Abstract
Gene clusters for biosynthesis of the fungal polyketides hypothemycin and radicicol from Hypomyces subiculosus and Pochonia chlamydosporia, respectively, were sequenced. Both clusters encode a reducing polyketide synthase (PKS) and a nonreducing PKS like those in the zearalenone cluster of Gibberella zeae, plus enzymes with putative post-PKS functions. Introduction of an O-methyltransferase (OMT) knockout construct into H. subiculosus resulted in a strain with increased production of 4-O-desmethylhypothemycin, but because transformation of H. subiculosus was very difficult, we opted to characterize hypothemycin biosynthesis using heterologous gene expression. In vitro, the OMT could methylate various substrates lacking a 4-O-methyl group, and the flavin-dependent monooxygenase (FMO) could epoxidate substrates with a 1′,2′ double bond. The glutathione S-transferase catalyzed cis-trans isomerization of the 7′,8′ double bond of hypothemycin. Expression of both hypothemycin PKS genes (but neither gene alone) in yeast resulted in production of trans-7′,8′-dehydrozearalenol (DHZ). Adding expression of OMT, expression of FMO, and expression of cytochrome P450 to the strain resulted in methylation, 1′,2′-epoxidation, and hydroxylation of DHZ, respectively. The radicicol gene cluster encodes halogenase and cytochrome P450 homologues that are presumed to catalyze chlorination and epoxidation, respectively. Schemes for biosynthesis of hypothemycin and radicicol are proposed. The PKSs encoded by the two clusters described above and those encoded by the zearalenone cluster all synthesize different products, yet they have significant sequence identity. These PKSs may provide a useful system for probing the mechanisms of fungal PKS programming.
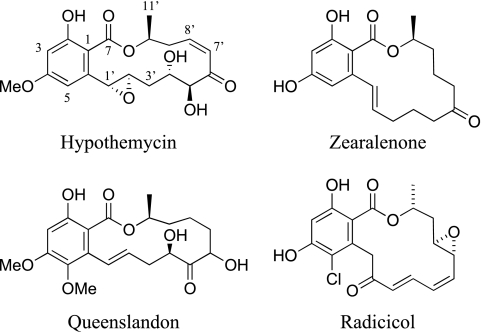
Hypothemycin and structurally related compounds are resorcylic acid lactones (RALs) that are produced by Hypomyces subiculosus and other fungi (1, 7, 14, 25, 26, 28, 43). The structures of some naturally occurring RALs are shown in Fig. 1. Hypothemycin irreversibly inhibits a subset of protein kinases with a conserved cysteine in the ATP binding site (e.g., Cys166 in human ERK2) (34, 43). Its pattern of activity makes it an attractive candidate for study as an anticancer agent. Radicicol (monorden), another RAL, is produced by Pochonia chlamydosporia and other fungi (11, 41), and it has a distinct activity: it inhibits the Hsp90 molecular chaperone, another important target for cancer chemotherapy (35, 36).
FIG. 1.
RALs. The unusual numbering scheme for these compounds is indicated for hypothemycin.
Fungal polyketides are synthesized by iterative multifunctional polyketide synthases (PKSs), which are classified by using phylogenetic and functional criteria as nonreducing PKSs (NR-PKSs), partially reducing PKSs (PR-PKSs), and reducing PKSs (R-PKSs) (6, 19). An NR-PKS and a PR-PKS produce aromatic products by cyclizing a polyketone intermediate. The 6-methylsalicylic acid synthase (3) is a PR-PKS, while the NR-PKSs include synthases for the precursor of aflatoxin (5) and for a spore pigment of Aspergillus nidulans (22). The R-PKSs have β-ketoreductase (KR), dehydratase, and (usually) enoyl reductase (ER) domains that are used after many of the iterations, leading to reduced products. Fungal PKSs can also have a variety of other catalytic domains, and cryptically encoded in their structure is a “program” for using (or skipping) specific catalytic activities after each condensation cycle (6, 16). Examples of R-PKSs include the two PKSs for lovastatin biosynthesis in Aspergillus terreus (12, 17) and one of the two PKSs for T-toxin biosynthesis in Cochliobolus heterostrophus (42). The mechanisms of fungal PKS programming are unknown and present a fascinating challenge to workers interested in engineering the production of novel polyketides.
Biosynthesis of zearalenone in Gibberella and Fusarium species is the best-studied RAL pathway. Early work showed that the carbon atoms are derived from acetate and that zearalenol is a precursor of zearalenone (4, 9, 10, 33). More recently, the zearalenone biosynthetic gene cluster was cloned and shown to encode an NR-PKS and an R-PKS, both of which are required for production of zearalenol, plus an alcohol oxidase (AOX) that converts zearalenol to zearalenone (8, 18, 21). There is strong evidence that the R-PKS transfers its reduced product to the NR-PKS, which then completes the synthesis (6). The structure of hypothemycin is similar to that of zearalenone, the synthesis differing by several apparent post-PKS reactions, including methylation of the C-4 hydroxyl, epoxidation of the 1′,2′ double bond, and hydroxylation at both C-4′ and C-5′. The origin of the cis-7′,8′ double bond of hypothemycin is less clear. The structure of radicicol differs in several ways from that of zearalenone, and studies of radicicol biosynthesis have not been reported.
Here we sequenced gene clusters from H. subiculosus and P. chlamydosporia that encode biosynthesis of hypothemycin and radicicol, respectively. Most of the post-PKS pathway for hypothemycin biosynthesis was elucidated using heterologous gene expression. We showed that both the R-PKS and the NR-PKS are required for synthesis of a product with a trans-7′,8′ double bond that is isomerized to the cis configuration in hypothemycin. Hypothetical pathways for biosynthesis of hypothemycin and radicicol are discussed in light of the data obtained.
MATERIALS AND METHODS
Strains and culture conditions.
H. subiculosus DSM 11931 and DSM 11932 were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH and were maintained on Difco potato dextrose agar (PDA). Media, growth conditions, and extraction procedures for analyzing hypothemycin congeners produced by these strains have been described previously (7, 40). P. chlamydosporia ATCC 16683 (previously Verticillium chlamydosporium var. catenulatum) was obtained from the American Type Culture Collection and was grown as previously described (38).
Escherichia coli strain DH5α was used for routine DNA manipulations, including propagation of cosmids, BL21(DE3) was used for expression of cDNAs, and EPI300 was used for propagation of fosmids in pCC1FOS (Epicentre Biotechnologies).
Compounds.
Hypothemycin, 4-O-desmethylhypothemycin, 7′,8′-dihydrohypothemycin, aigialomycin A, aigialomycin C, 4-O-desmethylaigialomycin A, 1′,2′-epoxyaigialomycin D, and 4-O-methyl-1′,2′-epoxyzearalenone were purified to >98% purity from culture broth of H. subiculosus strains as described previously (40). 1′,2′-Desepoxyhypothemycin was purified to >98% purity from a strain of Curvularia (39). Zearalenone, zearalenol, spinach ferredoxin, spinach ferredoxin reductase, and other reagents were purchased from Sigma-Aldrich, unless noted otherwise.
Genome library construction and screening.
Genomic DNA was isolated from fungal mycelia using a sodium dodecyl sulfate (SDS)-toluene extraction procedure (23, 37) with an additional equilibrium density centrifugation in CsCl with ethidium bromide in some cases. DNA was partially digested with Sau3AI, and fragments that were approximately 40 kb long were ligated into pAN26 (31) digested with BamHI and XbaI or into pCC1FOS (Epicentre) digested with BamHI. The ligation mixtures were packaged into λ phage using the GigaPak Gold packaging mixture (Stratagene) and transfected into E. coli DH5α or EPI300.
R-PKS ketoacyl synthase (KS) sequences were amplified from H. subiculosus DNA using the degenerate PCR primer pair described by Proctor et al. for isolation of the fumonisin PKS (30) and the primer pair described by Nicholson et al. from alignment of the T-toxin and lovastatin PKS sequences (27). KS sequences were amplified from P. chlamydosporia DNA using all four combinations of the KA degenerate primers described by Amnuaykanjanasin et al. for cloning PKSs from Xylaria sp. (2). The PCR conditions were optimized using a Failsafe PCR premix selection kit (Epicentre), and 700- to 800-bp products were gel isolated, cloned into pCR-Blunt (Invitrogen), and sequenced. Each unique KS sequence was used to probe cosmid (or fosmid) libraries by colony blot hybridization.
DNA sequencing and analysis.
Shotgun sequencing libraries were prepared from cosmids (or fosmids) either by mechanical shearing or by partial Sau3AI digestion, followed by ligation of 2- to 4-kb fragments into pCR-blunt (Invitrogen) cut with StuI or BamHI. DNA sequencing was performed with an ABI 3730 using BigDye chemistry (Applied Biosystems). Sequence assembly was done using Sequencher (GeneCodes), sequence analysis was done using MacVector (Accelerys), and BLAST analyses were done using the National Center for Biotechnology Information website.
Transformation of H. subiculosus.
The transformation procedure used was adapted from that developed for Fusarium verticilloides (30). Conidia were harvested from PDA plates after about 10 days of growth at 26°C and inoculated (2 × 106 conidia /ml) into YEPD broth (20 g of Bacto yeast extract per liter, 1 g of Bacto peptone per liter, 20 g of glucose per liter). After 8 h of incubation at 26°C, when the germ tubes were three to seven times longer than the conidia, the preparations were harvested and resuspended in 0.7 M NaCl (using one-fifth the culture volume) containing 10 mg/ml of lysing enzymes (L1412; Sigma-Aldrich). After 4 h of incubation at 30°C, undigested mycelium was removed by filtering with cotton wool, and the protoplasts were collected by centrifugation at 3,000 rpm for 10 min. The protoplasts were washed twice with 50 ml of 0.7 M NaCl and once with 10 ml of STC (1.4 M sorbitol, 10 mM Tris-HCl [pH 7.5], and 50 mM CaCl2, prepared fresh from stock solutions) and then diluted with STC to obtain 2 × 108 protoplasts/ml. An equal volume of a solution consisting of 8 parts of STC, 2 parts of 30% polyethylene glycol (PEG) (2.5 g of PEG 8000 [P5413; Sigma-Aldrich], 0.5 ml of 1 M CaCl2, 0.1 ml of 1 M Tris-HCl [pH 7.5], 7.5 ml of H2O), and 0.1 part of dimethyl sulfoxide was added. Aliquots (100 μl) of this protoplast suspension were stored at −80°C.
To transform protoplasts, the following were added in order to a 15-ml sterile conical tube with gentle mixing: 95 μl of STC, 5 μl of linearized plasmid DNA (about 5 to 10 μg), 100 μl of a protoplast solution, and 50 μl of 30% PEG. After 20 to 30 min of incubation at room temperature, 2 ml of 30% PEG was added with gentle mixing. After another 5 min of incubation at room temperature, 4 ml of STC was added, and protoplasts were plated by mixing 600-μl aliquots with 6.7 ml of molten regeneration medium (20 ml of PDA with 1.0 M sorbitol) and pouring the mixtures onto PDA. The transformation plates were incubated overnight at 26°C and overlaid with 2.5 ml of 1% agar containing phleomycin (final concentration, 25 μg/ml). The plates were incubated at 26°C, and colonies appearing after 5 to 10 days were streaked on PDA with 20 mg/ml of phleomycin (Cayla).
The plasmid used to disrupt the O-methyltransferase (OMT) gene was derived from one of the shotgun sequencing library clones by cutting it at a unique ScaI site near the center of the OMT gene and inserting a phleomycin resistance gene cassette consisting of an end-filled BglII-MluI fragment from pBC-phleo (Fungal Genetic Stock Center). The resulting plasmid (pKOS519-46B) was cut with SpeI and XbaI to release the insert from the pCRblunt vector and was transformed into DSM 11932 by the procedure described above. DNA isolated from phleomycin-resistant clones was checked by PCR using primers (CCTCGTCCTGTCACAACTAC and GGGTTCCGATTTAGTGCTTTAC) that amplified a 2.8-kb fragment from the phleomycin resistance cassette of pBC-phleo.
cDNA cloning.
RNA was isolated from H. subiculosus grown for 5 days in SCYS80 (80 g/liter sucrose, 10 g/liter soluble starch, 1 g/liter yeast extract, 1 g/liter corn gluten hydrolysate) using a Totally RNA kit (Ambion) with two LiCl precipitations. A First Choice 5′ RNA ligase-mediated random amplification of cDNA ends kit (Ambion) was used for reverse transcription (RT)-PCR with the gene-specific primers listed in Table 1. The 5′ and 3′ random amplification of cDNA ends primers were also used for direct sequencing of the untranslated regions. Gel-purified RT-PCR products treated with T4 polymerase and T4 polynucleotide kinase were cloned into the dephosphorylated EcoRV site of Litmus28 (New England Biolabs), and both orientations were obtained for most of the products. Full-length cDNA for each PKS was assembled from the sequence-verified cloned products shown in Table 1. The 2-kb NheI-EcoRI NR-PKS D fragment was isolated from pKOS518-82-B4A and ligated to the same sites of pKOS518-82-A6 carrying NR-PKS C to obtain pKOS518-92D. The 1.4-kb PstI-Acc65I NR-PKS B fragment was isolated from pKOS518-82-F4 and ligated to the PstI-BsiWI sites of pKOS518-82-E5B (NR-PKS A) to obtain pKOS518-104B. The 3.8-kb AseI-BsrGI fragment of pKOS518-104B was ligated to the 5-kb AseI-BsrGI fragment of pKOS518-92D to obtain pKOS518-117A carrying the assembled NR-PKS cDNA. The 2-kb HindIII-SpeI R-PKS D fragment from pKOS518-82-B2A was ligated between the HindIII-XbaI sites of pKOS518-82-C5 to obtain pKOS518-92E. The 2.2-kb EcoRI-Bsu36I R-PKS B fragment from pKOS518-96B was ligated between the same sites of pKOS518-92E to obtain pKOS518-104A. The 5.4-kb MluI-BssHII insert of pKOS518-104A was cloned back into the BssHII site of Litmus28 to obtain pKOS518-114C. The 5.3-kb SanDI-BamHI fragment of pKOS518-114C, the 1.9-kb NdeI-SanDI R-PKS A fragment of pKOS518-82-G4B, and pET16B (Novagen) cut with NdeI and BamHI were ligated to obtain pKOS518-115A carrying the assembled R-PKS cDNA in pET16B.
TABLE 1.
Primers used for RT-PCR cloning of hypothemycin cDNAs
| Primer | Sequence | Product (size of product [kb]) |
|---|---|---|
| P450-F(NdeI) | CCATATGGCAGTGTTACCCCAAG | P450 (1.5) |
| P450-B | TAGATGGCAGTCGAGACTACC | |
| GST-F(NdeI) | GCATATGGTTATCAAGCTGTACGGCTC | GST (0.7) |
| GST-B | AAACTTCATGTACAATCTTTCAGCTTC | |
| NR-PKS-F(NdeI) | CTCATATGGTGACTGTACCACAGAC | NR-PKS A (1.7) |
| NR-PKS-B1704 | CCAGGAGTGAGGATACCCG | |
| NR-PKS-F1239 | CGACATCAATGAGTTCTACTGTACC | NR-PKS B (1.6) |
| NR-PKS-B2848 | AGCGTGACAACAGCGAGCTG | |
| NR-PKS-F2691 | CGGGACAGGGCTCTCACTAC | NR-PKS C (1.8) |
| NR-PKS-B4441 | CGCTCTCACCCCAGTAAGG | |
| NR-PKS-F4239 | ATACTTCGTCAAGGCTCGTATG | NR-PKS D (2.0) |
| NR-PKS-B6238 | CAAATCGTCTCCCAAAAAGATACTC | |
| OMT-F(NdeI) | CCATATGAGTGCATCCAATGGCGAG | OMT (1.2) |
| OMT-B | GGTACTCCCGTCATCGTTGGG | |
| FMO-F(NcoI) | CCATGGCGAGAGCAATACTGCCAGAC | FMO (1.9) |
| FMO-B1897 | CGAGGGCTTCAAGGATAGG | |
| R-PKS-F(NdeI) | CCATATGCCTTCTACCAGCAATCC | R-PKS A (1.9) |
| R-PKS-B1777 | CTTGGAAAGCTCAACGATAGG | |
| R-PKS-F1782 | GCTGTGAATGGGACCCTATC | R-PKS B (2.8) |
| R-PKS-B4840 | TGCTCAAGCTCCAACAGAGAAATG | |
| R-PKS-F3743 | TTCCGAGCTTGATATGGATTC | R-PKS C (1.7) |
| R-PKS-B5448 | CACGGGAGTTGAAGATGTGC | |
| R-PKS-F5154 | CGTGTTTGCACCATGAACGTC | R-PKS D (2.1) |
| R-PKS-B7523 | CCCTCCGTAAACGCCTTAAACAG |
Expression of cDNA in E. coli and characterization of enzymes in vitro.
cDNA for the flavin monooxygenase (FMO) was cloned as an NcoI-HindIII fragment in pET30B to obtain pKOS518-92A. cDNAs for the cytochrome P450 and OMT were cloned as NdeI-BamHI fragments in pET16B to obtain pKOS518-92B and pKOS518-92C, respectively. The glutathione S-transferase (GST) cDNA was cloned as an NdeI-BamHI fragment in pET30B to obtain pKOS518-118B. Expression in E. coli BL21(DE3) was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and cultures were incubated overnight at 16°C. The cells were harvested, washed, and lysed by passage through a French pressure cell in a buffer containing 50 mM Tris-HCl (pH 7.8), 500 mM NaCl, 5% glycerol, and a cocktail of freshly added protease inhibitors (Roche). Centrifugation at 50,000 × g resulted in a crude lysate. His-tagged proteins were purified using an AKTA Explorer (GE Healthcare) programmed to bind and elute from a HiTrap 5-ml chelating HP column, desalt on a HiPrep 26/10 column, and then bind to and elute from a MonoQ 5/50 GL column with a salt gradient. Fractions (0.5 ml) from the MonoQ column having significant absorbance at 280 nm were analyzed by SDS-polyacrylamide gel electrophoresis (Novex NuPAGE; Invitrogen) with Coomassie blue staining. Fractions having the highest purity of the correct protein band were pooled, and the protein concentration was determined using a Bradford dye-binding assay (Pierce Thermo). After 20% glycerol was added, the purified enzymes were stored as aliquots at −80°C. The GST was purified by loading a GSTrap FF 1-ml column (GE Healthcare) with a syringe, washing it with 25 mM HEPES (pH 7.4), 20% glycerol, and eluting with the same buffer containing 10 mM reduced glutathione.
Enzymes were assayed using 200-μl reaction mixtures containing 25 mM HEPES (pH 7.4), 10 mM MgSO4, 0.1% bovine serum albumin, a RAL substrate at a concentration of 50 μM, and the appropriate cosubstrate (400 μM S-adenosylmethionine for OMT; 400 μM NADPH for FMO; 400 μM NADPH, 10 μM spinach ferredoxin, and 0.03 U spinach ferredoxin reductase for the hypothemycin P450). The reactions were initiated by adding 30 μg/ml enzyme, the mixtures were incubated at 30°C, and the reactions were stopped by extraction with an equal volume of ethyl acetate. After the ethyl acetate was evaporated, the reaction products were dissolved in 100 μl of 80% acetonitrile and analyzed with an Agilent 1100 high-performance liquid chromatography (HPLC) system with a diode array detector. The method was isocratic (Inertsil ODS-3 column; 5 μm; 2.1 by 150 mm; 70% acetonitrile-0.1% acetic acid at a rate of 1 ml/min for 15 min). Compounds were identified on the basis of their retention times (compared with purified standards when available) and UV spectra (which distinguishes compounds with a 1′,2′ double bond or an epoxide). Reactions were also analyzed by liquid chromatography-mass spectrometry (LC-MS) (see below).
Expression of cDNAs in Saccharomyces cerevisiae.
Hypothemycin cDNAs were expressed from the adh2 promoter in 2μ plasmid-based yeast-E. coli shuttle vectors (24) in an S. cerevisiae strain developed for fungal PKS expression (15). The R-PKS cDNA was cloned into pKOS187-98A (LEU2 marker) as an NdeI-SbfI fragment to obtain pKOS518-118A. The NR-PKS was cloned into pKOS247-14-3 (TRP1 marker) cut with NdeI and EcoRI in a three-piece ligation with the 0.75-kb NdeI-AlwNI and 5.4-kb AlwNI-EcoRI fragments from pKOS518-117A to obtain pKOS518-120A. The P450 and FMO cDNAs were cloned into pKOS187-98A as NdeI-AvrII fragments to obtain pKOS518-116 and pKOS518-137A, respectively. The OMT cDNA was cloned as a 1.6-kb NdeI-EcoRI fragment from pKOS518-92C into pKOS247-14-3 to obtain pKOS518-136A. Each cDNA expression cassette flanked by the ADH2 promoter and terminator was moved into pKOS187-4e (URA3 marker) (24) using the BssHII sites to obtain pKOS518-137B (P450), pKOS518-138A (FMO), and pKOS518-138D (OMT).
Plasmids were introduced into yeast by the LiCl-PEG procedure with plating on the appropriate complete minimal dropout agar plates (Teknova). Colonies were transferred to the appropriate liquid complete minimal dropout medium and grown at 30°C to generate inoculum for YPD medium containing 2% glucose and 20 g/liter of Amberlite XAD-1180 resin (Alfa Aesar). After 24 h of incubation at 30°C, the resin was collected and washed with water, and the products were eluted with methanol. The methanol was evaporated, and the residue was dissolved in 80% acetonitrile for HPLC analysis by the method described above for the in vitro enzyme reactions.
Additional analytical methods.
LC-MS analyses were performed with an Agilent 1100 system with an Inertsil ODS-3 column (5 μm; 2.1 by 150 mm), a diode array detector, and a Perseptive Biosystems Mariner mass spectrometer equipped with a turbo-ion spray source in negative-ion mode. The HPLC conditions included a linear 35 to 100% acetonitrile gradient (0.1% acetic acid) at a rate of 0.25 ml/min. 7′,8′-Dehydrozearalenol (DHZ) was purified by chromatography on a column (2.5 by 30 cm) of BakerBond C18 sorbent (40 μm). For column equilibration and sample loading 50% methanol-0.1% acetic acid was used. After the solution was changed to 80% methanol-0.1% acetic acid, 20-ml fractions were collected, and the desired fractions were identified by the isocratic HPLC method used for enzyme assays. Acetic acid was removed by C18 chromatography with 50% methanol, elution with 100% methanol, and rotary evaporation. High-resolution mass spectrometry was performed with an Applied Biosystems Mariner time of flight mass spectrometer configured with a turbo-ion spray source in negative-ion mode. Proton nuclear magnetic resonance (NMR) (400 MHz) and 13C NMR (100 MHz) were recorded in d6-acetone at 300 K with a Bruker DRX 400 spectrometer.
Nucleotide sequence accession numbers.
The DNA sequence data have been deposited in the GenBank database under accession numbers EU520417 (H. subiculosus DSM 11931 hpm cluster), EU520418 (H. subiculosus DSM 11932 hpm cluster), and EU520419 (P. chlamydosporia ATCC 16683 rdc cluster).
RESULTS
Cloning and analysis of the gene clusters.
The degenerate PCR primer pair previously used to clone the fumonisin PKS gene (30) amplified four distinct KS sequences from cDNA of H. subiculosus DSM 11932, and the relative clone frequencies were 10, 6, 74, and 10% for KS1, KS2, KS3, and KS4, respectively. Another pair of degenerate primers identical to the primers described previously (27) amplified KS1 and KS3 from cDNA (clone frequencies, 11 and 89%) and KS2 and KS3 from genomic DNA (clone frequencies, 55 and 45%), but not the KS4 sequence. Cosmids (or fosmids) for each of the four KS sequences were isolated by colony blot hybridization, and one of each type was fully sequenced. Features of the cosmids carrying the KS1, KS2, and KS3 sequences suggested that they were unlikely to represent the hypothemycin gene cluster. Briefly, the R-PKS with the KS1 sequence contained nonribosomal peptide synthetase and reductive chain release domains, the cosmid encoding the KS2 sequence encoded no oxidoreductase or OMT, and the R-PKS with the KS3 sequence contained a C-methyltransferase domain.
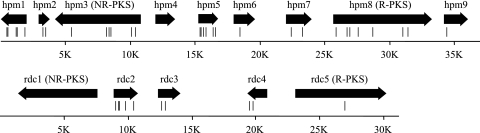
The KS4 cosmid encoded an R-PKS sharing 66% sequence identity with the R-PKS from the zearalenone cluster, an FMO, and a major facilitator superfamily transporter, encouraging isolation and sequencing of overlapping cosmids. This revealed genes for an NR-PKS and an AOX, both of which shared significant sequence identity with corresponding genes in the zearalenone cluster. There were also genes for an OMT, a cytochrome P450, and a GST. Gel-isolated and cloned RT-PCR products were sequenced, which identified transcription start sites and polyadenylation sites and verified the intron boundaries predicted from typical fungal 5′, 3′, and lariat splicing motifs (20). The gene cluster in H. subiculosus strain DSM 11931 was sequenced using PCR products amplified with primers designed by using the DSM 11932 sequence. The two clusters differed only by single nucleotide substitutions (most of which were silent) at a frequency of about 0.1%. The organization of the hypothemycin gene cluster is shown in Fig. 2, and the most significant P-BLAST hit for each gene product is shown in Table 2. Each gene product had many statistically significant P-BLAST hits to proteins with the putative or proven functions shown in Table 2.
FIG. 2.
Organization of the hypothemycin and radicicol gene clusters. The vertical lines beneath genes indicate the positions of introns.
TABLE 2.
Sequence similarities and predicted functions of genes in the clusters
| Gene | Best P-BLAST hit | % Similarity | Catalytic function |
|---|---|---|---|
| hpm1 | Trichothecene C-15 hydroxylase of Fusarium and Gibberella species | 61 | P450 hydroxylase |
| hpm2 | EAU37834 (Aspergillus terreus) | 81 | GST |
| hpm3 | XM_382571 (G. zeae PKS13) | 76 | Nonreducing PKS |
| hpm4 | None | Unknown | |
| hpm5 | XP_001550242 (Botryotinia fuckeliana) | 61 | O-methyltransferase |
| hpm6 | EAS29817 (Coccidioides immitis) | 66 | MFS transporter |
| hpm7 | EDO00156 (Sclerotinia sclerotiorum) | 66 | FMO epoxidase |
| hpm8 | XM_382572 (G. zeae PKS4) | 80 | Reducing PKS |
| hpm9 | XP_381981 (G. zeae) | 78 | AOX |
| rdc1 | XM_382571 (G. zeae PKS13) | 69 | Nonreducing PKS |
| rdc2 | XP_001225297 (Chaetomium globosum) | 62 | Halogenase |
| rdc3 | XP_960542 (Neurospora crassa) | 66 | MFS transporter |
| rdc4 | Benzoate 4-monooxygenase (Neosartorya fischeri) | 57 | P450 epoxidase |
| rdc5 | XM_382572 (G. zeae PKS4) | 73 | Reducing PKS |
The set of degenerate primers used to amplify the KS-acyl transferase region of R-PKSs (2) also amplified sequences from P. chlamydosporia genomic DNA, one of which was very similar to the R-PKS sequences in the zearalenone and hypothemycin clusters. Probing a P. chlamydosporia genomic DNA library with this PCR product resulted in two strongly hybridizing fosmids, which were found to overlap. Sequencing of these fosmids revealed a gene cluster that appeared likely to encode radicicol biosynthesis. The organization of this gene cluster is shown in Fig. 2, and the most significant P-BLAST hits for the gene products are shown in Table 2.
Targeted gene disruption of the OMT gene in H. subiculosus.
Transformation of H. subiculosus was quite difficult. The best procedure tried (see Materials and Methods) gave, on average, one colony per transformation for strain DSM 11932, while for strain DSM 11931 it gave no colonies. Six colonies were obtained using the linearized OMT gene disruption cassette, and PCR analysis showed that the phleomycin resistance cassette had integrated into the genomic DNA of four of them. LC-MS analysis showed that in these four colonies the production of hypothemycin was reduced and the production of 4-O-desmethylhypothemycin was increased compared to the wild-type strain. The transformant that produced the most 4-O-desmethylhypothemycin was reisolated from spores, and the reisolated strain produced ∼65 mg/liter of 4-O-desmethylhypothemycin, ∼45 mg/liter of 7′,8′-dihydrohypothemycin, and <1 mg/liter of hypothemycin when it was grown as described by Dombrowski et al. (7). By comparison, the wild-type strain grown under the same conditions produced 800 mg/liter of hypothemycin, 180 mg/liter of 7′,8′-dihydrohypothemycin, and 10 mg/liter of 4-O-desmethylhypothemycin (40). The significant shift to 4-O-desmethylhypothemycin production by this transformant was good evidence that we had cloned the gene cluster encoding hypothemycin biosynthesis. However, the difficulty with transformation led us to opt for heterologous gene expression to elucidate the biosynthetic pathway.
Activities of hypothemycin enzymes in vitro.
cDNAs expressed in E. coli gave inducible protein bands of the predicted size when SDS-polyacrylamide gel electrophoresis was performed, and soluble protein was obtained at 16°C. Enzymes were purified as described in Materials and Methods. Purified FMO was yellow, which was indicative of a tightly bound flavin cofactor.
In the presence of S-adenosylmethionine, the OMT catalyzed essentially complete conversion of available substrates lacking a 4-O-methyl group (4-O-desmethylhypothemycin, epoxyaigialomycin D, or zearalenone) to the corresponding methylated products (Table 3). All RAL compounds already having a 4-O-methyl group were unchanged when they were incubated with the OMT. In the presence of NADPH, the FMO efficiently converted zearalenone or desepoxyhypothemycin to the corresponding 1′,2′-epoxy product (Table 3). Thus, the OMT and FMO have broad substrate tolerance. We were unable to observe an in vitro reaction with the purified hypothemycin P450 in the presence of NADPH, spinach ferredoxin, spinach ferredoxin reductase, and either zearalenone or 4-O-methyl-1′,2′-epoxyzearalenone.
TABLE 3.
Conversion of RAL substrates by OMT or FMO in vitroa
| Enzyme | Substrate | Substrate peak retention time (min) | Retention time(s) of postreaction peak(s) (min) (area) | % Conversion |
|---|---|---|---|---|
| OMT | 4-O-Desmethylhypothemycin | 3.50 | 5.26 (1,212) | 100 |
| Zearalenone | 8.14 | 11.4 (310) | 100 | |
| 1′,2′-Epoxyaigialomycin D | 3.44 | 3.44 (31), 5.49 (1,354) | 98 | |
| FMO | Desepoxyhypothemycin | 5.95 | 5.26 (446), 5.95 (74) | 86 |
| Zearalenone | 8.14 | 7.02 (65), 8.14 (30) | 68 |
Peaks having the characteristic RAL UV absorbance profile are shown along with their retention times and peak areas at 267 nm. Reactions were performed as described in Materials and Methods, and the reaction mixtures were incubated for 60 min.
Adding purified GST to 100 μM hypothemycin or 100 μM aigialomycin A (the 7′,8′ trans isomer of hypothemycin) resulted in rapid equilibration to a mixture of 85% aigialomycin A and 15% hypothemycin, and this ratio remained the same for 4 h. Thus, the GST catalyzes cis-trans isomerization of the 7′,8′ double bond. Since the GST was purified by binding to glutathione affinity resin and elution with 10 mM glutathione, a low concentration of glutathione was also present in this in vitro reaction mixture.
Expression of hypothemycin cDNAs in yeast.
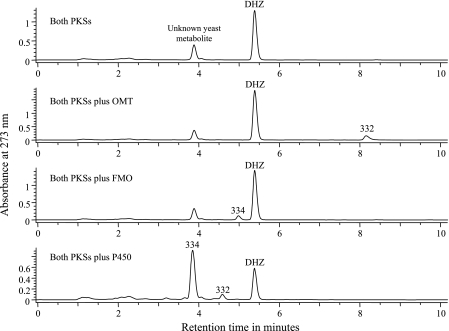
The hypothemycin PKS cDNAs on expression vectors with different auxotrophic markers (LEU2 and TRP1) were introduced into yeast. Eight clones able to grow without leucine and tryptophan were analyzed as described in Materials and Methods. Seven of these clones produced about 20 mg/liter of a compound (Fig. 3) with the mass and UV spectrum of zearalenone but a retention time close to that of β-zearalenol, suggesting that the compound was zearalenol with a second double bond. Upon purification to >98% purity, high-resolution mass spectrometry indicated that the mass was 317.13988 (M-H m/z), consistent with the molecular formula C18H22O5. NMR analysis (Table 4) indicated that the compound was DHZ. HMBC and TOCSY experiments verified that a trans double bond was in the 7′,8′ position. Yeast strains expressing either of the two PKSs alone gave chromatograms like those obtained for untransformed yeast, indicating that both PKSs are required for production of DHZ.
FIG. 3.
Production of compounds by yeast strains expressing combinations of hypothemycin genes. The numbers above peaks indicate the masses of the metabolites as determined by LC-MS. The peak at 334 amu (DHZ + 16 amu) eluted very close to the unknown yeast metabolite but had the signature UV spectrum of a RAL compound. Each chromatogram represents at least five nearly identical traces from independent clones after transformation with the constructs indicated.
TABLE 4.
NMR assignments for DHZ
| Position | δ 13C | δ 1H |
|---|---|---|
| C-1 | 104.9 | |
| C-2 | 162.9 | |
| C-3 | 102.4 | 6.27 (1H, d, J = 2.4 Hz) |
| C-4 | 165.3 | |
| C-5 | 107.8 | 6.49 (1H, d, J = 2.4 Hz) |
| C-6 | 144.2 | |
| C-7 | 172.0 | |
| C-1′ | 131.4 | 7.12 (1H, d, J = 15.6 Hz) |
| C-2′ | 133.4 | 5.97 (1H, ddd, J = 5.2, 6.4, 15.6 Hz) |
| C-3′ | 31.4 | 2.30 (Ha, m) |
| 2.18 (Hb, m) | ||
| C-4′ | 21.8 | 1.85 (Ha, m) |
| 1.55 (Hb, m) | ||
| C-5′ | 34.4 | 1.70 (Ha, m) |
| 1.55 (Hb, m) | ||
| C-6′ | 71.6 | 4.31 (1H, m) |
| C-7′ | 138.5 | 5.65-5.75 (2H, m) |
| C-8′ | 124.8 | 5.65-5.75 (2H, m) |
| C-9′ | 38.1 | 2.59 (Ha, ddd, J = 3.8, 5.0, 15.0 Hz) |
| 2.40 (Hb, dt, J = 15.0, 6.3 Hz) | ||
| C-10′ | 73.0 | 5.37 (1H, m) |
| C-11′ | 19.4 | 1.39 (3H, d, J = 6.4 Hz) |
A yeast strain expressing both hypothemycin PKSs was transformed with a third 2μ plasmid carrying a ura marker and expressing cDNA for either the OMT, FMO, or P450. The resulting strains were analyzed by LC-MS, and in each case a significant fraction of the DHZ was converted into a new compound with a retention time and mass consistent with methylation, epoxidation, and hydroxylation, respectively (Fig. 3). Three UV absorbance maxima at 219, 265, and 304 nm for the epoxy-DHZ peak (compared with maxima at 238, 272, and 311 nm for DHZ) indicated that there was epoxidation at the 1′,2′ position. The position of the hydroxyl group in the hydroxy-DHZ was not determined but was probably C-5′ because no congener that is hydroxylated at C-4′ and not at C-5′ has been reported, while several congeners are hydroxylated at C-5′ but not at C-4′.
DISCUSSION
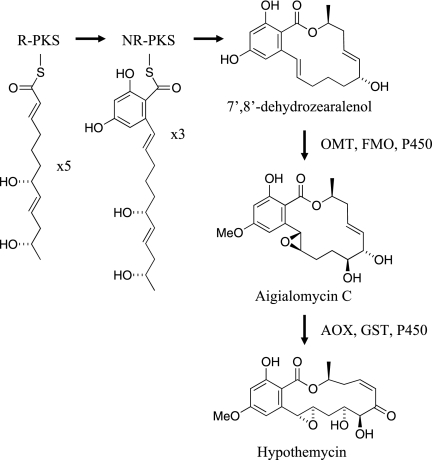
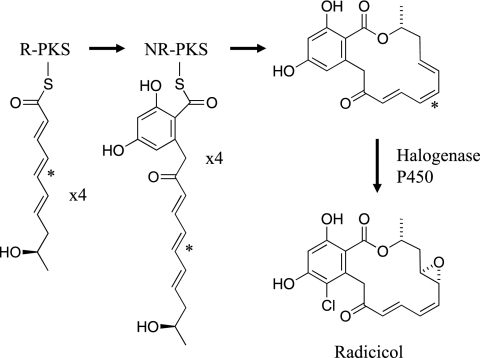
The data presented here strongly support the scheme for hypothemycin biosynthesis shown in Fig. 4 up to aigialomycin C. Both PKSs are required for DHZ biosynthesis, as was shown previously for zearalenol biosynthesis in Gibberella zeae (8, 18) The R-PKS apparently synthesizes the C-2′ to C-11′ portion of DHZ in five iterations and then transfers it to the NR-PKS, which after three iterations synthesizes the aromatic ring and releases DHZ with formation of the 14-member macrolactone ring. The 7′,8′ double bond of hypothemycin is generated by the R-PKS. Although the stereochemistry of the C-6′ hydroxyl of DHZ was not determined, it is very likely that established for β-zearalenol from G. zeae (8, 18) and queenslandon from Chrysosporium queenslandicum (13). The stereochemistry of the C-2′ and C-6′ hydroxyls is expected to be determined by the KR domain in the R-PKS. The wide substrate tolerance of the OMT and FMO in vitro implies that the reactions from DHZ to aigialomycin C can occur in any order. The yeast strain expressing both PKSs plus the P450 gave peaks with masses of 334 and 332 atomic mass units (amu) (Fig. 3); the first peak was consistent with hydroxylation of DHZ, and the second peak was consistent with additional loss of two hydrogen atoms. The retention time of the 332-amu peak suggests that one of the hydroxyl groups was oxidized to a ketone. The yeast strain has a native activity able to interconvert zearalenol and zearalenone (unpublished data), which probably accounts for the 332-amu compound.
FIG. 4.
Proposed scheme for hypothemycin biosynthesis. The reducing PKS passes its product to the nonreducing PKS, which finishes synthesis of DHZ. The post-PKS reactions that convert DHZ into aigialomycin C can probably occur in any order. See the text for details.
The steps from aigialomycin C to hypothemycin were less well established by this work and require further study. The AOX encoded in the hypothemycin gene cluster (Hpm9) has 59% sequence identity with the AOX encoded in the zearalenone cluster and presumably catalyzes oxidation of the C-6′ hydroxyl to a ketone (18). The timing of this oxidation is important, since the resulting enone functional group is a Michael acceptor that can react spontaneously with glutathione, an abundant metabolite in fungal cells (29). Hypothemycin or aigialomycin A incubated with 10 mM glutathione gave the thiol adduct with 50% conversion in about 8 min (unpublished data). As shown here, the GST catalyzes cis-trans isomerization of the 7′,8′ double bond with equilibrium favoring the trans isomer. How then is the equilibrium pulled toward the higher-energy cis isomer, such that hypothemycin is the major product? One possible answer is that the hpm6-encoded transporter preferentially pumps hypothemycin out of the cell relative to the trans isomer aigialomycin A. The cis-to-trans isomerization may be coupled with C-4′ hydroxylation, since all known hypothemycin analogues containing the enone functional group also have hydroxyl groups at both C-4′ and C-5′.
Some congeners with a fully saturated 7′,8′ bond are produced by H. subiculosus (40) and Aigialus parvus (14). These congeners could arise if the R-PKS sometimes catalyzes an enoyl reduction at the second iteration. However, only DHZ (and no zearalenol) was observed in culture broth of yeast expressing both hypothemycin PKS genes, suggesting that saturation of this double bond is due to an enzyme expressed in H. subiculosus but not in yeast.
Figure 5 shows a possible scheme for radicicol biosynthesis based only on the sequence of the putative radicicol gene cluster from P. chlamydosporia. The cluster encodes only two apparent post-PKS enzymes, a cytochrome P450 and a nonheme halogenase that could introduce the epoxide and the chlorine, respectively. If this cluster includes all the genes required for radicicol biosynthesis, the remaining structural features of radicicol are presumably generated by the PKS. The C-2′ ketone could arise if the R-PKS and NR-PKS each carry out four iterations, in contrast to the five iteration-three iteration split for the hypothemycin PKSs. The origin of the cis 5′,6′ double bond (indicated by an asterisk in Fig. 5) is not known. The minimal-energy conformation of the all-trans isomer was found to be 21.4 kcal mol−1 less stable than the cis-5′,6′ isomer, as estimated by quantum-chemical AM1 calculations in water solvent. This suggests that a 5′,6′ double bond in the PKS product is unlikely to have the trans configuration. The ER domain in the radicicol R-PKS has much greater sequence identity with the other two R-PKS ER domains than would be expected if it had no catalytic role. Thus, the radicicol R-PKS ER domain may catalyze either double bond isomerization or reduction in the third iteration. Radicicol with a saturated 5′,6′ bond is observed in P. chlamydosporia culture broth.
FIG. 5.
Possible scheme for radicicol biosynthesis. See the text for details, including the isomerization of the double bond indicated by an asterisk.
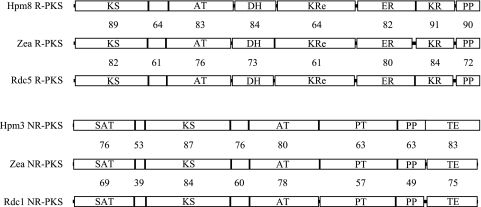
The R-PKS encoded in the hypothemycin and zearalenone gene clusters differs subtly in programming by either using the ER domain in the second iteration to reduce the 7′,8′ double bond or not using it. Radicicol biosynthesis seems to require significantly different PKS programming. Following the first condensation, the hydroxyl generated by the KR domain of the radicicol R-PKS (which is subsequently incorporated into the lactone) has stereochemistry opposite that of the corresponding hydroxyls of zearalenone and hypothemycin. A comparison of KR domain sequences from the RAL R-PKSs with KR domains from modular PKSs suggests that the RAL KR domains may be able to reduce from either side of the ketone to generate either hydroxyl stereochemistry (32). Another potential programming difference between the radicicol PKS and the other two characterized RAL PKSs is the apparent transfer from the R-PKS to the NR-PKS one cycle earlier. There is also the possible programming involved in generating the cis double bond, as discussed above. Given these programming differences, the level of end-to-end sequence identity between these RAL PKSs (Fig. 6) is remarkable.
FIG. 6.
Comparison of the PKSs of the hypothemycin, radicicol, and zearalenone clusters. The numbers between the PKSs are the percentages of identity plus similarity for the regions. Abbreviations for domains: KS, ketoacyl synthase; AT, acyl transferase; DH, dehydratase; ER, enoyl reductase; KR, ketoreductase; PP, phosphopantetheine region; KRe, extra ketoreductase (which also has an extra dehydratase sequence and has been called the “core”); SAT, starter unit acyl transferase; PT, product template; TE, thioesterase.
These fascinating biosynthetic pathways are clearly worthy of further study, as there appear to be several unprecedented enzymological features. The similarity between the PKSs in the clusters combined with the successful functional expression of the hypothemycin PKS genes in yeast indicates that this may be a tractable system for identifying structural features that control mechanisms of fungal PKS programming, which have been elusive thus far.
Acknowledgments
We thank John Carney for performing LC-MS and NMR analyses, Jinghua Tan for assisting with DNA sequencing and HPLC analyses, Giulio Rastelli for the molecular dynamics calculations, and Gary Ashley and Jonathan Kennedy for their intellectual input.
Footnotes
Published ahead of print on 20 June 2008.
REFERENCES
- 1.Agatsuma, T., A. Takahashi, C. Kabuto, and S. Nozoe. 1993. Revised structure and stereochemistry of hypothemycin. Chem. Pharm. Bull. 41:373-375. [Google Scholar]
- 2.Amnuaykanjanasin, A., J. Punya, P. Paungmoung, A. Rungrod, A. Tachaleat, S. Pongpattanakitshote, S. Cheevadhanarak, and M. Tanticharoen. 2005. Diversity of type I polyketide synthase genes in the wood-decay fungus Xylaria sp. BCC 1067. FEMS Microbiol. Lett. 251:125-136. [DOI] [PubMed] [Google Scholar]
- 3.Beck, J., S. Ripka, A. Siegner, E. Schiltz, and E. Schweizer. 1990. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur. J. Biochem. 192:487-498. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell, B. A., J. D. Miller, and R. Greenhalgh. 1985. 13C NMR study of the biosynthesis of toxins by Fusarium graminearum. J. Biol. Chem. 260:4243-4247. [PubMed] [Google Scholar]
- 5.Chang, P. K., J. W. Cary, J. Yu, D. Bhatnagar, and T. E. Cleveland. 1995. The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B1 biosynthesis. Mol. Gen. Genet. 248:270-277. [DOI] [PubMed] [Google Scholar]
- 6.Cox, R. J. 2007. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem. 5:2010-2026. [DOI] [PubMed] [Google Scholar]
- 7.Dombrowski, A., R. Jenkins, S. Raghoobar, G. Bills, J. Polishook, F. Pelaez, B. Burgess, A. Zhao, L. Huang, Y. Zhang, and M. Goetz. 1999. Production of a family of kinase-inhibiting lactones from fungal fermentations. J. Antibiot. (Tokyo) 52:1077-1085. [DOI] [PubMed] [Google Scholar]
- 8.Gaffoor, I., and F. Trail. 2006. Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl. Environ. Microbiol. 72:1793-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenhalgh, R., G. A. Neish, and J. D. Miller. 1983. Deoxynivalenol, acetyl deoxynivalenol, and zearalenone formation by Canadian isolates of Fusarium graminearum on solid substrates. Appl. Environ. Microbiol. 46:625-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagler, W. M., and C. J. Mirocha. 1980. Biosynthesis of [14C]zearalenone from [1-14C]acetate by Fusarium roseum ‘Gibbosum.’ Appl. Environ. Microbiol. 39:668-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellwig, V., A. Mayer-Bartschmid, H. Muller, G. Greif, G. Kleymann, W. Zitzmann, H. V. Tichy, and M. Stadler. 2003. Pochonins A-F, new antiviral and antiparasitic resorcylic acid lactones from Pochonia chlamydosporia var. catenulata. J. Nat. Prod. 66:829-837. [DOI] [PubMed] [Google Scholar]
- 12.Hendrickson, L., C. R. Davis, C. Roach, D. K. Nguyen, T. Aldrich, P. C. McAda, and C. D. Reeves. 1999. Lovastatin biosynthesis in Aspergillus terreus: characterization of blocked mutants, enzyme activities and a multifunctional polyketide synthase gene. Chem. Biol. 6:429-439. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino, Y., V. B. Ivanova, K. Yazawa, A. Ando, Y. Mikami, S. M. Zaki, A. Z. Karam, Y. A. Youssef, and U. Grafe. 2002. Queenslandon, a new antifungal compound produced by Chrysosporium queenslandicum: production, isolation and structure elucidation. J. Antibiot. (Tokyo) 55:516-519. [DOI] [PubMed] [Google Scholar]
- 14.Isaka, M., C. Suyarnsestakorn, M. Tanticharoen, P. Kongsaeree, and Y. Thebtaranonth. 2002. Aigialomycins A-E, new resorcylic macrolides from the marine mangrove fungus Aigialus parvus. J. Org. Chem. 67:1561-1566. [DOI] [PubMed] [Google Scholar]
- 15.Kealey, J. T., L. Liu, D. V. Santi, M. C. Betlach, and P. J. Barr. 1998. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. USA 95:505-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller, N. P., G. Turner, and J. W. Bennett. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3:937-947. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy, J., K. Auclair, S. G. Kendrew, C. Park, J. C. Vederas, and C. R. Hutchinson. 1999. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science 284:1368-1372. [DOI] [PubMed] [Google Scholar]
- 18.Kim, Y. T., Y. R. Lee, J. Jin, K. H. Han, H. Kim, J. C. Kim, T. Lee, S. H. Yun, and Y. W. Lee. 2005. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol. Microbiol. 58:1102-1113. [DOI] [PubMed] [Google Scholar]
- 19.Kroken, S., N. L. Glass, J. W. Taylor, O. C. Yoder, and B. G. Turgeon. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 100:15670-15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kupfer, D. M., S. D. Drabenstot, K. L. Buchanan, H. Lai, H. Zhu, D. W. Dyer, B. A. Roe, and J. W. Murphy. 2004. Introns and splicing elements of five diverse fungi. Eukaryot. Cell 3:1088-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lysoe, E., S. S. Klemsdal, K. R. Bone, R. J. Frandsen, T. Johansen, U. Thrane, and H. Giese. 2006. The PKS4 gene of Fusarium graminearum is essential for zearalenone production. Appl. Environ Microbiol. 72:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayorga, M. E., and W. E. Timberlake. 1992. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol. Gen. Genet. 235:205-212. [DOI] [PubMed] [Google Scholar]
- 23.Mueller, E., A. Bailey, A. Corran, A. J. Michael, and P. Bowyer. 2001. Ornithine decarboxylase knockout in Tapesia yallundae abolishes infection plaque formation in vitro but does not reduce virulence toward wheat. Mol. Plant-Microbe Interact. 14:1303-1311. [DOI] [PubMed] [Google Scholar]
- 24.Mutka, S. C., S. M. Bondi, J. R. Carney, N. A. Da Silva, and J. T. Kealey. 2006. Metabolic pathway engineering for complex polyketide biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res. 6:40-47. [DOI] [PubMed] [Google Scholar]
- 25.Nair, M. S. R., and S. T. Carey. 1980. Metabolites of Pyrenomyces. XIII. Structure of (+) hypothemycin, an antibiotic macrolide from Hypomyces trichothecoides. Tetrahedron Lett. 21:2011-2012. [Google Scholar]
- 26.Nair, M. S. R., S. T. Carey, and J. C. James. 1981. Metabolites of Pyrenomycetes. XIV. Structure and partial stereochemistry of the antibiotic macrolides hypothemycin and dihydrohypothemycin. Tetrahedron 37:2445-2449. [Google Scholar]
- 27.Nicholson, T. P., B. A. Rudd, M. Dawson, C. M. Lazarus, T. J. Simpson, and R. J. Cox. 2001. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 8:157-178. [DOI] [PubMed] [Google Scholar]
- 28.Ninomiya-Tsuji, J., T. Kajino, K. Ono, T. Ohtomo, M. Matsumoto, M. Shiina, M. Mihara, M. Tsuchiya, and K. Matsumoto. 2003. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 278:18485-18490. [DOI] [PubMed] [Google Scholar]
- 29.Pocsi, I., R. A. Prade, and M. J. Penninckx. 2004. Glutathione, altruistic metabolite in fungi. Adv. Microb. Physiol. 49:1-76. [DOI] [PubMed] [Google Scholar]
- 30.Proctor, R. H., A. E. Desjardins, R. D. Plattner, and T. M. Hohn. 1999. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol 27:100-112. [DOI] [PubMed] [Google Scholar]
- 31.Punt, P. J., and C. A. van den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216:447-457. [DOI] [PubMed] [Google Scholar]
- 32.Reid, R., M. Piagentini, E. Rodriguez, G. Ashley, N. Viswanathan, J. Carney, D. V. Santi, C. R. Hutchinson, and R. McDaniel. 2003. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42:72-79. [DOI] [PubMed] [Google Scholar]
- 33.Richardson, K. E., W. M. Hagler, Jr., and P. B. Hamilton. 1984. Bioconversion of alpha-[14C]zearalenol and beta-[14C]zearalenol into [14C]zearalenone by Fusarium roseum ‘Gibbosum.’ Appl. Environ. Microbiol. 47:1206-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schirmer, A., J. Kennedy, S. Murli, R. Reid, and D. V. Santi. 2006. Targeted covalent inactivation of protein kinases by resorcylic acid lactone polyketide. Proc. Natl. Acad. Sci. USA 103:4234-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulte, T. W., S. Akinaga, S. Soga, W. Sullivan, B. Stensgard, D. Toft, and L. M. Neckers. 1998. Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones 3:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma, S. V., T. Agatsuma, and H. Nakano. 1998. Targeting of the protein chaperone, HSP90, by the transformation suppressing agent, radicicol. Oncogene 16:2639-2645. [DOI] [PubMed] [Google Scholar]
- 37.Specht, C. A., C. C. DiRusso, C. P. Novotny, and R. C. Ullrich. 1982. A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Anal. Biochem. 119:158-163. [DOI] [PubMed] [Google Scholar]
- 38.Stadler, M., H.-V. Tichy, E. Katsiou, and V. Hellwig. 2003. Chemotaxonomy of Pochonia and other conidial fungi with Verticillium-like anamorphs. Mycol. Prog. 2:95-122. [Google Scholar]
- 39.Takehana, K., S. Sato, T. Kobayasi, and T. Maeda. 1999. A radicicol-related macrocyclic nonaketide compound, antibiotic LL-Z1640-2, inhibits the JNK/p38 pathways in signal-specific manner. Biochem. Biophys. Res. Commun. 257:19-23. [DOI] [PubMed] [Google Scholar]
- 40.Wee, J. L., K. Sundermann, P. Licari, and J. Galazzo. 2006. Cytotoxic hypothemycin analogues from Hypomyces subiculosus. J. Nat. Prod. 69:1456-1459. [DOI] [PubMed] [Google Scholar]
- 41.Wicklow, D. T., B. K. Joshi, W. R. Gamble, J. B. Gloer, and P. F. Dowd. 1998. Antifungal metabolites (monorden, monocillin IV, and cerebrosides) from Humicola fuscoatra traaen NRRL 22980, a mycoparasite of Aspergillus flavus sclerotia. Appl. Environ Microbiol. 64:4482-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, G., M. S. Rose, B. G. Turgeon, and O. C. Yoder. 1996. A polyketide synthase is required for fungal virulence and production of the polyketide T-toxin. Plant Cell 8:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, A., S. H. Lee, M. Mojena, R. G. Jenkins, D. R. Patrick, H. E. Huber, M. A. Goetz, O. D. Hensens, D. L. Zink, D. Vilella, A. W. Dombrowski, R. B. Lingham, and L. Huang. 1999. Resorcylic acid lactones: naturally occurring potent and selective inhibitors of MEK. J. Antibiot. (Tokyo) 52:1086-1094. [DOI] [PubMed] [Google Scholar]