Abstract
The microbial community diversity and composition of meromictic Soap Lake were studied using culture-dependent and culture-independent approaches. The water column and sediments were sampled monthly for a year. Denaturing gradient gel electrophoresis of bacterial and archaeal 16S rRNA genes showed an increase in diversity with depth for both groups. Late-summer samples harbored the highest prokaryotic diversity, and the bacteria exhibited less seasonal variability than the archaea. Most-probable-number assays targeting anaerobic microbial guilds were performed to compare summer and fall samples. In both seasons, the anoxic samples appeared to be dominated by lactate-oxidizing sulfate-reducing prokaryotes. High numbers of lactate- and acetate-oxidizing iron-reducing bacteria, as well as fermentative microorganisms, were also found, whereas the numbers of methanogens were low or methanogens were undetectable. The bacterial community composition of summer and fall samples was also assessed by constructing 16S rRNA gene clone libraries. A total of 508 sequences represented an estimated >1,100 unique operational taxonomic units, most of which were from the monimolimnion, and the summer samples were more diverse than the fall samples (Chao1 = 530 and Chao1 = 295, respectively). For both seasons, the mixolimnion sequences were dominated by Gammaproteobacteria, and the chemocline and monimolimnion libraries were dominated by members of the low-G+C-content group, followed by the Cytophaga-Flexibacter-Bacteroides (CFB) group; the mixolimnion sediments contained sequences related to uncultured members of the Chloroflexi and the CFB group. Community overlap and phylogenetic analyses, however, not only demonstrated that there was a high degree of spatial turnover but also suggested that there was a degree of temporal variability due to differences in the members and structures of the communities.
Soda lakes are highly alkaline and saline aquatic environments. These lakes are some of the most productive ecosystems in the world, and the daily primary production often exceeds that in many eutrophic lakes, presumably due to a virtually unlimited supply of CO2 and high daily light irradiance (29, 45). Because of the difference between the density of saline lake waters and the density of the external dilute input water, the widespread occurrence of soda lakes in endorheic basins, the absence of episodic mixing mechanisms, and the basins' morphometric features, these lakes have a propensity to become meromictic in response to both regional and local hydrologic events (19, 46).
One of the main sources of variation among inland lakes is the duration of meromixis (20). For instance, long-term meromixis at a high pH is often accompanied by an accumulation of toxic inorganic compounds—mainly sulfide and ammonia—in the anoxic monimolimnion (24, 60). Prolonged chemical stratification affects the flux of reduced metabolites into the upper layer from bottom water and lake bottom sediments (11). In the anoxic waters, high concentrations of sulfide may affect microbial processes directly (for instance, denitrification) (26). At high pH (i.e., pH > 8.5) one-half of the ammonia present is nondissociated and, therefore, in a toxic form, possibly inhibiting ecologically relevant activities like methane oxidation (29).
Historical interest in soda lake microbiology focused primarily on the isolation and characterization of individual microorganisms with potential industrial applications (16, 22, 28), although anaerobic strains with hypothesized ecological roles have also been described (48, 75, 76). Recent surveys of soda environments have indicated that there is elevated microbial phylogenetic diversity and have included surveys of the Wadi An Natrun lake system in Egypt (47), soda lakes in the Kenyan-Tanzanian Rift Valley (58), soda lakes in Inner Mongolia in China (42), saline, meromictic Lake Kaiike in Japan (33), saline Qinghai Lake in China (15), and athalassohaline Lake Chaka in China (27). In general, the studies examining the impact of meromixis on bodies of water have focused on saline-neutral gradients rather than saline-alkaline gradients, although the microbial ecology of Mono Lake, a transiently meromictic soda lake, has been extensively studied (24, 65).
This study focused on the abundance and distribution of prokaryotic assemblages in Soap Lake, a meromictic, alkaline (pH ∼9.8), and saline (∼15 to 140 g liter−1) lake situated in a semiarid area of eastern Washington State. This lake has two distinctive characteristics: (i) an estimated meromixis of at least 2,000 years (51) and (ii) extremely high sulfide concentrations (∼140 mM) in the monimolimnion. Owing to the lake's seasonal cycles (mainly blooms of Chlorella sp.-dominated algae [71]) and extended meromixis, we hypothesized that microbial assemblages would be both temporally and spatially segregated. In addition, we hypothesized that the lake's potential metabolic resources (for example, products of fermentation, sulfate, and H2/CO2) would be reflected in the occurrence of specific anaerobic microbial guilds. The first hypothesis was addressed by conducting denaturing gradient gel electrophoresis (DGGE) of bacterial and archaeal 16S rRNA genes and by constructing clone libraries of bacterial 16S rRNA genes present in the water and sediments. The second hypothesis was addressed using most-probable-number (MPN) assays of fermentative, sulfate-reducing, iron-reducing, and methanogenic populations.
Statistical and phylogenetic analyses of the clone libraries showed that there was a high degree of spatial turnover and, to a lesser extent, temporal turnover of prokaryotic communities, suggesting that these communities are tightly coupled with recurring environmental conditions related to long-term meromixis and to seasonal events.
MATERIALS AND METHODS
Site description and sampling.
Soap Lake (47°23′N, 119°30′W) is a closed-basin, meromictic lake in the lower Grand Coulee in eastern Washington State. It is the most saline and alkaline of a series of lakes occupying an old bed of the Columbia River. The main morphometric features were summarized by Edmondson and Anderson (17). Physical and chemical limnological attributes of the lake have been reported elsewhere (2, 70, 71), and some attributes were also determined during this study (Table 1).
TABLE 1.
Attributes of Soap Lake waters and sedimentsa
| Sample | NH3 concn (mM) | Na+ concn (g liter−1)b | H2S concn (mM) | SO42− concn (mM)b | Dissolved oxygen concn (mg liter−1) | DOC concn (mg liter−1) | Total organic carbon (%) | Total N concn (μg liter−1) | Total P concn (μg liter−1) | PO43− concn (μg liter−1) | Concn of NO3− + NO2− (μg liter−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| July | |||||||||||
| Mixolimnion | 0 | 4.7 | 0 | 30 | 4.35 | 17 | 803 | 463 | 476 | 33 | |
| Chemocline | 1.9 | 12.7 | 10 | 49 | 1.81 | 745 | 455 | 451 | 31 | ||
| Monimolimnion | 59.8 | 30.2 | 125 | 340 | 0 | 314 | 1,187 | 5,538 | 66 | ||
| Mixolimnion sediment | 3.7 | 1.7 | |||||||||
| Monimolimnion sediment | 145 | 9.1 | |||||||||
| October | |||||||||||
| Mixolimnion | 0 | 0 | 6.89 | 20 | 841 | 649 | 467 | <20 | |||
| Chemocline | 1.2 | 2.2 | 787 | 629 | 368 | <20 | |||||
| Monimolimnion | 65 | 131 | 0 | 341 | 1,210 | 4,736 | 33 | ||||
| Mixolimnion sediment | 2.9 | 1.3 | |||||||||
| Monimolimnion sediment | 140 | 9 |
The Secchi depth was 3.9 m in July and 5.2 m in October.
Data from reference 60.
The water and sediment samples used in this study were collected monthly between September 2002 and December 2003 from the central basin of Soap Lake. Water samples were collected from the oxic mixolimnion (0, 5, 10, 15, and 18 m), the chemocline (19, 20, 20.5, and 21 m), and the anoxic monimolimnion (21.5, 22, 23, 24, and 25 m) with a 2-liter Van Dorn bottle. Sediment samples were collected from the mixolimnion and the monimolimnion using a dredge. Sediment and water samples were placed in 500-ml sterile opaque polypropylene bottles and kept on ice until they were stored at 4°C for culture work or were frozen for DNA extraction. When required, anaerobic conditions were preserved by completely filling the bottles with water or sediment.
Limnological properties.
Dissolved oxygen, temperature, pH, and total dissolved solids were measured in situ by using a Hydrolab Datasonde 4 (Hydrolab Corp., Austin, TX). The ammonia content was determined by the indophenol blue method (68) using internal standards for each set of determinations. The sulfide content was determined spectrophotometrically by the methylene blue colorimetric method (7). Total nitrogen and concentrations of nitrate plus nitrite and orthophosphate, as well as total organic carbon and dissolved organic carbon (DOC) concentrations, were determined using standard methods (1).
MPN assays for anaerobic microbial populations.
Anaerobic microbial guilds were enumerated using a basal medium (SLBM) previously described by Dimitriu et al. (14), which was modified by adding yeast extract (1.0 g liter−1) and separately sterilizing the NaHCO3 and Na2CO3 solutions (final concentration, 1% [wt/vol]). Reducing conditions were achieved by boiling the medium and adding 1 ml of a sterile solution of cysteine hydrochloride and Na2S·9H2O (5 g liter−1 each) under an N2/H2 (90:10, vol/vol) atmosphere; methylene blue (0.004 g) was used as the redox indicator. NaCl was added at the following concentrations: 0.26 M (mixolimnion samples), 1.2 to 1.7 M (chemocline samples), and 2.41 M (monimolimnion samples). The NaCl concentrations selected reflect both historical values and values obtained in this study; for the chemocline samples, preliminary incubation experiments did not reveal appreciable differences in growth rates within the chosen salt range (data not shown). SLBM supplemented with 5 mM betaine or 10 mM glycerol was used to determine the number of fermentative bacteria. Sulfate-reducing prokaryotes (SRP) were enumerated by using 38 mM lactate or butyrate, 76 mM formate, or 10 mM glycerol as the electron donor and 3.3 mM FeSO4 as the electron acceptor; a black precipitate was used as an indicator of SRP growth. For iron-reducing bacteria (IRB), the medium was supplemented with 38 mM lactate and 100 mM amorphous Fe(III) oxyhydroxide (41). Separate sets of dilution tubes were inoculated with 20 mM Na2MoO4, an inhibitor of sulfate reduction (50). The appearance of a black precipitate was considered a signal that iron reduction was occurring. Methanogens were cultivated using either an H2/CO2 (80:20, vol/vol) headspace or 10 mM trimethylamine (TMA), while CH4 evolution was measured with an HP 3400 gas chromatograph (Hewlett-Packard). Twenty-milliliter serum bottles or Balch tubes with butyl rubber stoppers were used for three-vial dilution series, and all cultures were incubated anaerobically (4) for ∼30 days at room temperature in the dark.
DGGE analysis of prokaryotic communities.
DNA was extracted from water and sediment samples with a MoBio UltraClean DNA extraction kit (MoBio Inc., Solano Beach, CA) by following the manufacturer's recommended protocol. Because of the high salt and high sulfide contents, extraction of the monimolimnion samples was preceded by a dialysis step. Briefly, 5-ml samples were injected into dialysis cassettes and dialyzed against sterile, deionized water for 18 h; 1-ml portions of the resulting eluates were used as the material from which DNA was retrieved. Archaeal and bacterial small-subunit (16S) rRNA genes from extracted DNA were amplified using GC-clamped primers and PCR conditions described previously (7). Bacterial DNA was separated using an 8% polyacrylamide gel and a 50 to 80% urea-formamide denaturing gradient (100% denaturant was 7 M urea and 40% deionized formamide); archaeal DNA was separated using an 8% polyacrylamide gel with a 35 to 80% denaturing gradient. Both bacterial and archaeal gels were run in 1× Tris-acetate-EDTA buffer at 80 V for 15 h and a constant temperature of 60°C, using a Bio-Rad DCode electrophoresis system (Bio-Rad, Cambridge, MA). Gels were stained with ethidium bromide and visualized with a Bio-Rad FX molecular imager. The number of bands and the intensity of the bands, which indicated the approximate richness and phylotype abundance (18), respectively, were estimated for computation of Shannon's diversity indices.
Clone library construction.
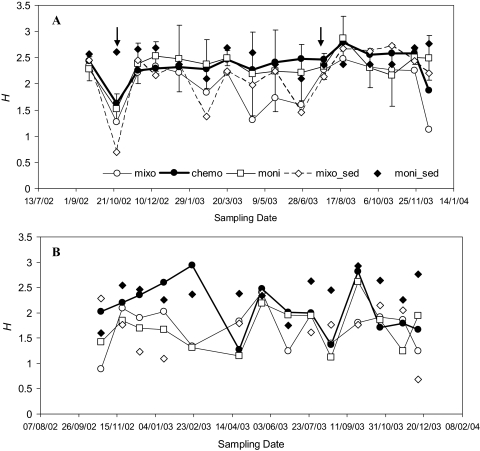
To obtain a finer-resolution picture of the microbial community composition, DNA was retrieved from fall (sampling date, 24 October 2002) and summer (sampling date, 26 July 2003) samples. We focused on these sampling dates to assess whether two samples with contrasting diversity levels, as indicated by DGGE, also exhibited shifts in community composition (Fig. 1).
FIG. 1.
Bacterial (A) and archaeal (B) diversity obtained from DGGE profiles. The Shannon-Weaver indices (H) are values averaged across layers. The arrows indicate sampling dates for which clone libraries were constructed. The error bars in panel A indicate one standard deviation (n = 5) for illustrative purposes; archaeal diversity fluctuations exhibited similar trends (data not shown). mixo, mixolimnion; chemo, chemocline; moni, monimolimnion; mixo_sed, mixolimnion sediment; moni_sed, monimolimnion sediment.
Small-subunit (16S) rRNA genes in samples were amplified by using the bacterium-specific forward primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′; positions 27 to 46) and universal reverse primer 907R (5′-CCGTCAATTCMTTTRAGTTT-3′; positions 907 to 926) (35). All 16S rRNA gene nucleotide positions reported here are based on the Escherichia coli numbering reported by Brosius et al. (6). Primers were synthesized at MWG Biotech. The PCR mixtures (total volume, 20 μl) contained PCR buffer (10 mM Tris-HCl, 50 mM KCl, 0.1% Triton X-100), 2.5 mM MgCl2, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 200 μM dTTP, 10 pmol of each primer, 1 U of Taq DNA polymerase, and 1 μl of template (extracted) DNA. The PCR conditions were as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation for 45 s at 94°C, annealing for 45 s at 50°C, and extension for 1 min at 72°C and then a final extension at 72°C for 10 min. The PCR products were ligated into the pGEM-T Easy vector (Promega Corp., Madison, WI) and transformed into competent E. coli JM109 cells by following the manufacturer's protocol. The transformed cells were plated on selective Luria-Bertani medium plates containing 100 μg of ampicillin ml−1, 80 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) ml−1, and 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) as recommended by the manufacturer and were incubated overnight at 37°C. Clones (i.e., white colonies) were randomly screened for inserts that were the correct length by PCR amplification with primers SP6 and T7. Amplification preparations containing products that were the expected length were purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA), and clean amplification products were sequenced at the MWG Biotech sequencing facility using primer 27F, which yielded readable sequences that were ∼800 bp long.
Phylogenetic analysis.
Small-subunit rRNA gene sequences were assigned to major subdivision- or division-level groups, and reference sequences for phylogenetic tree construction were chosen based on the 10 highest database sequence similarities obtained after BLAST searches. Sequences were tested for chimeras by using web-based Bellerophon (23). Nonchimeric sequences were aligned with Clustal X v.1.83 (69), and manual adjustment was performed with the alignment-visualizing tool of MEGA v.3.1 (34).
Phylogenetic structures were inferred with MEGA v.3.1 by using two optimality criteria. A distance matrix was constructed assuming Kimura's two-parameter model (32), and nucleotide positions containing gaps or missing data were deleted in a pairwise fashion. A neighbor-joining tree was inferred with pairwise deletion of gaps and with 1,000 bootstrap pseudoreplicates. A parsimony phylogeny was constructed by using the mini-mini heuristic search method, and sites containing gaps or missing data were completely deleted. The analysis was bootstrap pseudoreplicated 100 times.
Estimation of bacterial diversity.
Aligned sequences were used to construct Jukes-Cantor-corrected distance matrices, which were analyzed with DOTUR (62) to determine operational taxonomic units (OTUs) at several divergence cutoff values using the furthest-neighbor clustering algorithm. DOTUR was also used to generate lineage-per-time plots, to estimate richness (Chao1 and ACE [8, 9]), and to compute Shannon diversity indices for individual libraries and for combined data sets (i.e., sequences sorted into season and layer within the lake). Estimates were calculated using two genetic distance levels (D), 0.03 and 0.2, which, although arbitrary, are useful for delineating species-level and phylum-level OTUs, respectively (62). Collector's curves were plotted using the number of clones sampled against the Chao1 estimator of species richness.
Statistical analysis.
To compare the phylogenetic structures among libraries, a parsimony test (44) as implemented in TreeClimber (63) was performed using the following input files: (i) a parsimony tree containing all sequences and (ii) a list of the tree sequences coded according to community (sample) type (i.e., depth, sediment, and season). TreeClimber computes the number of changes (the parsimony score) for samples required to explain the observed tree, which are then compared to the distribution of changes on 1,000 random trees in order to generate a P value for the probability of the observed parsimony score. Therefore, the parsimony test determines whether phylogeny significantly covaries with the sample type.
Observed differences in community composition between pairs of libraries were statistically compared using ∫-LIBSHUFF (61). ∫-LIBSHUFF uses distance matrices (in this case, Jukes-Cantor matrices) to estimate the coverage values for the clone libraries being compared over a range of taxonomic levels, enabling detection of significantly different microbial communities defined by nonoverlapping coverage. Randomizations (10,000) were run to determine the significance (α = 0.05) of overlapping coverage after correction for multiple pairwise comparisons using the Bonferroni method. To obtain an indication of the degree of community overlap, we estimated the number of OTUs shared by two libraries (SA,B Chao), the fraction of sequences in shared OTUs (Uest and Vest), Ĵabd, an abundance-based Jaccard estimator (10, 64), and θ, a nonparametric maximum likelihood estimator of community structure similarity (73); all indices were computed with SONS (64). The pairwise θ values for the libraries were converted to distances and used to construct a dendrogram by the unweighted-pair group method with arithmetic means.
Nucleotide sequence accession numbers.
Partial 16S rRNA gene sequences have been deposited in the GenBank database under accession numbers EU644758 to EU645265.
RESULTS
Limnological properties.
Table 1 shows the main physicochemical attributes of Soap Lake water and sediment samples. Both NH3 and H2S were undetectable in the mixolimnion but were detectable in the chemocline (∼18.5 to 21 m), and they were present at high concentrations in the monimolimnion. The levels of DOC and SO42− were also higher below the chemocline. Dissolved oxygen showed the opposite pattern; the oxygen concentration decreased sharply until there was complete anoxia in the monimolimnion. With the exception of PO43−, for which there was a season-independent ∼12-fold increase in the concentration in the monimolimnion, total nitrogen, phosphorous, and mineral nitrogen levels showed little spatial or temporal variability. The average yearly pH values were 9.8 (standard error of the mean, 0.4) for the mixolimnion and 9.9 ± 0.3 for the monimolimnion; the mean temperatures were 16.3 ± 9.3 and 7.3 ± 1.1°C for the mixolimnion and monimolimnion, respectively; and the concentrations of total dissolved solids were 13.2 ± 3.2 g liter−1 for the mixolimnion and 140 ± 3.2 g liter−1 for the monimolimnion (data not shown).
MPN analysis.
Soap Lake samples supported growth of all anaerobic guilds analyzed, including SRP, IRB, methanogens, and fermentative microorganisms (Table 2).
TABLE 2.
MPN counts for anaerobic guilds from Soap Lake samples obtained in July and October
| Sample | MPN counts (105 cells ml−1 or 105 cells g−1)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SRP
|
Fermentative bacteria
|
IRB
|
Methanogens
|
||||||||
| Lactate | Formate | Butyrate | Glycerol | Glycerol | Betaine | Lactate | Lactate + molybdate | Acetate | TMA | H2 + CO2 | |
| July | |||||||||||
| Mixolimnion | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chemocline | 0.012 | 0.24 | 0.046 | 0.024 | 2.4 | 0 | 0 | 0.046 | 0 | 0 | 0 |
| Monimolimnion | 11 | 0.24 | 0.046 | 0.11 | 4.6 | 0.46 | 460 | 0.24 | 0.28 | 0.024 | 0.0093 |
| Mixolimnion sediment | 2.4 | 0.46 | 0.46 | 0.11 | 11 | 2.4 | 24 | 4.6 | 0.46 | 0.024 | 0.0093 |
| Monimolimnion sediment | 11 | 0.24 | 0.24 | 0.11 | 2.4 | 1.1 | 46 | 15 | 0.046 | 0.024 | 0.0023 |
| October | |||||||||||
| Mixolimnion | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chemocline | 0 | 0.24 | 0.046 | 0.024 | 0 | 0.24 | 0 | 0.046 | 0 | 0 | 0 |
| Monimolimnion | 24 | 0.093 | 0.11 | 0.24 | 11 | 4.6 | 15 | 240 | 1.1 | 0.024 | 0.0093 |
| Mixolimnion sediment | 2.4 | 0.46 | 0.046 | 2.4 | 24 | 11 | 46 | 24 | 0.024 | 0.024 | 0.0093 |
| Monimolimnion sediment | 0.24 | 0.24 | 1.1 | 1.1 | 4.6 | 0.24 | 150 | 1100 | 46 | 0.024 | 0.0023 |
(i) SRP.
Based on data obtained by the MPN technique, lactate was the electron donor that supported the highest number of SRP. In both seasons there were mixed patterns of sulfate-reducing growth when lactate was used as the carbon source. In the chemocline there was little (July) or no (October) SRP growth, while in October the monimolimnion supported the highest SRP MPN levels. Formate and butyrate also supported sulfate reduction, but acetate did not (data not shown). In July the butyrate-formate MPN counts were distinctively lower than the lactate counts. October was unique in that the butyrate-amended mixolimnion sediment values were about 1 order of magnitude lower (4.6 × 103 MPN ml−1) than the values obtained with the other electron donors (2.4 × 105 MPN ml−1). Glycerol also supported sulfate reduction, and the MPN counts were in the same ranges that were observed for butyrate and formate.
(ii) IRB.
To address the possibility that dissimilatory iron reduction is a potential terminal electron-accepting process that contributes to the overall carbon mineralization, an amorphous Fe(III) gel (100 mM) was added to the MPN assay medium, and the preparation was checked for the development of a black precipitate indicating that there was conversion of Fe(III) into Fe(II). After ∼14 days of incubation, we observed that all anoxic samples contained numbers of IRB that were often the same order of magnitude as the numbers of SRP. As shown in Table 2, with lactate as the electron donor, the July and October samples had similar IRB and SRP levels. With the exception of the July monimolimnion sample, there were no differences between molybdate-containing (20 mM) and molybdate-free incubations, implying that SRP were not involved in iron reduction.
(iii) Methanogens.
Consistent with the high-sulfate nature of Soap Lake, low numbers (i.e., ∼103 cells ml−1) of methanogens were observed in both seasons; TMA-amended cultures contained higher numbers of cells than H2/CO2-amended cultures (2.4 × 103 and 930 cells ml−1, respectively).
(iv) Fermentative bacteria.
In general, the numbers of fermentative bacteria paralleled the numbers of SRP. No differences were detected between betaine (a putative osmolyte in high-salt dwellers) and glycerol incubations. In addition, fermentative microorganisms were also present in the chemocline, albeit at numbers lower than those in deeper, oxygen-free samples.
Bacterial and archaeal DGGE profiles.
The DGGE banding patterns revealed a prokaryotic community with diversity levels dependent on both the sampling date and the layer (Fig. 1). The bacterial community exhibited marked mixolimnetic diversity fluctuations during the sampling period (note the error bars in Fig. 1), while relative seasonal stability for chemocline, monimolimnion, and monimolimnion sediment samples was observed. The archaeal diversity, in contrast, exhibited strong seasonal fluctuations. The bacterial diversity (Fig. 1A) appeared to peak in late summer samples, and the diversity of the archaeal community (Fig. 1B) peaked ∼1 month later (this was particularly true of samples retrieved from suboxic [chemocline] and anoxic [monimolimnion water column and sediment] locations). Prokaryotic diversity showed a moderate increase with depth on most sampling dates (Shannon indices versus depth, 0.48 < R2 < 0.84; P ≤ 0.009); these variables were strongly correlated in early spring samples (R2 = 0.92 and R2 = 0.95 for bacteria and archaea, respectively; P = 0.006). This was the only sampling date when a robust correlation (R2 = 0.89; P = 0.008) between bacterial and archaeal diversity was observed; the opposite relationship (i.e., slightly negative, nonsignificant coefficients) tended to be the case. When depths were averaged by stratum and compared across the entire sampling period, no significant bacterial diversity-archaeal diversity correlations were observed; notably, however, the average archaeal diversity for the chemocline and the monimolimnion was negatively correlated with the bacterial diversity for the same layers (R2 = −0.691 and R2 = −0.687, respectively; P ≤ 0.01).
Bacterial clone library analysis: general diversity patterns and extrapolated richness.
We generated 11 rRNA gene clone libraries for two representative months (July and October) and from each chemical layer, including the mixolimnion (depth, 5 m), the chemocline (depth, 20 m), the monimolimnion (depth, 23 m), the mixolimnion sediment, and the monimolimnion sediment, and a library from the 15-m sample for the October set. A total of 562 sequences were obtained. We conducted a thorough phylogenetic analysis with the complete sequence data set (see below); a ∫-LIBSHUFF analysis, however, revealed that the data for the 5- and 15-m samples from October were statistically nonsignificant (P > 0.05 for both reciprocal comparisons) when the OTUs were defined at D values of 0.03 and 0.2; therefore, in all additional analyses we focused on the 10 samples for which direct layer-to-layer comparisons could be made.
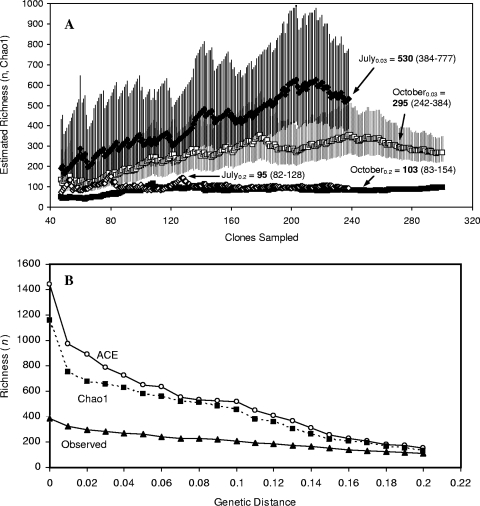
The 10 combined bacterial 16S rRNA gene libraries (n = 508) consisted of 482 unique sequences and 293 OTUs clustered by using a 3% cutoff criterion. Richness estimation resulted in a Chao1 of 635 OTUs (D = 0.03) for all sequences; the total Shannon diversity and evenness were 5.28 and 0.92, respectively (Table 3). Although the overall coverage value was relatively low (33%), collector's curves of estimated richness constructed on the basis of season (July or October) showed that the October sequences were well sampled at both genetic distances considered (Fig. 2A); the July Chao1, on the other hand, did not begin to stabilize with sampling effort, although the nonoverlapping lower 95% confidence interval of the end point estimate at the D value of 0.03 indicated, as also suggested by DGGE, that in this season the bacterial richness was higher (July Chao1 = 530; October Chao1 = 295). When OTUs were defined using a 20% divergence criterion, the seasonal estimated richness was indiscernible. The overall Chao1 and ACE richness estimates for OTUs sharing a cutoff level of 0.2 to 0 (a proxy for a lineage-per-time plot) produced comparable richness curves and indicated the potential presence of at least 1,100 unique sequences, with a ∼50% increase in richness for sequences sharing less than 1% similarity (Fig. 2B). For both seasons, diversity and evenness increased with depth (Table 3), again mirroring the DGGE results, and the former was higher for all July libraries (Table 3); this trend was reflected in higher estimated richness and lower coverage (July average coverage, 0.39; October average coverage, 0.60). Together, the richness estimates and diversity indices indicate that there was a high degree of undescribed diversity, a large fraction of which likely belonged to clusters containing sequences whose taxonomic resolution is finer than that defined for species.
TABLE 3.
Diversity parameters of Soap Lake sequences defined by a divergence cutoff of 0.03
| Sample | Chao1 (95% confidence interval) | Shannon diversity | Evennessa | Coverageb | No. of clones |
|---|---|---|---|---|---|
| July | |||||
| Mixolimnion | 88 (48-186) | 3.26 | 0.95 | 0.48 | 50 |
| Chemocline | 65 (43-128) | 3.32 | 0.96 | 0.54 | 53 |
| Monimolimnion | >290 (129-458)c | 3.70 | 0.98 | 0.20 | 45 |
| Mixolimnion sediment | >141 (89-214) | 3.83 | 0.98 | 0.33 | 56 |
| Monimolimnion sediment | >63 (39-132) | 3.24 | 0.97 | 0.43 | 48 |
| October | |||||
| Mixolimnion | 58 (37-127) | 2.96 | 0.88 | 0.69 | 46d |
| Chemocline | 32 (28-48) | 3.10 | 0.94 | 0.78 | 56 |
| Monimolimnion | 53 (38-97) | 3.25 | 0.95 | 0.51 | 47 |
| Mixolimnion sediment | >130 (75-253) | 3.58 | 0.96 | 0.43 | 50 |
| Monimolimnion sediment | 53 (41-88) | 3.32 | 0.94 | 0.63 | 57 |
| All | 653 (553-831) | 5.28 | 0.92 | 0.33 | 508 |
E = H/lnS0.03, where E is evenness and H is Shannon diversity.
C = 1 − n1/N, where C is coverage, n1 is the number of singletons, and N is the total number of sequences (49).
> indicates that the estimate did not stabilize with regard to sampling effort.
The results for only the 5-m sample are shown for October; the 15-m mixolimnion sample contained a total of 54 clones.
FIG. 2.
Bacterial diversity in Soap Lake. (A) Collector's curve of predicted richness (Chao1) for pooled July and October sequences. The subscript numbers indicate the genetic distance (divergence), and the bold numbers indicate the endpoint estimates and are followed by 95% confidence limits. The vertical lines indicate 95% confidence intervals. (B) Observed and estimated richness (Chao1 and ACE) with divergence levels set at 0 to 80%.
Phylogenetic distribution of the sequences.
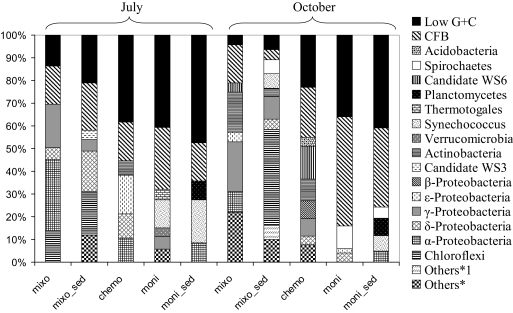
The frequencies of 16S rRNA sequences across different phylogenetic groups in the libraries were calculated for all samples (Fig. 3). For a detailed description of the phylogenetic affiliations of the sequences and corresponding phylogenetic trees, see the supplemental material. The phylogenetic compositions of our clone libraries shifted both with depth and with origin of the sediment. The mixolimnion (5-m) sample was dominated by sequences affiliated with Alpha- and Gammaproteobacteria (30 and 18% of the sequences, respectively), followed by low-G+C-content gram-positive group and Chloroflexi-related sequences (both 13% of the sequences); 36, 42, and 45% of the sequences obtained from the chemocline (20-m), the monimolimnion (23-m), and the monimolimnnion sediment, respectively, were related to sequences belonging to the low-G+C-content group. Sequences related to the Cytophaga-Flexibacter-Bacteroides (CFB) group were also abundant; a total of 16% of the sequences from the mixolimnion, the chemocline, and the monimolimnion sediment libraries were affiliated with this clade, whereas in the mixolimnion sediment and the monimolimnion 28 and 21% of the sequences, respectively, were affiliated with this group. The Gammaproteobacteria were especially prevalent in the mixolimnion library, Chloroflexi-related sequences were present only in the mixolimnion and the mixolimnion sediment, and Epsilonproteobacteria-like sequences were found only in the chemocline library. The other groups detected in the libraries included Deltaproteobacteria, Cyanobacteria (Synechococcus), and various candidate divisions.
FIG. 3.
Frequencies of sequences affiliated with major phylogenetic groups in libraries from the water column and sediments for July and October. Others*, Synechococcus, chloroplast DNA, Alphaproteobacteria, candidate taxon OD1, Actinobacteria, Planctomycetes, and Spirochaetes; Others*1, Planctomycetes, candidate taxon OP1, candidate taxon NKB19, and candidate taxon OD1.
Although not on a sample-to-sample basis, the overall quantitative composition of the libraries from October was different than that of the July libraries. Altogether, 25 and 26% of the sequences retrieved from the 15- and 5-m libraries, respectively, were affiliated with the Gammaproteobacteria. The populations of sequences obtained from the chemocline and the monimolimnion sediment were dominated by sequences related to low-G+C-content gram-positive bacteria (24 and 41%, respectively). The monimolimnion was dominated by sequences related to the CFB group (47%), and the mixolimnion sediment contained mainly sequences distantly related to Chloroflexi-like sequences (47%). Sequences related to the Betaproteobacteria were found only in the chemocline library.
Clone library comparisons.
In order to compare the bacterial communities present in each layer, we conducted a ∫-LIBSHUFF analysis using the individual libraries. For pairwise comparisons between sequences belonging to the mixolimnion and chemocline libraries and between these libraries and the monimolimnion libraries, the P values were <0.001 (data not shown); for comparisons between the monimolimnion libraries within each season, the P values were 0.16 (water versus sediment) and 0.0018 (sediment versus water) for October and 0.013 and 0.087 for July (data not shown). Although it is generally assumed (61, 66) that nonsignificant LIBSHUFF comparisons can reveal whether the community structure of one library is a subset of another library, recent simulations suggest that no conclusions can be drawn when one comparison is significant and the reciprocal comparison is not significant; this probably indicates that the libraries are different (P. D. Schloss, personal communication). Thus, the reported marginally significant P values for monimolimnion water and sediment comparisons indicate that these areas contain significantly different bacterial communities.
A parsimony tree containing all the sequences was used for a parsimony test. The comparison of July with October yielded a P value of 0.034, while for the interlayer (pooled across season) and interlibrary comparisons the P values were <0.001, indicating that there was significant covariation between phylogeny and community type.
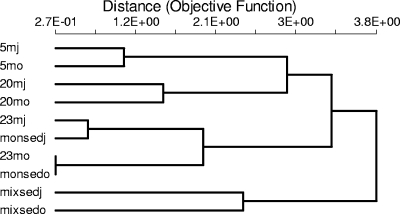
To evaluate the memberships of the communities with predefined OTU definitions, we first applied a SONS community structure similarity metric, θ, using a divergence level of 0.03. A dendrogram of the estimated values supported the notion that communities were endemic to the chemically demarcated layers from which they were obtained (Fig. 4). Season, however, seemed to exert some influence on the differentiation of communities, particularly the communities from the mixolimnion sediments, as suggested by the lengths of the branches joining these communities in the dendrogram (Fig. 4). We then pooled the sequences according to layer for a more detailed analysis of the overlaps between the communities' members. Table 4 shows the estimated calculations at the species and phylum levels. The July and October communities shared a richness (SA,B Chao) of 115 species and 98 phyla. At a D value of 0.03, the estimated fraction of sequences from July that was shared (Uest) was ∼46%, while for October the estimated percentage of sequences belonging to shared species (Vest) was ∼72%. The overall community overlap (Ĵabd) was 0.39, which implies that, if libraries were very large (i.e., exhibited almost complete coverage), about 40% of the sequences in both communities would belong to shared species-level OTUs (56). In general, the overlaps between pairs of layers were low; the highest Ĵabd values were observed for the mixolimnion and the chemocline and for the monimolimnion water and sediment, which was consistent with the θ dendrogram.
FIG. 4.
Dendrogram (constructed by the unweighted-pair group method with arithmetic means) of the pairwise nonparametric estimates of community structure similarity (θ) between Soap Lake samples obtained from the mixolimnion (5m), the chemocline (20m), the monimolimnion (23m), and sediments (mixsed and monsed). The suffixes “j” and “o” indicate July and October, respectively. Distance = 1 − θ.
TABLE 4.
Estimated community overlap of interseason and intersample comparisons for OTUs defined by 0.03 and 0.2 Jukes-Cantor distances units
| Comparison |
Uesta
|
Vesta
|
Ĵabd
|
SA,B Chao
|
||||
|---|---|---|---|---|---|---|---|---|
| D = 0.03 | D = 0.2 | D = 0.03 | D = 0.2 | D = 0.03 | D = 0.2 | D = 0.03 | D = 0.2 | |
| Between seasons (July and October) | 0.46 | 0.87 | 0.72 | 1 | 0.39 | 0.87 | 115 | 98 |
| Between layers within the lake | ||||||||
| Mixolimnion and chemocline | 0.30 | 0.62 | 0.69 | 0.56 | 0.26 | 0.42 | 33 | 16 |
| Mixolimnion and monimolimnion | 0.17 | 0.98 | 0.027 | 0.20 | 0.024 | 0.20 | 5 | 22 |
| Mixolimnion and mixolimnion sediment | 0.043 | 0.20 | 0.35 | 0.40 | 0.040 | 0.15 | 13 | 13 |
| Mixolimnion and monimolimnion sediment | 0.12 | 0.54 | 0.048 | 0.18 | 0.036 | 0.16 | 6 | 14 |
| Chemocline and monimolimnion | 0.19 | 0.61 | 0.17 | 0.93 | 0.10 | 0.58 | 22 | 38 |
| Chemocline and mixolimnion sediment | 0.009 | 0.32 | 0.025 | 0.35 | 0.007 | 0.20 | 1 | 11 |
| Chemocline and monimolimnion sediment | 0.15 | 0.49 | 0.63 | 1.00 | 0.14 | 0.49 | 25 | 28 |
| Monimolimnion and mixolimnion sediment | 0.00 | 0.31 | 0.00 | 0.28 | 0.00 | 0.17 | 0 | 9 |
| Monimolimnion and monimolimnion sediment | 0.29 | 0.77 | 0.64 | 0.80 | 0.25 | 0.65 | 32 | 19 |
| Mixolimnion sediment and monimolimnion sediment | 0.037 | 0.28 | 0.010 | 0.47 | 0.008 | 0.21 | 1 | 28 |
Uest and Vest are the fractions of sequences from the first and second seasons or layers indicated, respectively, that belong to a shared OTU (64).
DISCUSSION
Previous work on Soap Lake's microbiological attributes has emphasized the description of cultivable isolates, including aerobic (14), microaerophilic (67), or phototrophic (3) isolates. Here, we integrated two approaches that not only corroborate what is known about the microbiology of similar ecosystems but also provide evidence of the significance of anaerobic processes as key contributors to prokaryotic productivity (71).
MPN study.
The observed spatial variability corresponded well with the lake's redox gradient; no anaerobic bacteria were found in the mixolimnion, some anaerobic bacteria occurred in the chemocline, and the largest numbers of anaerobic bacteria were observed in anoxic regions, including the mixolimnion sediments. The dominant process underlying terminal carbon mineralization appeared to be sulfate reduction driven by the oxidation of lactate and other organic acids. Indeed, methanogenic growth was not relieved even when we enriched for TMA-consuming cells. These results agree with the results of a study performed by Oremland and Miller (51), who observed little methane production from TMA- or CO2-amended sediments. The authors concluded, as we did, that sulfate reduction is the main anaerobic carbon mineralization process, even for a noncompetitive substrate like TMA. Taken as a whole, the functional groups that we analyzed seemed to display seasonal homogeneity, which suggests that their growth patterns are not limited by phytoplankton “bloom dynamics” in the oxygen-rich mixolimnion. Since allochthonous inputs of organic matter are unlikely, most of the organic carbon supporting anaerobic processes (mainly sulfate reduction, but also iron reduction) may emanate from phototrophically derived DOC pools that are maintained at constant levels throughout the year (Table 1). Although we focused on the culturable fraction, this independence may suggest that there is a decoupling between phytoplankton growth and bacterioplankton production resulting from nutrient limitation (e.g., a low N/P ratio) (38). Alternatively, although not mutually exclusively, anaerobic carbon cycling may be connected to light-independent chemosynthetic growth in anoxic compartments (71).
DGGE analysis.
We are aware of the potential risks of drawing “diversity” conclusions based on DGGE (13). However, the inherent limitations of the technique did not prevent a comparative assessment; DGGE detected abundant members of the targeted communities.
Our results seem to support two conclusions. First, vertical changes in environmental conditions (salinity, redox potential, accumulation of reduced compounds) are associated with an increase in prokaryotic diversity. In contrast, using a DGGE approach, Ovreas et al. (52) and Koizumi et al. (33) reported higher diversity in oxic surface water than in anoxic waters. A possible explanation for this discrepancy is that most of the sequences that these workers found in anoxic waters were present at a very low relative abundances and formed faint bands that were not detected by DGGE, although methodological considerations, such as different primer sets or running conditions, should also be considered. Second, the negative diversity correlations between archaeal and bacterial populations in anaerobic compartments suggest that their relative abundances are tightly linked (for instance, through competition or cross-feeding) and/or are controlled by a set of environmental constraints whose effects are predictable and constant over time (30). Although our knowledge concerning the ecological significance of archaea in lakes is limited, archaeal populations from nonmeromictic soda systems appear to be dominated by Halobacteriales (47), and archaeal nitrification is thought to be quantitatively important in stratified aquatic ecosystems (59).
Clone library analysis.
In agreement with previously reported findings for other meromictic soda lakes (24, 74), an important outcome of our clone library analysis is the shift in the qualitative and quantitative compositions of the bacterial assemblage across the mixolimnion, the chemocline, and the monimolimnion. The divergence in the layers was primarily due to the distribution of two major clades. The clone libraries from the mixolimnion were dominated by sequences belonging to the Proteobacteria, while the clone libraries from the monimolimnion were dominated by sequences affiliated with the low-G+C-content gram-positive bacteria and the CFB group. This trend was observed for both seasons. It is doubtful that the difference in the compositions that we detected was simply due to PCR bias, cloning bias, or other analytical errors, as the changes seemed to be reflected in the DGGE analysis, if only through changes in diversity, and are consistent with well-studied properties of these groups. Not only was the discrepancy due to differences in community structure (LIBSHUFF and parsimony test analyses), but it was also due to the presence of abundant (and perhaps rare for some undersampled groups) OTUs. Although inadequate sampling is a matter of concern with the cloning-sequencing approach, our SONS analyses showed that the number of sequences that we sampled was sufficient to obtain stable estimates. A caveat for SONS-based analyses, however, is that it is not possible to ascertain which clades are responsible for the segregation among samples (64). Selectively deleting data sets to obtain such information is not justified, as it could reduce the statistical power, unless more detailed a priori knowledge about the correspondence between phylogeny and function is obtained (64).
The bottom water of Soap Lake might be considered to be more “extreme” than the surface water because of the potential toxicity of the high concentrations of sulfide and ammonia that have accumulated in the deep, cold water after extended meromixis. Hence, it would be expected that this milieu has relatively limited diversity. The results show, however, that anoxic deep waters were, in both seasons, more diverse than oxic surface waters, which was congruent with our DGGE results. This has also been found to be the case for various cold marine environments, such as the Arctic Ocean (57), saline meromictic lakes of eastern Antarctica (5), and permanently stratified Lake Pavin (39) and Mono Lake (24). Habitats that experience spatially structured changes in redox conditions are often associated with concomitant shifts in diversity (for instance, hypersaline microbial mats [40] and wet alpine soils [12]).
Although the strong physicochemical gradients in Soap Lake may contribute to the overall diversity of the bacterioplankton in the lake, it is unlikely that this component of habitat diversity is responsible for the elevated diversity in the monimolimnion, as the gradients there are weak relative to those in other locations in the water column (the oxycline, for example). It is possible, however, that downward metabolite fluxes across layers contribute to the generation of chemical complexity (43), which, coupled with old, stagnant waters, would allow the maintenance of high diversity levels; indeed, ∼30% of the total estimated richness was comprised of monimolimnion sequences, suggesting that closely related taxa in anaerobic waters represent multiple unique ecotypes with diverse ecophysiological capabilities (25). Visual as well as microscopic inspections indicated that there were elevated concentrations of particles in monimolimnion sediments, which may provide an additional range of potential physical niches for specialized bacterial groups.
Lehours et al. (39), invoking Humayoun et al. (24), proposed that anoxic environments maintain a higher diversity of energetic pathways (for which we obtained preliminary support from the MPN study) and that this complexity permits the retention of higher metabolic and thus ecological diversity. Such a scenario, however, implies, at least, a concordance between functional potential (as determined by the presence of an energetic pathway) and phylogenetic diversity. Because 16S rRNA genes and functional genes evolve at different rates (37), such thesis is untenable. As has been observed in soil (21, 77), it is likely that interspecific and intraspecific competitive interactions are the principal factors that govern the maintenance of diversity in anoxic samples, while oxic samples are subject to a different set of ecological forces, which may include grazing pressure and marked fluctuations in resource supply.
Whereas shifts in bacterial community composition across space were apparent, the temporal variability was less marked. According to our SONS analysis, the July and October sequence collections shared a core set of an estimated 115 species-level OTUs, while phylum-level phylotypes were indistinguishable. The predicted overlap at a genetic distance of 0.03, however, was low (Table 4), which suggested that temporal turnover is also an important vehicle for the generation of diverse “accessory” populations (64) with season-specific ecological roles.
Succession in the phytoplankton community can affect the concentration and biochemical composition of autochthonous organic matter available to bacteria (31, 55). Compositional shifts associated with different phytoplankton-derived DOC compounds have been documented using 16S rRNA gene-based analyses (54). This implies that the primary producers interact with bacteria by introducing variations in the qualitative properties of the compounds that they secrete and thus select for subsets of the prokaryotic community; i.e., different, closely related populations may mediate the turnover of different compounds (53). In Soap Lake, sinking detritus derived from seasonal (spring and fall) blooms of Chlorella sp. and from other phototrophs (71) may provide an important fraction of fermentable or readily utilizable carbon sources, as suggested by the high DOC concentrations found in both the mixolimnion and the monimolimnion (Table 1) and by the presence of fermentative microorganisms (Table 2). If this is the case, it is not known which member(s) of the bacterioplankton is closely coupled to Chlorella production. Under this interpretational framework, the higher prokaryotic diversity observed in July samples may have been associated with a higher diversity (or increase in quantity) of phototrophically derived chemical species; conversely, populations following the progression of phytoplankton blooms are likely exposed to additional short-term constraints (for instance, diel changes [36]) that may become more intense during the summer. Interestingly, the finding that the mean bacterial Shannon index (DGGE), although not the archaeal Shannon index (DGGE), in the chemocline was positively correlated (R2 = 0.71; P = 0.006) with Secchi depth (a measure of overall lake productivity) may indeed suggest that there is seasonal repetition of abundant bacterial assemblages endemic to the chemocline linked to phytoplankton bloom dynamics, to productivity per se, or to both factors. The fact that a significant relationship was only found for the chemocline community is not unexpected since (i) it is known that bacterial groups respond differently to seasonal cycles (for instance, cycles of primary production [72]) and (ii) the chemocline lies in the transitional zone between productivity due to dark carbon assimilation and productivity due to light carbon assimilation (71). Although we did not systematically measure other environmental parameters that could potentially limit the seasonal distribution of prokaryotic assemblages (i.e., the parameters in Table 1), the data that we do have suggest that there is little variation over time.
Interestingly, around 30% of the sequences were closely related (≥95% similarity) to sequences retrieved from Mono Lake, an alkaline (pH 9.8), saline, meromictic soda lake located in California (24). Additionally, although their sequences are not closely related to Soap Lake sequences, previous studies have shown that marinelike, nonalkaliphilic microorganisms tend to dominate libraries retrieved from oxic portions of soda lakes (39, 42, 58). Humayoun et al. (24) found that many of the sequences that they retrieved from anoxic samples clustered with the low-G+C-content group. The cultured representatives that closely matched our sequences, such as Thialkalivibrio, Belliella, Algoriphagus, Tanella, Tindallia, and Rhodobaca, were isolated from environments characterized by physical and chemical parameters that coincide with parameters found in Soap Lake. In general, we found that our sequences were frequently related to sequences found in ecosystems such as other soda lakes, hypersaline and/or cold meromictic lakes, marine and brackish waters, marine sediments, and deep-sea sediments.
Supplementary Material
Acknowledgments
This work was supported by the Microbial Observatories Program of the United States National Science Foundation (grant MCB-0132158).
Footnotes
Published ahead of print on 13 June 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.American Water Works Association, Water Pollution Control Federation, and American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, DC.
- 2.Anderson, G. C. 1958. Seasonal characteristics of two saline lakes in Washington. Limnol. Oceanogr. 3:51-68. [Google Scholar]
- 3.Asao, M., S. Takaichi, and M. T. Madigan. 2007. Thiocapsa imhoffii, sp. nov., an alkaliphilic purple sulfur bacterium of the family Chromatiaceae from Soap Lake, Washington (USA). Arch. Microbiol. 188:665-675. [DOI] [PubMed] [Google Scholar]
- 4.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, J. P., S. A. McCammon, and J. H. Skerratt. 1997. Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology 143:1451-1459. [DOI] [PubMed] [Google Scholar]
- 6.Brosius, J., M. L. Palmer, J. K. Poindexter, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casamayor, E., H. Schafer, L. Baneras, C. Pedros-Alio, and G. Muyzer. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 9.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 10.Chao, A., R. L. Chazdon, R. K. Colwell, and T. J. Shen. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8:148-159. [Google Scholar]
- 11.Cloern, J. E., B. E. Cole, and S. W. Wienke. 1987. Big Soda Lake (Nevada). 4. Vertical fluxes of particulate matter: seasonality and variations across the chemocline. Limnol. Oceanogr. 32:815-824. [Google Scholar]
- 12.Costello, E. K., and S. K. Schmidt. 2006. Microbial diversity in alpine tundra wet meadow soil: novel Chloroflexi from a cold, water-saturated environment. Environ. Microbiol. 8:1471-1486. [DOI] [PubMed] [Google Scholar]
- 13.Crosby, L. D., and C. S. Criddle. 2003. Understanding the bias in microbial community analysis due to rnn operon copy number heterogeneity. BioTechniques 34:790-802. [DOI] [PubMed] [Google Scholar]
- 14.Dimitriu, P. A., S. K. Shukla, J. Conradt, M. C. Márquez, A. Ventosa, A. Maglia, B. M. Peyton, H. C. Pinkart, and M. R. Mormile. 2005. Nitrincola lacisaponensis gen. nov., sp. nov., a novel alkaliphilic bacterium isolated from an alkaline, saline lake. Int. J. Syst. Evol. Microbiol. 55:2273-2278. [DOI] [PubMed] [Google Scholar]
- 15.Dong, H., G. Zhang, H. Jiang, B. Yu, L. R. Chapman, C. R. Lucas, and M. W. Fields. 2006. Microbial diversity in sediments of saline Qinghai Lake, China: linking geochemical controls to microbial ecology. Microb. Ecol. 51:65-82. [DOI] [PubMed] [Google Scholar]
- 16.Duckworth, A. W., W. D. Grant, B. E. Jones, and R. van Steenbergen. 1996. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol. Ecol. 19:181-191. [Google Scholar]
- 17.Edmondson, W. T., and G. C. Anderson. 1965. Some features of saline lakes in central Washington. Limnol. Oceanogr. 10(Suppl.):R87-R96. [Google Scholar]
- 18.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 19.Hakala, A., K. Sarmaja-Korjonen, and A. Miettinen. 2004. The origin and evolution of Lake Vähä-Pitkusta, SW Finland—a multi-proxy study of a meromictic lake. Hydrobiologia 527:85-97. [Google Scholar]
- 20.Hakala, A. 2004. Meromixis as a part of lake evolution—observations and a revised classification of true meromictic lakes in Finland. Boreal Environ. Res. 9:37-53. [Google Scholar]
- 21.Hansel, C. M., S. Fendorf, P. M. Jardine, and C. A. Francis. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 74:1620-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horikoshi, K. 1999. Alkaliphiles: some applications of their products for biotechnology. Microbiol. Mol. Biol. Rev. 63:735-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon, a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 24.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaspers, E., and J. Overmann. 2004. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 70:4831-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen, K. M., and R. P. Cox. 1992. Effects of sulfide and low redox potential on the inhibition of nitrous-oxide reduction by acetylene in Pseudomonas nautica. FEMS Microbiol. Lett. 96:13-17. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, H., H. Dong, G. Zhang, B. Yu, L. R. Chapman, and M. W. Fields. 2006. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl. Environ. Microbiol. 72:3832-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, B. E., W. D. Grant, A. W. Duckworth, and G. G. Owenson. 1998. Microbial diversity of soda lakes. Extremophiles 2:191-200. [DOI] [PubMed] [Google Scholar]
- 29.Joye, S. B., T. L. Connell, L. G. Miller, R. S. Oremland, and J. R. Jellison. 1999. Oxidation of ammonia and methane in an alkaline, saline lake. Limnol. Oceanogr. 44:178-188. [Google Scholar]
- 30.Kassen, R., and P. B. Rainey. 2004. The ecology and genetics of microbial diversity. Annu. Rev. Microbiol. 58:207-231. [DOI] [PubMed] [Google Scholar]
- 31.Kent, A. D., A. C. Yannarell, J. A. Rusak, E. W. Triplett, and K. D. McMahon. 2007. Synchrony in aquatic microbial community dynamics. ISME J. 1:38-47. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 33.Koizumi, Y., H. Kojima, K. Oguri, H. Kitazato, and M. Fukui. 2004. Vertical and temporal shifts in microbial communities in the water column and sediment of saline meromictic Lake Kaiike (Japan), as determined by a 16S rDNA-based analysis, and related to physicochemical gradients. Environ. Microbiol. 6:622-637. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, S., K. Tamura, and N. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 35.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, NY.
- 36.Lau, W. W. Y., R. G. Keil, and E. V. Armbrust. 2007. Succession and diel transcriptional response of the glycolate-utilizing component of the bacterial community during a spring phytoplankton bloom. Appl. Environ. Microbiol. 73:2440-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence, J. G. 2001. Catalyzing bacterial speciation: correlating lateral transfer with genetic headroom. Syst. Biol. 50:479-496. [DOI] [PubMed] [Google Scholar]
- 38.Le, J., J. D. Wehr, and L. Campbell. 1994. Uncoupling of bacterioplankton and phytoplankton production in fresh waters is affected by inorganic nutrient limitation. Appl. Environ. Microbiol. 60:2086-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehours, A.-C., C. Bardot, A. Thenot, D. Debroas, and G. Fonty. 2005. Anaerobic microbial communities in Lake Pavin, a unique meromictic lake in France. Appl. Environ. Microbiol. 71:7389-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ley, R. E., J. K. Harris, J. Wilcox, J. R. Spear, S. R. Miller, B. M. Bebout, J. A. Maresca, D. A. Bryant, M. L Sogin, and N. R. Pace. 2006. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovley, D. R., and E. J. P. Phillips. 1986. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma, Y., W. Zhang, Y. Xue, P. Zhou, A. Ventosa, and W. D. Grant. 2004. Bacterial diversity of the inner Mongolian Baer Soda Lake as revealed by 16S rRNA gene sequence analyses. Extremophiles 8:45-51. [DOI] [PubMed] [Google Scholar]
- 43.MacIntyre, S., K. M. Flynn, R. Jellison, and J. R. Romero. 1999. Boundary mixing and nutrient fluxes in Mono Lake, California. Limnol. Oceanogr. 44:512-524. [Google Scholar]
- 44.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melack, J. M., and P. Kilham. 1974. Photosynthetic rates of phytoplankton in East African alkaline, saline lakes. Limnol. Oceanogr. 19:743-755. [Google Scholar]
- 46.Melack, J. M., and R. Jellison. 1998. Limnological conditions in Mono Lake: contrasting monomixis and meromixis in the 1990s. Hydrobiologia 384:21-39. [Google Scholar]
- 47.Mesbah, N. M., S. H. Abou-El-Ela, and J. Wiegel. 2007. Novel and unexpected prokaryotic diversity in water and sediments of the alkaline, hypersaline lakes of the Wadi An Natrun, Egypt. Microb. Ecol. 54:598-617. [DOI] [PubMed] [Google Scholar]
- 48.Moser, D. P., T. C. Onstott, N. Spoelstra, S. M. Pfiffner, A. Dohnalkova, and J. K. Fredrickson. 2001. Alkaliphilus transvaalensis gen. nov., sp. nov., an extremely alkaliphilic bacterium isolated from a deep South African gold mine. Int. J. Syst. Evol. Microbiol. 51:1245-1256. [DOI] [PubMed] [Google Scholar]
- 49.Mullins, T. D., T. B. Britschgi, R. L. Krest, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:148-158. [Google Scholar]
- 50.Oremland, R. S., and D. G. Capone. 1988. Use of “specific” inhibitors in biogeochemistry and microbial ecology. Adv. Microb. Ecol. 10:285-383. [Google Scholar]
- 51.Oremland, R. S., and L. G. Miller. 1993. Biogeochemistry of natural gases in three alkaline, permanently stratified (meromictic) lakes, p. 439-452. In D. G. Howell (ed.), The future of energy gases (USGS Professional Paper 1570). United States Geological Survey, Washington, DC.
- 52.Ovreas, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pernthaler, J., and R. Amann. 2005. Fate of heterotrophic microbes in pelagic habitats: focus on populations. Microbiol. Mol. Biol. Rev. 69:440-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinhassi, J., F. Azam, J. Hemphälä, R. A. Long, J. Martinez, U. L. Zweifel, and Å. Hagström. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13-26. [Google Scholar]
- 55.Pinhassi, J., M. M. Sala, H. Havskum, F. Peters, O. Guadayol, A. Malits, and C. Marrasé. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Popp, N., M. Schlömann, and M. Mau. 2006. Bacterial diversity in the active stage of a bioremediation system for mineral oil hydrocarbon-contaminated soils. Microbiology 152:3291-3304. [DOI] [PubMed] [Google Scholar]
- 57.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rees, H. C., W. D. Grant, B. E. Jones, and S. Heaphy. 2004. Diversity of Kenyan soda lake alkaliphiles. Extremophiles 8:63-71. [DOI] [PubMed] [Google Scholar]
- 59.Revsbech, N. P., N. Risgaard-Petersen, A. Schramm, and L. P. Nielsen. 2006. Nitrogen transformations in stratified aquatic microbial ecosystems. Antonie van Leeuwenhoek 90:361-375. [DOI] [PubMed] [Google Scholar]
- 60.Rice, C. A., M. L. Tuttle, and P. H. Briggs. 1988. Sulfur speciation, sulfur isotopy, and elemental analyses of water-column, pore water, and sediment samples from Soap Lake, Washington. USGS Open File Report 88-22. United States Geological Survey, Washington, DC.
- 61.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schloss, P. D., and J. Handelsman. 2006. Introducing TreeClimber, a test to compare microbial community structures. Appl. Environ. Microbiol. 72:2379-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scholten, J. C. M., S. B. Joye, J. T. Hollibaugh, and J. C. Murrell. 2005. Molecular analysis of the sulfate reducing and archaeal community in a meromictic soda lake (Mono Lake, California) by targeting 16S rRNA, mcrA, apsA, and dsrAB genes. Microb. Ecol. 50:29-39. [DOI] [PubMed] [Google Scholar]
- 66.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sorokin, D. Y., M. Foti, H. C. Pinkart, and G. Muyzer. 2007. Sulfur-oxidizing bacteria in Soap Lake (Washington State), a meromictic, haloalkaline lake with an unprecedented high sulfide content. Appl. Environ. Microbiol. 73:451-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strickland, J. D. H., and T. R. Parsons. 1972. A practical handbook of seawater analysis, 2nd ed. Bulletin 167. Fisheries Research Board of Canada, Ottawa, Ontario.
- 69.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and G. M. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker, K. F. 1974. The stability of meromictic lakes in central Washington. Limnol. Oceanogr. 19:209-222. [Google Scholar]
- 71.Walker, K. F. 1975. The seasonal phytoplankton cycles of two saline lakes in central Washington. Limnol. Oceanogr. 20:40-53. [Google Scholar]
- 72.Wu, Q. L., and M. W. Hahn. 2006. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ. Microbiol. 8:1660-1666. [DOI] [PubMed] [Google Scholar]
- 73.Yue, J. C., and M. K. Clayton. 2005. A similarity measure based on species proportions. Commun. Stat. Theor. Methods 34:2123-2131. [Google Scholar]
- 74.Zehr, J. P., R. W. Harvey, R. S. Oremland, J. E. Cloern, L. H. George, and J. L. Lane. 1987. Big Soda Lake (Nevada). 1. Pelagic bacterial heterotrophy and biomass. Limnol. Oceanogr. 32:781-793. [Google Scholar]
- 75.Zhilina, T. N., G. A. Zavarzin, F. A. Rainey, E. N. Pikuta, G. A. Osipov, and N. A. Kostrikina. 1997. Desulfonatronovibrio hydrogenovorans gen. nov., sp. nov., an alkaliphilic, sulfate-reducing bacterium. Int. J. Syst. Bacteriol. 47:144-149. [DOI] [PubMed] [Google Scholar]
- 76.Zhilina, T. N., G. A. Zavarzin, F. A. Rainey, V. V. Kevbrin, N. A. Kostrikina, and A. M. Lysenko. 1996. Spirochaeta alkalica sp. nov., Spirochaeta africana sp. nov., and Spirochaeta asiatica sp. nov., alkaliphilic anaerobes from continental soda lakes in central Asia and the East African Rift. Int. J. Syst. Bacteriol. 46:305-312. [DOI] [PubMed] [Google Scholar]
- 77.Zhou, J., B. Xia, D. S. Treves, L.-Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.