Abstract
Of 2,692 sera screened for dengue virus immunoglobulin M by using a μ-capture enzyme-linked immunosorbent assay (ELISA), 954 had equivocal (index from 0.90 to 1.10) or positive (index of >1.10) results and were retested using a background subtraction (BS) ELISA that identifies screen false positives. No false positives were found among 427 sera with screen ELISA indices of >6.00; thus, retesting this specimen subset by BS ELISA is unnecessary.
Dengue viruses are flaviviruses transmitted among humans by Aedes mosquitoes (mainly Aedes aegypti) in tropical and subtropical areas worldwide (5, 14). Dengue virus infections are associated with significant morbidity, ranging from a nonspecific febrile illness to severe hemorrhagic fever, and in rare cases are fatal (5-7).
A major laboratory tool used in diagnosing dengue virus infections is measurement of the level of dengue virus immunoglobulin M (IgM) in serum (5, 6). Most dengue virus IgM tests utilize the μ-capture enzyme-linked immunosorbent assay (ELISA) format (1, 3, 4, 9, 14), which employs capture wells coated with anti-human IgM, inactivated dengue virus antigen, and enzyme-conjugated murine anti-flavivirus monoclonal antibody (reporter reagent). Because captured IgM with heterophilic antibody activity may yield false-positive results by directly binding the reporter reagent (8, 10), we employ a two-step testing algorithm for dengue virus IgM detection. Sera are first screened using a μ-capture ELISA (one well per sample); samples with equivocal or positive results are then retested using a background subtraction (BS) modification of this same ELISA (two wells per sample), designed to identify false-positive screening assay reactivity due to heterophilic antibodies (8, 12, 13). Because the expected screening ELISA reactivity rate is <50%, using this algorithm consumes fewer capture wells and less reporter reagent than testing all samples using the BS ELISA.
In conjunction with a surge in the number of dengue cases in Mexico and the Caribbean during the summer and fall of 2007 (2, 11), our reference laboratory tested 2,692 consecutive sera for dengue virus IgM. We capitalized on this large number of dengue virus IgM results to evaluate the efficiency of our testing algorithm. Specifically, we asked if there is a screening ELISA value above which false-positive results are not observed; if such a value exists, then sera with screening ELISA results above this value can be interpreted as dengue virus IgM positive without performing the BS ELISA, thus improving the algorithm's efficiency.
Sera submitted for dengue virus antibody testing were evaluated using an in-house-developed dengue virus IgM screening ELISA; this assay utilizes many of the components found in the dengue virus IgM ELISA kit manufactured by the diagnostic products division of Focus Diagnostics, Cypress, CA (the kit is for research use only in the United States) (1, 9). Each assay included negative control serum, positive control serum, and calibrator serum (Focus Diagnostics). Control, calibrator, and patient sera were diluted 1:101 in specimen diluent (Focus Diagnostics), and 0.1 ml of diluted sample was added to an assigned microtiter well coated with rabbit anti-human IgM (Focus Diagnostics). After incubation for 1 h at room temperature (RT), the wells were washed three times and then received inactivated dengue virus antigen (containing all four dengue virus serotypes [Focus Diagnostics]). After incubation for 2 h at RT and washing, the wells received horseradish peroxidase-conjugated 6B6C anti-flavivirus monoclonal antibody (Focus Diagnostics). After incubation for 30 min at RT and washing, the wells received tetramethylbenzidine (enhanced K-blue; Neogen Corp., Lexington, KY); after 10 min, the color reaction was stopped by adding sulfuric acid (Ricca Chemicals, Arlington, TX). Absorbance at 450 nm was measured using an ELISA reader (BioTek, Winooski, VT). The results were expressed as an index, calculated by dividing the specimen absorbance value by the calibrator absorbance value; indices of <0.90 were considered negative, indices from 0.90 to 1.10 were considered equivocal, and indices of >1.10 were considered positive.
Sera with equivocal or positive results in the dengue virus IgM screening ELISA were tested using the BS ELISA (8, 12). In this modified screening ELISA, all control, calibrator, and patient sera were added to two capture wells. After incubation for 1 h at RT and washing, dengue virus antigen was added to one well and specimen diluent was added to the other well. The assay was then finished per the screening ELISA procedure. For each specimen (including controls and calibrator), the absorbance value of the well receiving specimen diluent was subtracted from the absorbance value of the well receiving dengue virus antigen. This corrected absorbance value was then used to calculate the index. As with the screening ELISA, BS ELISA indices of <0.90 were considered negative, indices from 0.90 to 1.10 were considered equivocal, and indices >1.10 were considered positive.
Table 1 presents representative results for the screening ELISA and BS ELISA, demonstrating how index values were calculated. Data for calibrator sera, controls, one screen false-positive patient sample, and two true-positive patient samples are shown.
TABLE 1.
Results from a representative dengue virus IgM screening ELISA and subsequent BS ELISA
| Specimen | Screening ELISA
|
BS ELISA
|
||||
|---|---|---|---|---|---|---|
| Absorbance | Index | Antigen well absorbance | Diluent well absorbance | Corrected absorbance | Index | |
| Calibrator | 0.254 | NCa | 0.289 | 0.107 | 0.182 | NC |
| Negative control | 0.097 | 0.38 | 0.086 | 0.071 | 0.015 | 0.08 |
| Positive control | 1.153 | 4.54 | 0.887 | 0.080 | 0.807 | 4.43 |
| Screen false positive | 0.664 | 2.61 | 0.534 | 0.471 | 0.063 | 0.35 |
| True positive | 0.500 | 1.97 | 0.413 | 0.157 | 0.256 | 1.41 |
| True positive | 2.745 | 10.81 | 2.839 | 0.089 | 2.750 | 15.11 |
NC, not calculated.
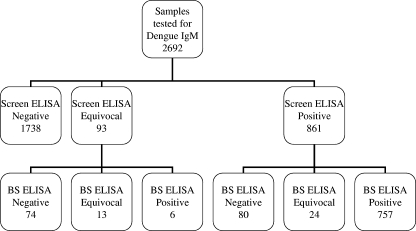
Figure 1 summarizes the findings for 2,692 sera tested for dengue virus IgM; 954 sera (35%) had equivocal (n = 93) or positive (n = 861) results in the results dengue virus IgM screening ELISA and were further tested using the BS ELISA. Most samples with screen equivocal results (74/93 [80%]) were negative for dengue virus IgM based on the BS ELISA results. In contrast, most screen positive samples (757/861 [88%]) were also positive for dengue virus IgM in the BS ELISA; only 9% (80/861) of the screen positive samples were identified as false positives by the BS ELISA. The overall false-equivocal/positive rate for the screening ELISA was 5.7% (154/2,692).
FIG. 1.
Summary of dengue virus IgM screening ELISA and BS ELISA results for 2,962 consecutive sera submitted for dengue virus IgM testing. For both assays, indices of <0.90 were considered negative, indices from 0.90 to 1.10 were considered equivocal, and indices of >1.10 were considered positive.
Table 2 demonstrates the relationship between screening ELISA indices and the proportion of samples positive in the BS ELISA. Of the 592 sera with indices of >3.00 in the screening ELISA, 587 (99%) were positive in the BS ELISA. All 427 sera with indices of >6.00 in the screening ELISA were positive in the BS ELISA.
TABLE 2.
Relationship of dengue virus IgM BS ELISA results to screening ELISA index values
| Screen ELISA index | Total no. of samples | No. (%) of samples with indicated BS ELISA result
|
||
|---|---|---|---|---|
| Negative | Equivocal | Positive | ||
| 0.90-1.10 | 93 | 74 (80) | 13 (14) | 6 (6) |
| 1.11-2.00 | 180 | 66 (37) | 22 (12) | 92 (51) |
| 2.01-3.00 | 89 | 9 (10) | 2 (2) | 78 (88) |
| 3.01-4.00 | 53 | 2 (4) | 0 (0) | 51 (96) |
| 4.01-5.00 | 54 | 2 (4) | 0 (0) | 52 (96) |
| 5.01-6.00 | 58 | 1 (2) | 0 (0) | 57 (98) |
| 6.01-8.00 | 97 | 0 (0) | 0 (0) | 97 (100) |
| 8.01-10.00 | 103 | 0 (0) | 0 (0) | 103 (100) |
| 10.01-14.00 | 130 | 0 (0) | 0 (0) | 130 (100) |
| >14.00 | 97 | 0 (0) | 0 (0) | 97 (100) |
| Total | 954 | 154 (16) | 37 (4) | 763 (80) |
These findings demonstrate that all sera with strong reactivity (index of >6.00) in the dengue virus IgM screening ELISA are also positive in the BS ELISA; thus, our dengue virus IgM testing algorithm can be modified to eliminate further testing of such sera in the BS ELISA. The application of this modified algorithm to the current data set would have reduced the number of samples evaluated using the BS ELISA from 954 to 527, a reduction of 45%. If a laboratory's quality assurance program allows an overall false-positive rate of <0.5%, the algorithm could be further modified to eliminate BS ELISA testing of sera with screening ELISA indices of >3.00; the application of this algorithm to the current data set would have reduced the number of samples tested by BS ELISA from 954 to 362 (a reduction of 62%), with only 5 of 2,692 sera (0.19%) exhibiting false-positive dengue virus IgM results.
The BS approach for identifying false-positive reactivity is routinely applied to other screening μ-capture ELISA systems besides dengue virus IgM (e.g., West Nile virus IgM) (8, 13). Thus, it stands to reason that these other screening assays, like the dengue virus IgM assay, may also have a characteristic high index cut point above which BS ELISA performance is not necessary. This cut point will undoubtedly vary among different assays, depending on the absorbance value of the screening ELISA calibrator and the dynamic range of the assay. Each laboratory must therefore define its own reflex testing algorithms for analytes measured by μ-capture ELISA.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Branch, S. L., and P. N. Levett. 1999. Evaluation of four methods for detection of immunoglobulin M antibodies to dengue virus. Clin. Diagn. Lab. Immunol. 6:555-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2007. Weekly dengue surveillance report. CDC Dengue Branch and Puerto Rico Department of Health. http://www.cdc.gov/ncidod/dvbid/dengue/documents/weeklyreport.pdf. Accessed 30 January 2008.
- 3.Chanama, S., S. Anantapreecha, A. A-nueggonpipat, A. Sa-gnasang, I. Kurane, and P. Sawanpanyalert. 2004. Analysis of specific IgM responses in secondary dengue virus infections: levels and positive rates in comparison with primary infections. J. Clin. Virol. 31:185-189. [DOI] [PubMed] [Google Scholar]
- 4.Falconar, A. K. I., E. de Plata, and C. M. E. Romero-Vivas. 2006. Altered enzyme-linked immunosorbent assay immunoglobulin M (IgM)/IgG optical density ratios can correctly classify all primary or secondary dengue virus infections 1 day after the onset of symptoms, when all of the viruses can be isolated. Clin. Vaccine Immunol. 13:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guzman, M. G., and G. Kouri. 1996. Advances in dengue diagnosis. Clin. Diagn. Lab. Immunol. 3:621-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes, E. B., and D. J. Gubler. 1992. Dengue and dengue hemorrhagic fever. Pediatr. Infect. Dis. 11:311-317. [DOI] [PubMed] [Google Scholar]
- 8.Hogrefe, W. R., R. Moore, M. Lape-Nixon, M. Wagner, and H. E. Prince. 2004. Performance of immunoglobulin G (IgG) and IgM enzyme-linked immunosorbent assays using a West Nile virus recombinant antigen (preM/E) for detection of West Nile virus- and other flavivirus-specific antibodies. J. Clin. Microbiol. 42:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koraka, P., C. Suharti, T. E. Setiati, A. T. A. Mairuhu, E. Van Gorp, C. E. Hack, M. Juffrie, J. Sutaryo, G. M. Van Der Meer, J. Groen, and A. D. M. E. Osterhaus. 2001. Kinetics of dengue virus-specific serum immunoglobulin classes and subclasses correlate with clinical outcome of infection. J. Clin. Microbiol. 39:4332-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levinson, S. S., and J. J. Miller. 2002. Toward a better understanding of heterophile (and the like) antibody interferences with modern immunoassays. Clin. Chim. Acta 325:1-15. [DOI] [PubMed] [Google Scholar]
- 11.Pan American Health Organization. 2008. 2007: number of reported cases of dengue and dengue hemorrhagic fever (DHF), region of the Americas (by country and subregion). http://www.paho.org/English/AD/DPC/CD/dengue-cases-2007.htm. Accessed 30 January 2008.
- 12.Prince, H. E., and W. R. Hogrefe. 2003. Detection of West Nile virus (WNV)-specific immunoglobulin M in a reference laboratory setting during the 2002 WNV season in the United States. Clin. Diagn. Lab. Immunol. 10:764-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rawlins, M. L., E. M. Swenson, H. R. Hill, and C. M. Litwin. 2007. Evaluation of an enzyme immunoassay for detection of immunoglobulin M antibodies to West Nile virus and the importance of background subtraction in detecting nonspecific reactivity. Clin. Vaccine Immunol. 14:665-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigau-Perez, J. G., G. G. Clark, D. J. Gubler, P. Reiter, E. J. Sanders, and A. V. Vorndam. 1998. Dengue and dengue haemorrhagic fever. Lancet 352:971-977. [DOI] [PubMed] [Google Scholar]