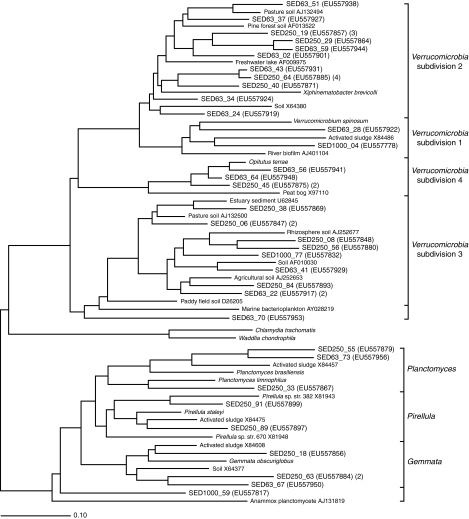
The Planctomycetes and Verrucomicrobia form a superphylum with the Chlamydiae (20). Planctomycetes-affiliated sequences recovered from 63- to 250-μm and 250- to 1,000-μm particles could be placed in subgroups related to Gemmata, Pirellula, and Planctomyces, whereas the sole sequence recovered from the coarser particles represented a more deeply branching lineage (Fig. 2), suggesting that differences between particles also occurred at finer phylogenetic scales. Almost all of the Verrucomicrobia sequences fell into subdivisions 1 to 4, lineages that are predominantly found in freshwater or soil (15). Subdivision 2 is a particularly clear example of particle size differences. This group accounted for 8 and 11% of the ribotypes identified for the A-63 and A-250 clone libraries, respectively, but was absent in the sample from the A-1000 library. The libraries from the two sizes of finer particles also shared similar ribotypes (e.g., SED250_29 and SED63_59, as well as SED250_64 and SED63_43), suggesting that similar populations were present (Fig. 2). Thus, the A-63 and A-250 libraries appeared to have quite similar structures, while the A-1000 library was significantly different.
FIG. 2.
Phylogenetic tree of representatives of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum found on 63- to 125-μm (SED63), 250- to 500-μm (SED250), and 1,000- to 2,000-μm (SED1000) particles in stream sediments. GenBank accession numbers are indicated in parentheses, as are the numbers of clones containing the ribotypes when the number was more than 1. Related sequences are shown for reference. Scale bar = 0.10 change per base pair.
Fluorescent in situ hybridization studies suggested that there were only small differences between bacterial assemblages on 125- to 800-μm particles and bacterial assemblages on 1,180- to 4,000-μm particles (16). However, these studies were limited to using established probes for the Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes. Our sequencing approach confirmed that these lineages do not show particle size differences at a broad phylogenetic level, but differences are apparent in the Verrucomicrobia-Planctomycetes lineage. The dominance of this lineage in 16S rRNA gene clone libraries on finer particles (where they accounted for 20 to 30% of the ribotypes detected) compared to their reduced presence (less than 4% of ribotypes) on coarser particles is similar to observations that we have made with wetland sediments (19) and suggests that the structures of these communities are different. Considering different lineages of bacteria is important in ecological studies since, as shown here, different phylogenetic groups are likely to have different distribution patterns. Furthermore, studies that survey soils or sediments without considering particle size may be missing an important component of spatial heterogeneity in bacterial community structure.
Nucleotide sequence accession numbers.
Sequences obtained in this study have been deposited in the GenBank database under accession numbers EU557775 to EU557969.
Acknowledgments
This work was supported in part by the Sally McDonnell Barksdale Honors College at The University of Mississippi.
Footnotes
Published ahead of print on 20 June 2008.
REFERENCES
- 1.Allgaier, M., and H.-P. Grossart. 2006. Seasonal dynamics and phylogenetic diversity of free-living and particle-associated bacterial communities in four lakes in northeastern Germany. Aquat. Microb. Ecol. 45:115-128. [Google Scholar]
- 2.Beier, S., K.-P. Witzel, and J. Marxsen. 2008. Bacterial community composition in central European running waters examined by temperature gradient gel electrophoresis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 74:188-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besemer, K., M. M. Moeseneder, J. M. Arrieta, G. J. Herndl, and P. Peduzzi. 2005. Complexity of bacterial communities in a river-floodplain system (Danube, Austria). Appl. Environ. Microbiol. 71:609-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLong, E. F., D. G. Franks, and A. L. Alldredge. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924-934. [Google Scholar]
- 5.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeSantis, T. Z., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson, C. R., H. W. Langner, J. Donahoe-Christiansen, W. P. Inskeep, and T. R. McDermott. 2001. Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ. Microbiol. 3:532-542. [DOI] [PubMed] [Google Scholar]
- 8.Jackson, E. F., H. L. Echlin, and C. R. Jackson. 2006. Changes in the phyllosphere community of the resurrection fern, Polypodium polypodioides, associated with rainfall and wetting. FEMS Microbiol. Ecol. 58:236-246. [DOI] [PubMed] [Google Scholar]
- 9.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozupone, C. A., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozupone, C. A., M. Hamady, and R. Knight. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyautey, E., C. R. Jackson, J. Cayrou, J.-L. Rols, and F. Garabétian. 2005. Bacterial community succession in natural river biofilm assemblages. Microb. Ecol. 50:589-601. [DOI] [PubMed] [Google Scholar]
- 14.Nold, S. C., and G. Zwart. 1998. Patterns and governing forces in aquatic microbial communities. Aquat. Ecol. 32:17-35. [Google Scholar]
- 15.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 16.Santmire, J. A., and L. G. Leff. 2007. The influence of stream sediment particle size on bacterial abundance and community composition. Aquat. Ecol. 41:153-160. [Google Scholar]
- 17.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fraction of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinsabaugh, R. L., T. Weiland, T., and A. E. Linkins. 1992. Enzymic and molecular analysis of microbial communities associated with lotic particulate organic matter. Freshw. Biol. 28:393-404. [Google Scholar]
- 19.Smart, K. A., H. L. Smart, and C. R. Jackson. 2008. The effects of fine scale environmental variation on microbial community structure and function in aquatic environments, p. 167-190. In G. V. Kurladze (ed.), Environmental microbiology research trends. Nova Science Publishers, Hauppauge, NY.
- 20.Wagner, M., and M. Horn. 2006. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17:241-249. [DOI] [PubMed] [Google Scholar]
- 21.Yeager, P. E., and R. L. Sinsabaugh. 1998. Microbial diversity along a sediment detrital particle size gradient. Aquat. Ecol. 32:281-289. [Google Scholar]