Abstract
Multipathogen detection on a single-assay platform not only reduces the cost for testing but also provides data on the presence of pathogens in a single experiment. To achieve this detection, a multipathogen selective enrichment medium is essential to allow the concurrent growth of pathogens. SEL broth was formulated to allow the simultaneous growth of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes. The results were compared to those obtained with the respective individual selective enrichment broths, Rappaport-Vassiliadis (RV) for S. enterica, modified E. coli broth with 20 mg of novobiocin/liter for E. coli O157:H7, and Fraser broth for L. monocytogenes, and a currently used universal preenrichment broth (UPB). The growth of each pathogen in SEL inoculated at 101 or 103 CFU/ml was superior to that in the respective individual enrichment broth, except in the case of RV, in which Salmonella cells inoculated at both concentrations grew equally well. In mixed-culture experiments with cells of the three species present in equal concentrations or at a 1:10:1,000 ratio, the overall growth was proportional to the initial inoculation levels; however, the growth of L. monocytogenes was markedly suppressed when cells of this species were present at lower concentrations than those of the other two species. Further, SEL was able to resuscitate acid- and cold-stressed cells, and recovery was comparable to that in nonselective tryptic soy broth containing 6% yeast extract but superior to that in the respective individual selective broths. SEL promoted the growth of all three pathogens in a mixture in ready-to-eat salami and in turkey meat samples. Moreover, each pathogen was readily detected by a pathogen-specific immunochromatographic lateral-flow or multiplex PCR assay. Even though the growth of each pathogen in SEL was comparable to that in UPB, SEL inhibited greater numbers of nontarget organisms than did UPB. In summary, SEL was demonstrated to be a promising new multiplex selective enrichment broth for the detection of the three most prominent food-borne pathogens by antibody- or nucleic acid-based methods.
Every year, up to 81 million people in the United States suffer from food-borne diseases, and food-borne pathogens continue to be a major public health concern (37, 40). Among the known food-borne pathogens, Salmonella enterica, Listeria monocytogenes, and Escherichia coli O157:H7 are of major concern because of their continued association with highly popular foods such as poultry products, ready-to-eat meats, dairy products, and fruits and vegetables. Above all, these pathogens have very high incidence and mortality rates and have been involved in several recent outbreaks (37). Therefore, the control and prevention of these pathogens are of high priority to improve the safety of the U.S. food supply. The accurate and rapid detection of these three pathogens is essential, and testing is sometimes mandatory before certain food items can be distributed for retail sale for human consumption.
Though the sensitivities of many of the modern detection methods, such as antibody-, nucleic acid-, and biosensor-based methods, have improved significantly (6, 23, 38, 46), an enrichment step is still needed. This step is required not only to increase the target-pathogen concentration in a sample but also to resuscitate physiologically stressed or injured cells. Selective enrichment is also necessary to suppress the natural background microorganisms so as to improve detection efficiency and to avoid false results. However, the drawbacks of some of the selective enrichment broths are that the selective agents can be inhibitory or can delay the recovery and growth of healthy or stressed target pathogens (26) and may also down regulate antigen expression, thus affecting the detection of pathogens (21, 24, 34).
Current research trends emphasize the development of multipathogen platforms in a single-assay format. For example, multiplex PCR assays (5, 20, 30, 42), protein/antibody microarray biosensors (35, 50), array-based immunosorbent assays (14), and DNA microarray methods (15) continue to be developed. The multipathogen detection approach is attractive and economically favorable since it can reduce the total space requirement for handling a large number of samples, as well as the bench space, supplies, reagents, and labor needed, thus reducing the overall cost of testing per pathogen. Furthermore, multiplex detection is a rational approach since many foods, such as milk and dairy products (1), meat and poultry (16, 45), and fruits and vegetables (4, 10), are common carriers of S. enterica, E. coli O157:H7, and L. monocytogenes. Moreover, multipathogen detection can mitigate the industry and regulatory needs for the testing of foods that have a high risk of contamination with these pathogens.
To facilitate multipathogen detection in a single-assay format, a suitable enrichment medium is urgently needed. A universal preenrichment broth (UPB) for multipathogen enrichment (2) is commercially available from Difco Lab, Sparks, MD; however, this medium lacks inhibitory agents to provide selectivity for target pathogens and, thus, may not be suitable for samples with high levels of background microflora, such as raw or unprocessed samples from animal and plant origins. Thus, the objectives of this study were to formulate a single medium that can support the simultaneous growth primarily of three food-borne pathogens, S. enterica, E. coli O157:H7, and L. monocytogenes, if present in a single sample and to demonstrate the performance of the medium by employing an antibody-based immunochromatographic lateral-flow assay (ICLFA) and a multiplex PCR assay. The multipathogen medium, designated SEL (for Salmonella, Escherichia, and Listeria), was developed in this study, and its performance as an enrichment broth was verified by growing three pathogens in various proportions and detecting the bacteria by using ICLFA and multiplex PCR. The spectra of growth of target and nontarget bacteria obtained from our collection, as well as natural isolates from food, in SEL were also determined. Next, the ability of SEL to resuscitate acid- or cold-stressed bacteria was investigated. Finally, the performance of SEL was examined and compared with that of UPB by testing pathogen-inoculated meat samples.
MATERIALS AND METHODS
Bacterial cultures and growth conditions.
E. coli O157:H7 EDL 933, L. monocytogenes V7 (serotype 1/2a), and S. enterica serovar Enteritidis phage type 1 (PT1) cultures were used as standard reference cultures in all the studies and were maintained on brain heart infusion (BHI; Accumedia, Lansing, MI) agar plates at 4°C. Fresh cultures were prepared by inoculating tryptic soy broth containing 0.6% yeast extract (TSBYE; Difco Lab, Sparks, MD) at 37°C. The other organisms used in this study are listed in Table 1 and were maintained similarly, except for lactic acid bacteria, which were grown and maintained in deMann Rogosa Sharpe (MRS) broth and on MRS agar (both from Difco).
TABLE 1.
Growth spectra of food-borne bacteria in multipathogen enrichment broth SEL
| Organism | Sourcea | Ribotyping result | Growth in SEL (mean OD595 ± SD) at:
|
Growth in UPB (mean OD595 ± SD) at:
|
||||
|---|---|---|---|---|---|---|---|---|
| 12 h | 16 hc | 24 h | 12 h | 16 hc | 24 h | |||
| Target pathogens | ||||||||
| S. enterica | ||||||||
| Serovar Enteritidis PT1 | Our collection | DUP-2035 | 1.05 ± 0.02 | 1.09 ± 0.03 A | 1.09 ± 0.03 | 0.62 ± 0.02 | 0.80 ± 0.07 B | 0.93 ± 0.07 |
| Serovar Kentucky 1271-94 | Our collection | NTb | 0.95 ± 0.04 | 1.05 ± 0.02 A | 1.07 ± 0.01 | 0.50 ± 0.04 | 0.70 ± 0.07 B | 0.85 ± 0.11 |
| Serovar Tennessee 825-94 | Our collection | NT | 0.99 ± 0.01 | 1.12 ± 0.01 A | 1.19 ± 0.03 | 0.47 ± 0.05 | 0.73 ± 0.05 B | 0.88 ± 0.05 |
| Serovar Typhimurium | Our collection | DUP-1167 | 1.01 ± 0.03 | 1.10 ± 0.01 A | 1.15 ± 0.01 | 0.72 ± 0.02 | 0.84 ± 0.04 B | 1.00 ± 0.06 |
| E. coli | ||||||||
| O157:H7 G5303 (EHEC) | CDC | NT | 0.98 ± 0.04 | 1.01 ± 0.04 A | 0.96 ± 0.03 | 0.61 ± 0.00 | 0.80 ± 0.02 B | 0.86 ± 0.06 |
| O157:H7 G5324 (EHEC) | CDC | NT | 0.98 ± 0.09 | 1.08 ± 0.04 A | 0.97 ± 0.09 | 0.65 ± 0.05 | 0.77 ± 0.05 B | 0.95 ± 0.02 |
| O157:H7 C7927 (EHEC) | Apple cider | NT | 1.03 ± 0.04 | 1.04 ± 0.06 A | 1.03 ± 0.06 | 0.63 ± 0.01 | 0.76 ± 0.06 B | 0.89 ± 0.02 |
| O25:K98:NM (ETEC) | M. Donnenberg | DUP-18656 | 0.04 ± 0.02 | 0.37 ± 0.04 B | 0.52 ± 0.06 | 0.62 ± 0.06 | 0.77 ± 0.08 A | 0.88 ± 0.13 |
| O78:H11 (ETEC) | M. Donnenberg | DUP-19199 | 0 | 0 B | 0 | 0.63 ± 0.06 | 0.82 ± 0.00 A | 0.94 ± 0.12 |
| O127:H6 ATCC 35401 (EPEC) | ATCC | DUP-3017 | 0.96 ± 0.09 | 1.03 ± 0.06 A | 1.00 ± 0.10 | 0.67 ± 0.06 | 0.75 ± 0.09 B | 0.87 ± 0.06 |
| O142:H6 ATCC 43886 (EPEC) | ATCC | NT | 0.09 ± 0.02 | 0.29 ± 0.13 B | 0.40 ± 0.06 | 0.63 ± 0.00 | 0.71 ± 0.00 A | 0.87 ± 0.00 |
| K-12 (nonpathogenic) | Our collection | NT | 1.05 ± 0.07 | 1.13 ± 0.05 A | 1.11 ± 0.09 | 0.63 ± 0.10 | 0.80 ± 0.08 B | 0.86 ± 0.09 |
| L. monocytogenes | ||||||||
| V7 (1/2a) | FDA (dairy) | DUP-1039 | 0.09 ± 0.02 | 0.69 ± 0.05 A | 0.91 ± 0.01 | 0.13 ± 0.01 | 0.47 ± 0.01 B | 0.42 ± 0.01 |
| Scott A (4b) | FDA (human) | DUP-1042 | 0.07 ± 0.00 | 0.73 ± 0.01 A | 1.03 ± 0.00 | 0.40 ± 0.02 | 0.46 ± 0.01 B | 0.44 ± 0.01 |
| F4244 (4b) | CDC (human) | DUP-1044 | 0.03 ± 0.00 | 0.59 ± 0.01 A | 0.94 ± 0.05 | 0.36 ± 0.08 | 0.36 ± 0.05 B | 0.31 ± 0.05 |
| F4260 (1/2b) | CDC (human) | DUP-1042 | 0.13 ± 0.00 | 0.94 ± 0.01 A | 1.09 ± 0.02 | 0.43 ± 0.00 | 0.42 ± 0.01 B | 0.41 ± 0.00 |
| Listeria innocua F4248 | CDC | DUP-1006 | 0.19 ± 0.02 | 0.97 ± 0.02 A | 1.19 ± 0.02 | 0.43 ± 0.00 | 0.43 ± 0.00 B | 0.42 ± 0.00 |
| Nontarget bacteria | ||||||||
| Bacillus cereus MS1-9 | J. Handelsman | DUP-12561 | 0 | 0 | 0 | 0.40 ± 0.01 | 0.43 ± 0.00 A | 0.48 ± 0.00 |
| Bacillus megaterium ATCC 6633 | ATCC | DUP-12551 | 0 | 0 | 0 | 0.39 ± 0..00 | 0.47 ± 0.00 A | 0.57 ± 0.01 |
| Bacillus subtilis ATCC 9885 | ATCC | DUP-16973 | 0 | 0 | 0 | 0.21 ± 0.02 | 0.18 ± 0.00 A | 0.18 ± 0.00 |
| Enterobacter aerogenes | Our collection | DUP-14591 | 1.11 ± 0.04 | 1.17 ± 0.03 A | 1.23 ± 0.01 | 0.82 ± 0.03 | 0.93 ± 0.01 B | 1.14 ± 0.10 |
| Enterococcus faecalis ATCC 344 | ATCC | NT | 0 | 0.03 ± 0.01 B | 0.03 ± 0.01 | 0.45 ± 0.01 | 0.54 ± 0.01 A | 0.48 ± 0.00 |
| Hafnia alvei | Our collection | DUP-18066 | 0.57 ± 0.11 | 0.09 ± 0.06 B | 0.80 ± 0.07 | 0.62 ± 0.06 | 0.74 ± 0.06 A | 0.86 ± 0.08 |
| Streptococcus mutans ATCC 25175 | ATCC | NT | 0.32 ± 0.01 | 0.89 ± 0.10 A | 0.84 ± 0.07 | 0.51 ± 0.01 | 0.66 ± 0.01 B | 0.80 ± 0.01 |
| Pseudomonas aeruginosa ATCC 10145 | ATCC | DUP-11042 | 0.15 ± 0.04 | 0.29 ± 0.02 A | 0.37 ± 0.06 | 0.09 ± 0.05 | 0.28 ± 0.01 A | 0.56 ± 0.01 |
| Proteus vulgaris | Our collection | DUP-10074 | 0 | 0 B | 0 | 0.34 ± 0.00 | 0.54 ± 0.04 A | 0.65 ± 0.03 |
| Brochothrix thermosphacta | Our collection | NT | 0 | 0 | 0 | 0 | 0 | 0 |
| Serratia marcescens | Our collection | NT | 0.03 ± 0.02 | 0.21 ± 0.06 B | 0.83 ± 0.06 | 0.51 ± 0.01 | 0.70 ± 0.07 A | 0.71 ± 0.05 |
| Lactobacillus acidophilus ATCC 4356 | ATCC | NT | NT | NT | 0 | NT | NT | 0 |
| Lactobacillus casei KCTC 3109 | KCTC | NT | NT | NT | 0 | NT | NT | 0.068 ± 0.00 |
| Lactobacillus rhamnosus GG ATCC 53103 | ATCC | NT | NT | NT | 0 | NT | NT | 0.07 ± 0.00 |
| Lactobacillus plantarum NCDO955 | NCDO | NT | NT | NT | 0 | NT | NT | 0.07 ± 0.00 |
| Lactococcus lactis subsp. lactis ATCC 11454 | ATCC | NT | NT | NT | 0 | NT | NT | 0.14 ± 0.00 |
| Pediococcus sp. | Our collection | NT | NT | NT | 0 | NT | NT | 0.00 ± 0.00 |
| Leuconostoc mesenteroides | Our collection | NT | NT | NT | 0 | NT | NT | 0.03 ± 0.00 |
| Natural food isolates | ||||||||
| Bacillus megaterium HK1 | This study | DUP-6058 | 0 | 0 B | 0 | 0.40 ± 0.00 | 0.50 ± 0.04 A | 0.59 ± 0.06 |
| Staphylococcus epidermidis HK7 | This study | DUP-4123 | 0.21 ± 0.04 | 0.74 ± 0.04 A | 0.65 ± 0.00 | 0.48 ± 0.09 | 0.69 ± 0.00 A | 0.82 ± 0.02 |
| Enterobacter cloacae HK8 | This study | DUP-15301 | 1.06 ± 0.04 | 1.15 ± 0.05 A | 1.13 ± 0.07 | 0.75 ± 0.01 | 0.85 ± 0.00 B | 0.98 ± 0.00 |
| Lactococcus lactis subsp. lactis HK21 | This study | DUP-12773 | 0 | 0 B | 0 | 0.36 ± 0.00 | 0.33 ± 0.00 A | 0.32 ± 0.00 |
| Pediococcus acidilactici HK32 | This study | DUP-5600 | 0 | 0 B | 0 | 0.11 ± 0.01 | 0.10 ± 0.00 A | 0.21 ± 0.00 |
FDA, Food and Drug Administration; CDC, Centers for Disease Control and Prevention; ATCC, American Type Culture Collection; KCTC, Korean Culture Type Collection; NCDO, National Collection of Dairy Organisms; J. Handelsman, University of Wisconsin, Madison; M. Donnenberg, University of Maryland, Baltimore.
NT, not tested.
OD595 readings for growth in SEL and UPB at the 16-h time point that are in the same row and labeled with the same letter (A or B) are not significantly different at P of <0.05.
The individual selective enrichment broths and plating agars (all purchased from Difco) used in this study included modified E. coli broth with 20 mg of novobiocin/liter (mEC+n) and sorbitol MacConkey agar with cefixime-tellurite (CT-SMAC) for E. coli O157:H7, Rappaport-Vassiliadis (RV) broth and xylose-lysine-deoxycholate (XLD) agar for Salmonella serovars Enteritidis and Typhimurium, and Fraser broth (FB) and modified Oxford agar (MOX) for L. monocytogenes. The cefixime-tellurite supplement was purchased from BioMerieux (Hazelwood, MO), and UPB was purchased from Difco Lab.
Formulation of multipathogen selective enrichment broth SEL.
Commercially available buffered Listeria enrichment broth base (BLEB; Difco Laboratories) without an antibiotic supplement was used as a base medium for the development of the multipathogen enrichment broth SEL. Four antimicrobial agents, acriflavine (ICN Biomedical Inc., Aurora, OH) and cycloheximide, fosfomycin, and nalidixic acid (all purchased from Sigma, St. Louis, MO), were used as selective agents. The concentration of each to be used for SEL formulation was optimized by growing all three pathogens separately in a series of growth curve experiments (31). The final composition of the SEL medium is presented in Table 2.
TABLE 2.
Composition of SEL (Salmonella, Escherichia, Listeria) broth
| Ingredient | Amt (g per liter) | Comment |
|---|---|---|
| Pancreatic digest of casein | 17 | Same as in BLEB |
| Yeast extract | 6 | Same as in BLEB |
| Dextrose | 2.5 | Same as in BLEB |
| Soytone | 3 | Same as in BLEB |
| Sodium chloride | 5 | Same as in BLEB |
| Monopotassium phosphate | 1.35 | Same as in BLEB |
| Dipotassium phosphate | 2.5 | Same as in BLEB |
| Disodium phosphate | 9.6 | Same as in BLEB |
| Sodium pyruvate | 1.1 | Same as in BLEB |
| Acriflavine | 0.01 | Modification of BLEB recipe |
| Cycloheximide | 0.05 | Modification of BLEB recipe |
| Fosfomycin | 0.05 | Newly added (not in BLEB) |
| Nalidixic acid | 0.002 | Modification of BLEB recipe |
Antibody-based ICFLA.
Widely used antibody-based ICLFA kits were employed to verify the antibody-based detection of target pathogens following enrichment in SEL. Reveal kits (Neogen Corp., Lansing, MI) for Salmonella, E. coli O157:H7, and Listeria were used for verification. The Reveal kits for E. coli O157:H7 and Salmonella allow the testing of samples without heat treatments, while the Reveal kit for Listeria recommends a heat treatment (80°C for 20 min) prior to testing. Briefly, following the growth of test organisms in SEL, 120-μl aliquots of E. coli and Salmonella samples and 135-μl heat-inactivated L. monocytogenes samples were dispensed into the sample ports of the ICLFA strips, and the strips were incubated at room temperature for 15 to 20 min. Positive antibody reactions (indicated by the appearance of a dark band in the viewing window) were recorded by capturing digital images, and the reaction intensities were quantified by using a densitometer software program (Scion Corp., Frederick, MD). As controls, the procedures recommended by the manufacturer (Neogen Corp.) for each pathogen were used.
Multiplex PCR.
Multiplex PCR assays were employed to verify whether SEL could be used as an enrichment broth for PCR-based detection of pathogens. DNA was extracted from 1 ml of each culture by using DNA extraction kits (DNeasy tissue kits; catalog no. 69506) per the instructions of the manufacturer (Qiagen, Valencia, CA). The primer sequences and the putative product sizes for each amplicon are listed in Table 3. The PuReTaq ready-to-go PCR beads (GE Healthcare, Piscataway, NJ) were used for PCR amplification (39). PCR mixtures (25 μl) each contained 1 μg of each DNA template, 15 pmol of each primer, and one PuReTaq PCR bead containing 2.5 U of PuReTaq polymerase, 200 μmol of each deoxynucleoside triphosphate, 10 mM Tris-HCl, 50 mM KCl, and 1.5 nM MgCl2. After the initial DNA denaturation at 94°C for 3 min, 40 amplification cycles consisting of 1 min of denaturation at 94°C, 1.5 min of annealing at 60°C, and 1.5 min of elongation at 72°C were done in a thermal cycler (MJ Research, Watertown, MA). Amplified DNA products were detected in agarose gel (1.5%, wt/vol) containing 1 μg of ethidium bromide/ml.
TABLE 3.
Oligonucleotide primers used in the multiplex PCR
| Pathogen | Target virulence factor | Target gene | Primerb | Sequence (5′ to 3′) | Product size (bp) | Reference |
|---|---|---|---|---|---|---|
| Salmonella serovar | Salmonella serovar Enteritidis | sefA | F | GCAGCGGTTACTATTGCAGC | 310 | 53 |
| Enteritidis | fimbrial antigen | R | TGTGACAGGGACATTTAGCG | |||
| Salmonella plasmid virulence | spva | F | GCCGTACACGAGCTTATAGA | 250 | 48 | |
| factor | R | ACCTACAGGGGCACAATAAC | ||||
| E. coli O157:H7 | Attachment and effacement | eaeA | F | TCAATGCAGTTCCGTTATCAGTT | 482 | 52 |
| R | GTAAAGTCCGTTACCCCAACCTG | |||||
| Shiga-like toxin 1 | stx1 | F | CAGTTAATGTGGTGGCGAAGG | 348 | 13 | |
| R | CACCAGACAATGTAACCGCTG | |||||
| Shiga-like toxin 2 | stx2 | F | ATCCTATTCCCGGGAGTTTACG | 584 | 13 | |
| R | GCGTCATCGTATACACAGGAGC | |||||
| L. monocytogenes | Actin polymerization protein | actA | F | GACGAAAATCCCGAAGTGAA | 385 | 27 |
| R | CTAGCGAAGGTGCTGTTTCC | |||||
| Internalin B | inlB | F | AAAGCACGATTTCATGGGAG | 146 | 17 | |
| R | ACATAGCCTTGTTTGGTCGG |
spv-specific primer sets are designated S1 and S4 in reference 50.
F, forward; R, reverse.
Growth kinetics of individual target pathogens in SEL.
To examine the growth of target pathogens in SEL, two inoculation levels, 101 and 103 CFU/ml, were chosen. Volumes of 100 ml of SEL were inoculated with freshly grown Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes and incubated at 37°C in a shaking incubator (Edison, New Brunswick, NJ) set at 150 rpm. The growth rates were determined by enumerating bacterial cells at every 2-h interval by plating the cells onto BHI agar plates. At the same time, the growth of target pathogens in the respective specific selective enrichment broths, RV broth for Salmonella serovar Enteritidis, mEC+n for E. coli O157:H7, and FB for L. monocytogenes, was also evaluated. The Gompertz equation (47) was used to compare the growth kinetics of the different pathogens in SEL. ICLFA and PCR assays were used to evaluate the medium performance. These experiments were repeated twice.
Growth kinetics of target pathogens in a mixture.
Four different combinations of initial cell numbers were used to examine the growth kinetics of each pathogen in SEL. In experiment I, equal concentrations of Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes cultures (ca. 3 × 102 CFU of each pathogen/ml) were inoculated into 100 ml of SEL. In experiments II through IV, the ratio of the cultures used as inocula was set at 1:10:1,000, with the proportion of each culture varying in order throughout the different experiments. In experiment II, the inocula contained Salmonella serovar Enteritidis cells at a mean concentration ± a standard deviation (SD) of 13.5 ± 1.1 CFU/ml, E. coli at 1,327 ± 166 CFU/ml, and L. monocytogenes at 1.3 ± 0.6 CFU/ml. In experiment III, the mixture consisted of Salmonella serovar Enteritidis at 1.4 ± 0.1 CFU/ml, E. coli at 14.6 ± 1.6 CFU/ml, and L. monocytogenes at 1,180 ± 125 CFU/ml, and in experiment IV, Salmonella serovar Enteritidis at 1,178 ± 124 CFU/ml, E. coli at 1.3 ± 0.1 CFU/ml, and L. monocytogenes at 11.5 ± 1.9 CFU/ml were used. The inoculated SEL broths (100 ml each) were incubated at 37°C for 24 h in a shaking incubator, and samples were withdrawn every 2 h. The cell counts for each pathogen were determined by plating the cells onto plates with the appropriate selective agar: XLD agar for Salmonella serovar Enteritidis, CT-SMAC for E. coli O157:H7, and MOX for L. monocytogenes. The lateral-flow immunoassay and multiplex PCR assays were performed with culture samples taken at 16 to 18 h of growth to determine if a pathogen-specific detection assay could be employed for the detection of individual pathogens from a mixed culture. These experiments were repeated three times with two replicates per trial.
In a separate experiment, several samples of ready-to-eat sliced turkey meat (25 g each; see below for details on meat sample procurement and the preparation procedure) were inoculated with bacterial mixtures as listed above for experiments I to IV, enriched with 225 ml of SEL for 24 h, and analyzed by multiplex PCR as described above.
Isolation of resident bacteria from food.
Bacterial isolates were obtained from ready-to-eat meats. A total of two pieces of roasted turkey breast and three Genoa salamis (218 g each) were procured from several different local grocery stores (West Lafayette, IN). Each meat sample (25 g) was homogenized in 225 ml of 20 mM phosphate-buffered saline (pH 7.0), dilutions were plated onto BHI or MRS agar plates, and the plates were incubated at 37°C. Colonies were randomly picked and identified by metabolic fingerprinting using the BioLog culture identification system (Hayward, CA) or by ribotyping (27) employing an automated RiboPrinter (Qualicon, Wilmington, DE). Five selected isolates were used in this study (Table 1).
Growth profiles of food-borne microorganisms in SEL.
To investigate the spectra of bacterial growth in SEL, several found food-borne pathogens and spoilage and resident bacterial isolates (Table 1) were inoculated (ca. 103 CFU/ml) into 10 ml of SEL and incubated at 30 or 37°C with agitation (100 rpm). Aliquots of 1.0 ml of each culture were withdrawn at 12, 16, and 24 h into polystyrene disposable cuvettes, and the growth was monitored by measuring the absorbance at 595 nm in a DU-640 spectrophotometer (Beckman-Coulter). This experiment was performed three times with six replicates per trial. At the same time, bacterial growth in UPB and the respective specific selective enrichment broths, RV broth, mEC+n, and FB, under similar conditions was examined.
Recovery of cold- or acid-stressed bacteria in SEL.
The abilities of SEL to resuscitate stress-exposed bacterial cells and enrich samples were investigated. The two most common stress conditions, exposure to acid (pH 4.5 and 5.5) and cold (4°C) (24), were examined. Each freshly grown culture of Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes was inoculated into 30 ml of TSBYE (1%, vol/vol) and then incubated at 37°C in a shaker incubator (150 rpm) to mid-log phase (see Fig. 1): 2 h for E. coli O157:H7, 4 h for L. monocytogenes, and 2.5 h for Salmonella serovar Enteritidis. Aliquots (5 ml each) were centrifuged (5,000 × g for 10 min) and washed once with 30 ml of phosphate-buffered saline, and the cell pellets were resuspended and held for 3 h in 5 ml of TSBYE with the appropriate stressors: (i) TSBYE adjusted to pH 4.5 and (ii) TSBYE adjusted to pH 5.5 by using 1 M lactic acid and (iii) TSBYE at 4°C (precooled TSBYE was used). The cells exposed to acid stress were incubated at 37°C, and cold-stressed cells were incubated at 4°C. Each stress-exposed culture (1%, vol/vol) was transferred into SEL, TSBYE, or the corresponding individual selective enrichment broth and incubated for 3 h (short recovery) and 6 h (long recovery) at 37°C in a shaking incubator. Bacterial cell counts immediately after the exposure to stress and after 3 and 6 h of recovery in different media were determined by surface plating of cells onto BHI agar plates (1, 29).
FIG. 1.
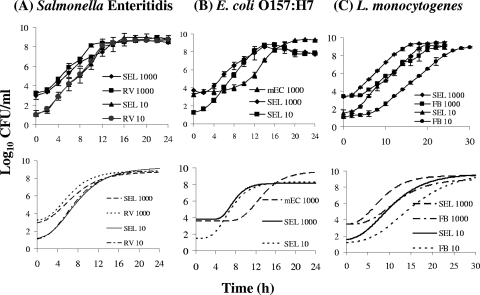
Growth curves for the individual pathogens Salmonella serovar Enteritidis (A), E. coli O157:H7 (B), and L. monocytogenes (C) in SEL inoculated at two different concentrations (10 and 1,000 CFU/ml). The growth of each pathogen in SEL was compared with that in the respective individual selective enrichment broth: RV broth for Salmonella, mEC+n for E. coli, and FB for Listeria. The broths were inoculated at the indicated concentrations, and the cultures were incubated at 37°C in a shaker incubator. The top panels show the actual growth curves, and the plots in the bottom panels are corresponding Gompertz fitted curves.
Comparative enrichment of artificially inoculated meat samples with pathogens in SEL and UPB broth and subsequent detection by ICLFA and PCR.
Several 218-g portions of ready-to-eat deli meats (roasted turkey breast and Genoa salami) were purchased from local grocery stores in West Lafayette, IN. The turkey breast samples had 5% fat and 15 g of protein per 56-g serving, and the salami samples had 28% fat and 21 g of protein per 56-g serving. The absence of Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes in each meat sample was confirmed by using standard procedures as outlined in the Bacteriological Analytical Manual (18) before the initiation of the challenge study. Twenty-five grams of each meat sample was placed into a stomacher bag containing an inner filter lining (Whirl-Pak [catalog no. B01318; Nasco Fort Atkinson, WI]). The meat samples were inoculated with approximately 3 × 102 CFU of each culture/g and held at room temperature for 15 min to allow bacterial adsorption. Then, a 225-ml volume of SEL or UPB was added to each bag, and the samples were blended for 2 min by using a Stomacher 400 (Seward, Norfolk, United Kingdom). The homogenized meat samples were incubated at 37°C for 24 h. Uninoculated meat samples (25 g of meat in 225 ml of SEL) served as negative controls. After 8, 10, 12, 16, and 24 h of incubation, 5-ml aliquots were collected from each bag, serially diluted in 0.1% sterile peptone water, and analyzed for microbial counts by being plated onto the corresponding selective agar plates. Samples were also tested by PCR and lateral-flow immunoassay as described above. In a separate experiment, the influence of different meat samples (two different brands of salami and two brands of turkey) on pathogen enrichment in SEL was evaluated as described above.
Gompertz equation and statistical analyses.
To determine the exponential growth rate (EGR), generation time (GT), lag-phase duration (LPD), and maximum population density (MPD), the growth of each bacterium in SEL was modeled with the Gompertz equation (47) by using a nonlinear mixed model with PROC NLMIXED in software version 9.1 for Windows (SAS Institute Inc., Cary, NC). To test for differences among the broths in the comparison experiments, the statistical significance was assessed by a t test; a P value of <0.05 was considered significant.
RESULTS
Growth kinetics of individual target pathogens in SEL. (i) Salmonella serovar Enteritidis.
The growth of Salmonella serovar Enteritidis in SEL was compared with that in RV broth. Both media were inoculated with 101 and 103 CFU/ml. Data extrapolated from the fitted Gompertz curves indicated that the average EGRs, GTs, LPDs, and MPDs for the two broths at the two inoculation levels were comparable (Fig. 1A; see Table S1 in the supplemental material), suggesting that the performance of SEL was equivalent to that of RV broth.
(ii) E. coli O157:H7.
The E. coli O157:H7 growth rate in SEL was also examined and compared with that in mEC+n after both media were inoculated with 101 and 103 CFU/ml. The first distinguishable result was that no growth of E. coli O157:H7 inoculated at 101 CFU/ml into mEC+n was observed, whereas SEL supported growth at that inoculation level (Fig. 1B). Data extrapolated from the fitted Gompertz curves indicated that the average EGR in SEL inoculated with 103 CFU/ml (0.89 log10 CFU/ml/h) was significantly (P < 0.05) higher than that in mEC+n inoculated with the same concentration (0.73 log10 CFU/ml/h) and that the GT and LPD in SEL were shorter than those in mEC+n. However, the MPD in mEC+n was greater than that in SEL (see Table S1 in the supplemental material). Overall, these data indicate that E. coli O157:H7 had a higher growth rate but reached a lower maximum cell population in SEL than in mEC+n.
(iii) L. monocytogenes.
At both inoculation levels (101 and 103 CFU/ml), L. monocytogenes growth in SEL was significantly better than that in FB (Fig. 1C). Though the EGRs and MPDs in the two media were comparable, the GT and LPD in SEL were significantly shorter than those in FB (see Table S1 in the supplemental material).
Growth of the three target pathogens in a mixture in SEL. (i) Experiment I: Salmonella serovar Enteritidis/E. coli O157:H7/L. monocytogenes culture ratio, 1:1:1.
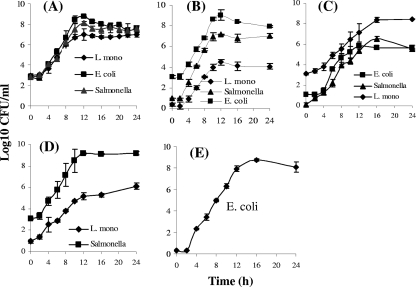
In a mixture (containing ca. 3 × 102 CFU of each pathogen/ml), the three pathogens grew well and showed similar growth patterns (Fig. 2A). The values extrapolated from Gompertz fitted curves indicated that the EGR of L. monocytogenes (0.72 CFU/ml/h) was the lowest, followed by those of Salmonella serovar Enteritidis (0.82 CFU/ml/h) and E. coli O157:H7 (1.10 CFU/ml/h). Of the three pathogens, E. coli O157:H7 exhibited the shortest GT and LPD, 0.68 and 3.21 h, compared to 0.84 and 3.64 h for Salmonella serovar Enteritidis and 0.96 and 3.48 h for L. monocytogenes, respectively (Table 4). Furthermore, E. coli cells had a higher MPD than Salmonella serovar Enteritidis and L. monocytogenes cells (Table 4). In summary, these data indicate that SEL is capable of supporting the concurrent growth of Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes but with a lower growth rate for L. monocytogenes than for the other species when cells of the three species are present at equal initial concentrations.
FIG. 2.
(A to D) Growth curves for the three pathogens Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes (L. mono) mixed at various ratios in SEL: Salmonella CFU/E. coli CFU/L. monocytogenes culture ratios, 1:1:1 (A), 10:1,000:1 (B), and 1:10:1,000 (C), and Salmonella CFU/L. monocytogenes culture ratio, 1,000:10 (D). (E) Growth curve for E. coli O157:H7 alone after inoculation at ∼1 CFU/ml.
TABLE 4.
Growth kinetics valuesa for Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes in mixed cultures set up in SEL
| Expt | Organism (CFU/ml)b | EGR (log10 CFU/ml/h) | GT (h) | LPD (h) | MPD (log10 CFU/ml) |
|---|---|---|---|---|---|
| I | Salmonella serovar Enteritidis (1) | 0.82 ± 0.10 | 0.84 ± 0.10 | 3.64 ± 0.58 | 7.66 ± 0.08 |
| E. coli O157:H7 (1) | 1.10 ± 0.11 | 0.68 ± 0.07 | 3.21 ± 0.42 | 8.01 ± 0.10 | |
| L. monocytogenes (1) | 0.72 ± 0.06 | 0.96 ± 0.08 | 3.48 ± 0.33 | 6.89 ± 0.05 | |
| II | Salmonella serovar Enteritidis (10) | 1.04 ± 0.06 | 0.67 ± 0.04 | 2.70 ± 0.24 | 7.11 ± 0.09 |
| E. coli O157:H7 (1,000) | 0.96 ± 0.09 | 0.72 ± 0.07 | 3.02 ± 0.41 | 8.58 ± 0.14 | |
| L. monocytogenes (1) | 0.71 ± 0.07 | 0.98 ± 0.10 | 3.70 ± 0.38 | 4.28 ± 0.09 | |
| III | Salmonella serovar Enteritidis (1) | 0.65 ± 0.05 | 1.07 ± 0.08 | 2.67 ± 0.53 | 6.25 ± 0.19 |
| E. coli O157:H7 (10) | 1.00 ± 0.09 | 0.69 ± 0.06 | 3.56 ± 0.24 | 5.71 ± 0.09 | |
| L. monocytogenes (1,000) | 0.52 ± 0.04 | 1.33 ± 0.10 | 3.01 ± 0.61 | 8.44 ± 0.09 | |
| IV | Salmonella serovar Enteritidis (1,000) | 0.79 ± 0.07 | 0.88 ± 0.08 | 2.17 ± 0.54 | 9.33 ± 0.18 |
| E. coli O157:H7 (1) | NDc | NAd | NA | NA | |
| L. monocytogenes (10) | 0.47 ± 0.02 | 1.46 ± 0.08 | 1.75 ± 0.49 | 6.00 ± 0.06 |
The growth kinetics values for the three pathogens in mixtures were extrapolated from data from Fig. 2 by using the Gompertz equation (47). Values are expressed as means ± SDs.
The numbers in parentheses indicate the initial inoculation levels for the given experiment.
ND, not determined. Counts of E. coli CFU in the mixture could not be determined because of the overgrowth of Salmonella on E. coli-selective CT-SMAC plates (see the text for a detailed explanation).
NA, not applicable.
(ii) Experiment II: Salmonella serovar Enteritidis/E. coli O157:H7/L. monocytogenes culture ratio, 10:1,000:1.
The overall growth profiles of the three cultures were highly proportional to their initial inoculation levels (Salmonella serovar Enteritidis at 13.5 ± 1.1 CFU/ml, E. coli at 1,327 ± 166 CFU/ml, and L. monocytogenes at 1.3 ± 0.6 CFU/ml). The growth rate and the MPD of Salmonella serovar Enteritidis cells were the highest of those of the pathogens in SEL (Fig. 2B). In this mixture, L. monocytogenes, inoculated at the lowest concentration (1 CFU/ml), grew in the presence of two other bacterial species inoculated at higher initial cell concentrations. The GT of L. monocytogenes was the longest of those of the three pathogens, and the EGR of L. monocytogenes was the lowest. Furthermore, the MPD of L. monocytogenes cells was 4.28 log10 CFU/ml, while E. coli O157:H7 and Salmonella serovar Enteritidis reached 8.58 and 7.11 log10 CFU/ml, respectively (Table 4). This result indicates that the fast-growing Salmonella and E. coli possibly utilized the most nutrients and that, thus, the depleted nutrient levels probably resulted in a lower growth rate for Listeria, normally a slow-growing bacterium.
(iii) Experiment III: Salmonella serovar Enteritidis/E. coli O157:H7/L. monocytogenes culture ratio, 1:10:1,000.
When the inoculation level of L. monocytogenes cells (1,180 ± 125 CFU/ml) was greater than those of Salmonella serovar Enteritidis (1.4 ± 0.1 CFU/ml) and E. coli (14.6 ± 1.6 CFU/ml) cells, L. monocytogenes showed better growth than the other two pathogens (Fig. 2C). Interestingly, Salmonella serovar Enteritidis cells exhibited a shorter LPD and a higher MPD than E. coli O157:H7 cells, although the initial number of Salmonella serovar Enteritidis cells was lower than that of E. coli O157:H7 cells (Table 4). Additionally, the MPDs of Salmonella serovar Enteritidis and E. coli O157:H7 cells approached barely 5 to 6 log10 CFU/ml, while that of L. monocytogenes cells reached about 8.5 log10 CFU/ml.
(iv) Experiment IV: Salmonella serovar Enteritidis/E. coli O157:H7/L. monocytogenes culture ratio, 1,000:1:10.
In experiment IV, the inoculation level for Salmonella serovar Enteritidis was 1,178 ± 124 CFU/ml, that for E. coli was 1.3 ± 0.1 CFU/ml, and that for L. monocytogenes was 11.5 ± 1.9 CFU/ml. We were able to determine the growth curves for Salmonella and Listeria but not for E. coli (Fig. 2D). As expected, Salmonella cells had a significantly higher growth rate than Listeria cells, with a shorter GT (0.88 versus 1.46 h) and a higher MPD (9.33 versus 6.0 log10 CFU/ml) (Table 4). E. coli O157:H7 cells could not be enumerated after inoculation at 1 CFU/ml because of the overgrowth of Salmonella (for which the initial inoculation level was 1,000 times higher than that for E. coli) on the CT-SMAC plate, which hindered the E. coli colonies. E. coli growth in this mixture was further confirmed by obtaining positive reactions in the ICLFA (data not shown) and PCR assay (see Fig. 4C). In a separate experiment, we demonstrated that E. coli inoculated at 1 CFU/ml was indeed capable of growing in SEL (Fig. 2E). Though CT-SMAC is a selective medium for E. coli O157:H7, Salmonella was able to grow on CT-SMAC, producing opaque pink colonies, while E. coli O157:H7 produced colorless gray-white colonies because of its inability to ferment sorbitol. This result indicates that improvement in differential plating media is necessary for separations of E. coli O157:H7 and Salmonella serovar Enteritidis from the same sample during the plating procedure.
FIG. 4.
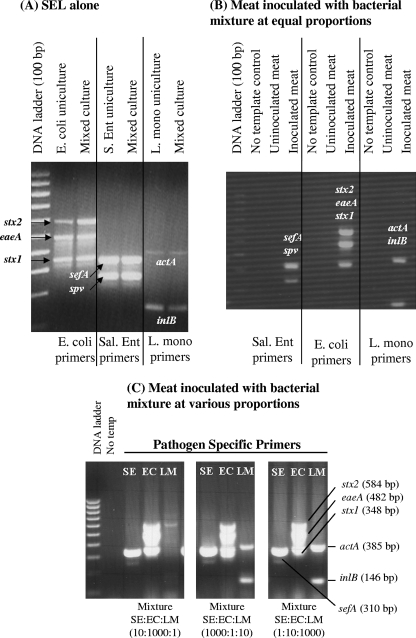
Results from multiplex PCR assays for the detection of Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes bacteria grown individually (ca. 3 × 102 CFU/ml) in SEL broth (A) or in mixtures in meat (B and C). (A) Cultures were incubated at 37°C for 16 to 18 h in a shaker incubator and analyzed by PCR assays using species-specific primer sets: primers targeting sefA (310 bp) and spv (250 bp) for Salmonella serovar Enteritidis (S. Ent, or Sal. Ent), actA (395 bp) and inlB (146 bp) for L. monocytogenes (L. mono), and stx2 (584 bp), eaeA (482 bp), and stx1 (348 bp) for E. coli O157:H7. (B) Ready-to-eat sliced turkey meat samples were inoculated with Salmonella serovar Enteritidis (SE), E. coli O157:H7 (EC), and L. monocytogenes (LM) cultures at equal concentrations (ca. 3 × 102 CFU/ml) and analyzed by PCR after 18 h of enrichment in SEL broth. (C) Meat samples were inoculated with three mixtures of Salmonella serovar Enteritidis, E. coli, and L. monocytogenes CFU prepared with the bacteria at various ratios as indicated, enriched for 18 h, and assayed by multiplex PCR using Salmonella serovar Enteritidis-, E. coli-, or L. monocytogenes-specific primers. No temp, no-template DNA control.
Growth patterns of food-borne bacteria in SEL.
The growth patterns of different microorganisms that are commonly associated with food, along with those of additional species and other strains or serovars of the three target pathogens, in SEL were investigated and compared with those in UPB (Table 1) and mEC+n, RV broth, and FB (see Table S2 in the supplemental material) by measuring optical densities at 595 nm (OD595) at 12, 16, and 24 h. Seven enterohemorrhagic E. coli (EHEC) strains, two enteropathogenic E. coli (EPEC) strains, and one enterotoxigenic E. coli (ETEC) strain showed significantly higher levels of growth (P < 0.05; 16 h of growth) in SEL than in UPB; another strain of ETEC (O78:H11) failed to grow in SEL but showed good growth in UPB. Four strains of L. monocytogenes belonging to serovars 1/2a, 1/2b, and 4b and a strain of Listeria innocua exhibited better growth in SEL than in UPB. Likewise, four serovars of S. enterica showed improved growth in SEL compared to that in UPB. Among the nontarget bacteria, three Bacillus species, three Lactobacillus species, and one strain each of Enterococcus faecalis, Proteus vulgaris, Lactococcus lactis, and Leuconostoc mesenteroides did not show any growth in SEL but did grow in UPB. Among the five natural food isolates (obtained in this study), Bacillus megaterium HK1, Lactococcus lactis HK21, and Pediococcus acidilactici HK32 did not grow in SEL but showed some growth in UPB (Table 1). Among the test organisms, only three (Brochothrix thermosphacta, Lactobacillus acidophilus, and Pediococcus sp.) did not show any detectable growth in either medium. We also observed that several nontarget bacteria, including Streptococcus mutans, Staphylococcus epidermidis, Pseudomonas aeruginosa, Serratia marcescens, Enterobacter aerogenes, Enterobacter cloacae, and Hafnia alvei, showed good growth in SEL and that these organisms also grew well in UPB and certain selective media (Table 1; see also Table S2 in the supplemental material). Overall, these data indicate that the levels of growth of most desirable target pathogens in SEL were higher than those in UPB and that SEL was more inhibitory to several food-borne organisms than UPB (Table 1).
Furthermore, bacterial growth in SEL was superior to that in individual selective enrichment broths for the respective target pathogens, such as RV broth for Salmonella, mEC+n for E. coli, and FB for Listeria, when analyzed after 24 h of enrichment (see Table S2 in the supplemental material).
Recovery of acid- and cold-stressed cells in SEL.
The ability of SEL to resuscitate acid- and cold-injured cells was evaluated and compared with the recovery ability of TSBYE, a commonly used nonselective enrichment broth, as well as those of the respective individual selective enrichment broths. As expected, the stress conditions caused the inhibition of cell growth (Table 5), resulting in 0.5- to ∼2.1-log reductions in cell counts for target pathogens compared with those in the control (incubated at 37°C). The pH 4.5 stress caused the highest numbers of cell deaths among all three pathogens, reducing populations by more than 2 logs (2.07 and 2.08 logs) for Salmonella serovar Enteritidis and E. coli O157:H7, respectively, and 1.15 logs for L. monocytogenes. Cold stress resulted in moderate cell injury: a reduction in the bacterial population of about 1.6 logs for E. coli O157:H7, 1 log for Salmonella serovar Enteritidis, and 0.99 log for L. monocytogenes. Finally, cultures under the pH 5.5 stress condition showed the least cell injury (population reductions of less than 1 log for all three pathogens) among those exposed to the three stress conditions (Table 5). Stress-exposed cells were allowed to recover in TSBYE, SEL, and their respective selective enrichment broths for 3 and 6 h. Data for each pathogen are presented below. An increase in bacterial cell counts of ≥1 log was considered to indicate significant recovery in the corresponding medium (Table 5).
TABLE 5.
Comparisons of recovery rates for stress-exposed Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes cells in TSBYE, SEL, and the respective individual selective enrichment brothsa
| Stress or condition | Organism | Log10 CFU/ml (mean ± SD) at 0 h | Log10 CFU/ml (mean ± SD) at 3 h in:
|
Log10 CFU/ml (mean ± SD) at 6 h in:
|
||||
|---|---|---|---|---|---|---|---|---|
| TSBYE | Selective enrichment brothb | SEL | TSBYE | Selective enrichment brothb | SEL | |||
| Temp (°C) | Salmonella serovar | |||||||
| 37 | Enteritidis | 7.27 ± 0.24 | 8.81 ± 0.20* | 8.35 ± 0.15 | 8.29 ± 0.28 | 9.23 ± 0.10* | 8.41 ± 0.29 | 9.27 ± 0.14 |
| 4 | 6.25 ± 0.14 | 8.31 ± 0.34* | 7.35 ± 0.48* | 7.66 ± 0.22* | 9.10 ± 0.05* | 8.40 ± 0.05* | 9.21 ± 0.14* | |
| pH | ||||||||
| 4.5 | 5.20 ± 0.54 | 7.22 ± 0.02* | 5.39 ± 0.68 | 5.07 ± 0.00 | 7.33 ± 0.07* | 5.83 ± 1.05 | 4.89 ± 1.06 | |
| 5.5 | 6.69 ± 0.02 | 8.71 ± 0.26* | 8.25 ± 0.36* | 8.28 ± 0.28* | 9.05 ± 0.06* | 8.53 ± 0.09* | 9.35 ± 0.17* | |
| Temp (°C) | E. coli O157:H7 | |||||||
| 37 | 7.16. ± 0.11 | 8.63 ± 0.11* | 7.53 ± 0.10 | 7.84 ± 0.07 | 9.02 ± 0.11* | 8.08 ± 0.18 | 8.73 ± 0.17 | |
| 4 | 5.57 ± 0.09 | 8.02 ± 0.14* | 5.02 ± 0.73 | 5.77 ± 0.91 | 8.86 ± 0.07* | 4.90 ± 0.49 | 7.80 ± 0.23* | |
| pH | ||||||||
| 4.5 | 5.08 ± 0.36 | 6.43 ± 0.11* | 3.21 ± 0.28 | 3.54 ± 0.28 | 8.56 ± 0.49* | 3.66 ± 0.68 | 5.24 ± 0.37 | |
| 5.5 | 6.69 ± 0.15 | 8.43 ± 0.11* | 6.89 ± 0.13 | 7.43 ± 0.21 | 9.02 ± 0.13* | 6.99 ± 0.60 | 8.66 ± 0.20* | |
| Temp (°C) | L. monocytogenes | |||||||
| 37 | 7.56 ± 0.04 | 8.72 ± 0.03* | 8.20 ± 0.10 | 8.38 ± 0.12 | 9.60 ± 0.10* | 8.78 ± 0.10 | 9.17 ± 0.16 | |
| 4 | 6.57 ± 0.08 | 8.04 ± 0.08* | 7.28 ± 0.10 | 7.67 ± 0.08* | 9.06 ± 0.10* | 8.36 ± 0.07* | 8.88 ± 0.09* | |
| pH | ||||||||
| 4.5 | 6.42 ± 0.17 | 7.60 ± 0.18* | 6.74 ± 0.08 | 7.33 ± 0.11* | 9.22 ± 0.49* | 7.75 ± 0.09* | 8.06 ± 0.64* | |
| 5.5 | 6.71 ± 0.04 | 8.23 ± 0.10* | 7.46 ± 0.10 | 7.91 ± 0.16* | 9.56 ± 0.03* | 8.41 ± 0.06* | 8.99 ± 0.09* | |
Each culture was inoculated at 3 × 102 CFU/ml. Values marked with * indicate the recovery of stressed cells as defined by an increase in cell numbers of ≥1 log in the given medium compared to the initial numbers after stress.
The selective enrichment broths were RV broth for Salmonella serovar Enteritidis, mEC+n for E. coli, and FB for L. monocytogenes.
(i) Salmonella serovar Enteritidis.
All three media (TSBYE, SEL, and RV broth) resuscitated cold (4°C)- and pH 5.5-stressed Salmonella serovar Enteritidis; however, RV broth and SEL failed to resuscitate pH 4.5-stressed cells after 3 and 6 h of recovery (Table 5). Overall, TSBYE yielded the best recovery at 3 h, while SEL supported the best recovery at 6 h. RV broth showed the lowest recovery rates for all stress conditions at both time points.
(ii) E. coli O157:H7.
SEL and TSBYE allowed injured E. coli O157:H7 cells to recover; however, mEC+n failed to allow recovery (Table 5). Stress-exposed E. coli O157:H7 grown in TSBYE showed recovery at both time points (3 and 6 h); however, SEL helped to resuscitate cold (4°C)- and pH 5.5-stressed cells only after 6 h. SEL was unable to show any resuscitation of pH 4.5-stressed cells.
(iii) L. monocytogenes.
SEL and TSBYE resuscitated injured L. monocytogenes after 3 and 6 h of recovery, while FB did so only after 6 h (Table 5). The recovery rates in order from highest to lowest were those for TSBYE, SEL, and FB.
In summary, the stress recovery studies indicate that SEL supported the recovery of stress-exposed cells and that its performance was equivalent to that of TSBYE when a 6-h recovery period was allowed, with the exception of pH 4.5-induced stress for E. coli O157:H7 and Salmonella serovar Enteritidis. As a selective enrichment broth, SEL demonstrated performance superior to that of the respective individual selective enrichment broths, RV broth, mEC+n, and FB.
Detection of pathogens by antibody-based ICLFA and multiplex PCR. (i) ICLFA.
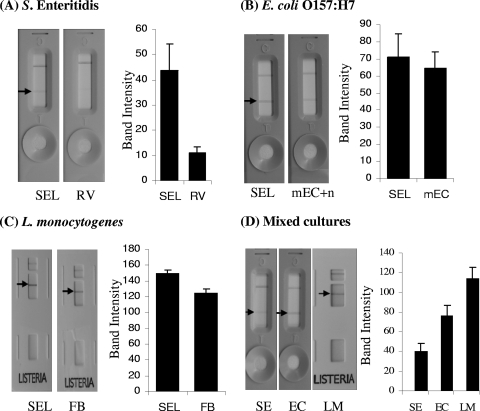
Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes grown in SEL gave positive reactions in the ICFLA, and the reaction intensities for Salmonella serovar Enteritidis and L. monocytogenes grown in SEL were significantly higher (P < 0.05) than those for the same pathogens grown in their respective individual selective enrichment broths, RV broth and FB (Fig. 3A and C). The antibody reaction intensity for E. coli grown in SEL was slightly higher than but comparable to that for mEC+n-grown cells (Fig. 3B). When all three pathogens grown in SEL were inoculated at equal cell concentrations as a mixture, they all gave positive reactions with their respective Reveal kits; however, the band intensity for L. monocytogenes was the strongest, followed by those for E. coli O157:H7 and Salmonella serovar Enteritidis (Fig. 3D). The overall reaction of Salmonella serovar Enteritidis grown either in SEL or RV broth with the Salmonella Reveal kit was relatively weaker than those of the other two pathogens with their kits. It was later confirmed that the ICLFA kit (Neogen Corp.) is intended primarily for the detection of Salmonella serovar Typhimurium; however, we used Salmonella serovar Enteritidis as the test organism, thus obtaining a weaker reaction. Altogether, ICLFA data demonstrated that SEL is suitable for enrichment with the three pathogens individually or in a mixture for subsequent detection by the antibody-based ICLFA method.
FIG. 3.
Results from ICLFA showing the reaction patterns of cells of the pathogens Salmonella serovar Enteritidis (SE) (A and D), E. coli O157:H7 (EC) (B and D), and L. monocytogenes (LM) (C and D) grown individually (with each pathogen inoculated at 103 CFU/ml) (A to C) or in mixed cultures (with each pathogen inoculated at ca. 3 × 102 CFU/ml) set up in SEL (D). The ICLFA reaction patterns were also compared with those of cells grown in the respective selective enrichment broths (RV broth, mEC+n, and FB). Cultures were incubated at 37°C for 16 to 18 h in a shaker incubator. The lateral-flow strips were loaded with 120-μl samples of Salmonella serovar Enteritidis and E. coli O157:H7 live cultures or 135 μl of heat-killed L. monocytogenes cells, and the antibody reaction intensities (band densities in pixels) were quantified by using software (Scion Crop., Frederick, MD) and presented as bar graphs.
(ii) Multiplex PCR.
Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes grown individually or in mixtures (experiments I to IV) were also tested in a multiplex PCR assay. As expected, E. coli grown individually or in a mixture in SEL alone or in the presence of meat (ready-to-eat turkey) showed three amplified products, those for stx2 (584 bp), eaeA (482 bp), and stx1 (348 bp) (Fig. 4). Similarly, Salmonella serovar Enteritidis gave amplified products for sefA (310 bp) and/or spv (250 bp) and L. monocytogenes gave products for actA (385 bp) and inlB (146 bp) without any nonspecific amplification when grown individually or in a mixture (Fig. 4). PCR analysis of uninoculated control meat did not yield any DNA amplification with species-specific primers, thus confirming the absence of background target pathogens in the product (Fig. 4B). When Salmonella, E. coli O157:H7, and L. monocytogenes cells were inoculated in equal (Fig. 4B) or variable (Fig. 4C) proportions into turkey meat, all gave positive amplified PCR products, except in experiment II, in which L. monocytogenes was undetectable. In experiment II, L. monocytogenes cells were inoculated at about 1.5 CFU/g together with Salmonella (∼13 CFU/g) and E. coli (∼1,300 CFU/g) cells (Fig. 4C). The lack of amplification is attributed to the poor growth of L. monocytogenes in the mixture compared to that of the other two organisms (Fig. 2B). Nevertheless, these data demonstrate that all three pathogens could be readily detected when grown in SEL individually or in a mixture in a meat sample by using species-specific PCR primer sets, suggesting that SEL is a suitable enrichment broth for PCR-based detection. However, some situations in which L. monocytogenes cells are present in low numbers (∼1 CFU/g) along with a large number of other microbes, as described above, may yield negative PCR results.
Selective enrichment of artificially inoculated meat samples with pathogens in SEL broth and subsequent detection using viable-cell counting lateral-flow and PCR assays.
In artificially inoculated ready-to-eat turkey and salami samples, the detection of three target pathogens grown in SEL after 12 and 24 h of enrichment was demonstrated. The growth patterns of Salmonella serovar Enteritidis and E. coli O157:H7 were similar, and cell numbers in all meat samples reached 8 to 9 log10 CFU/ml at 12 and 24 h (see Table S3 in the supplemental material). The level of growth of L. monocytogenes (which reached 5 to 7 log10 CFU/ml) was lower than those of the other two target pathogens. Numbers of Salmonella serovar Enteritidis and L. monocytogenes cells in turkey samples were higher than those in salami samples. The growth of E. coli O157:H7 was not affected by the food type. Although growth behaviors varied among the target pathogens, SEL supported the growth of these pathogens concurrently in the artificially inoculated meat samples.
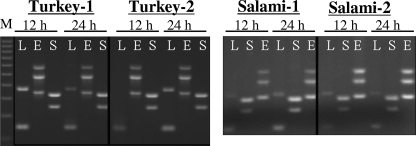
In the lateral-flow immunoassay, E. coli O157:H7 gave strong positive reactions after both 12 and 24 h of enrichment (see Fig. S1 in the supplemental material). After 12 h of enrichment in SEL, Salmonella serovar Enteritidis showed a weak positive reaction, but the reaction was slightly improved after 24 h of enrichment in the salami sample. No reaction was observed for L. monocytogenes in salami after 12 h of enrichment, but the reaction intensities were high after 24 h of incubation. These data indicate that SEL could be used as an enrichment broth for antibody-based detection by ICLFA; however, the duration of enrichment time is critical to obtain a strong reaction. PCR assays of the same inoculated turkey and salami samples after 12 and 24 h of enrichment showed positive PCR-amplified products for the target pathogens (Fig. 5), confirming that SEL could potentially be used as an enrichment broth for PCR-based detection.
FIG. 5.
Results from PCR assays of Salmonella serovar Enteritidis-, L. monocytogenes-, and E. coli O157:H7-inoculated ready-to-eat turkey and salami samples. The meat samples (25 g each) were inoculated (ca. 3 × 102 CFU of each pathogen/g), mixed with 225 ml of SEL, and incubated for 12 and 24 h with agitation. In panel A, PCR lanes are as follows, from left to right: M, 100-bp ladder DNA marker; L, Listeria primers targeting genes actA (395 bp) and inlB (146 bp); E, E. coli O157:H7 primers targeting genes stx2 (584 bp), eaeA (482 bp), and stx1 (348 bp); and S, Salmonella primers targeting genes sefA (310 bp) and spv (250 bp). An ICLFA also showed positive reactions with Salmonella serovar Enteritidis, E. coli O157:H7, and L. monocytogenes antigens for corresponding samples (see Fig. S1 in the supplemental material).
Comparison of SEL performance with UPB performance for sample enrichment and detection of three pathogens in artificially inoculated meat samples.
Turkey and salami samples were inoculated with three pathogens at equal cell concentrations and subjected to enrichment in SEL and UPB for up to 24 h. The performance of SEL was compared with that of UPB by determining bacterial cell counts in each medium and by performing ICLFA and PCR assays.
In turkey samples, the overall growth of E. coli O157:H7 in UPB was better than that in SEL, while the growth of Salmonella serovar Enteritidis and that of L. monocytogenes were better in SEL (Table 6). During the early periods of meat enrichment (8 to 10 h), the numbers of cells of the three pathogens grown in UPB were higher than those grown in SEL. However, 12 h postenrichment, sharply accelerated growth in SEL was observed. In the lateral-flow immunoassay, there were no differences between the two broths in the reaction intensities for Salmonella serovar Enteritidis or E. coli O157:H7; however, L. monocytogenes was detected after as little as 8 h of growth in SEL compared to 10 h of growth in UPB (see Table S3 in the supplemental material). PCR showed positive amplifications for the three pathogens at all incubation time points and in both media (see Fig. S2 in the supplemental material).
TABLE 6.
Comparison of viable-cell counts for pathogens inoculated concurrently into turkey or salami and subjected to enrichment in UPB and SEL
| Meat and organism | Mean log10 CFU/ml ± SDa at:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 8 h
|
10 h
|
12 h
|
16 h
|
24 h
|
||||||
| UPB | SEL | UPB | SEL | UPB | SEL | UPB | SEL | UPB | SEL | |
| Turkey | ||||||||||
| Salmonella serovar Enteritidis | 5.52 ± 0.07 A | 5.59 ± 0.32 A | 6.98 ± 0.07 A | 6.95 ± 0.08 A | 7.24 ± 0.12 B | 8.81 ± 0.02 A | 7.50 ± 0.15 B | 8.57 ± 0.06 A | 8.05 ± 0.13 A | 6.40 ± 0.17 B |
| E. coli O157:H7 | 6.56 ± 0.04 A | 4.74 ± 0.23 B | 8.48 ± 0.04 A | 5.56 ± 0.07 B | 8.20 ± 0.03 A | 7.15 ± 0.09 B | 7.76 ± 0.21 A | 7.19 ± 0.06 B | 6.06 ± 0.20 B | 7.73 ± 0.05 A |
| L. monocytogenes | 5.46 ± 0.03 A | 5.08 ± 0.15 B | 6.05 ± 0.08 A | 6.21 ± 0.16 A | 6.02 ± 0.06 B | 7.30 ± 0.04 A | 6.14 ± 0.17 B | 7.24 ± 0.06 A | 6.83 ± 0.13 B | 7.47 ± 0.04 A |
| Salami | ||||||||||
| Salmonella serovar Enteritidis | 4.63 ± 0.06 B | 4.84 ± 0.06 B | 5.95 ± 0.07 A | 5.70 ± 0.07 B | 6.62 ± 0.15 B | 7.04 ± 0.17 A | 7.02 ± 0.06 B | 7.56 ± 0.04 A | 7.28 ± 0.07 B | 8.47 ± 0.98 A |
| E. coli O157:H7 | 5.76 ± 0.15 A | 5.75 ± 0.05 A | 8.39 ± 0.07 A | 7.09 ± 0.13 B | 8.70 ± 0.02 A | 8.70 ± 0.07 A | 8.77 ± 0.01 B | 9.20 ± 0.18 A | 8.78 ± 0.16 A | 8.90 ± 0.05 A |
| L. monocytogenes | 4.48 ± 0.06 A | 4.02 ± 0.21 B | 5.49 ± 0.08 A | 4.55 ± 0.12 B | 5.65 ± 0.03 A | 4.52 ± 0.07 B | 6.10 ± 0.05 A | 5.95 ± 0.10 A | 6.67 ± 0.06 A | 5.88 ± 0.03 B |
Counts in the same row labeled with the same letter (A or B) and corresponding to growth in UPB and SEL at a given time point are not significantly different at P of <0.05. Meat samples (25 g/225 ml of SEL) were inoculated with Salmonella, E. coli, and Listeria (ca. 3 × 102 CFU of each pathogen/ml) and assayed at 8, 10, 12, 16, and 24 h. Bacterial cell counts were determined by plating Salmonella cells onto XLD agar, E. coli cells onto CT-SMAC, and Listeria cells onto MOX.
In salami samples, generally, bacterial counts were lower than those in turkey samples, due possibly to the presence of preservatives and bacterial inhibitors. The growth of L. monocytogenes in salami in SEL, in particular, was reduced by almost 2 logs compared to that in turkey in SEL (Table 6). Similarly, the growth of Salmonella serovar Enteritidis in salami in UPB showed a l-log reduction compared to that in turkey in UPB. Overall, the levels of growth of all three pathogens in salami in SEL were comparable to those in salami in UPB. Lateral-flow immunoassay results for each pathogen in SEL and UPB were similar; however, L. monocytogenes was detected after as little as 12 h when subjected to enrichment in SEL, compared to the 16 h of incubation needed in UPB. PCR analysis showed unambiguous positive amplified bands for the three target pathogens at all time points, except for L. monocytogenes, which produced very faint amplified bands in SEL after 8 h of enrichment (see Fig. S2 in the supplemental material).
DISCUSSION
Current industry trends emphasize the need for the detection of multiple pathogens on a single-assay platform to reduce the cost of testing per pathogen. Moreover, a multipathogen detection scheme could mitigate the industry and regulatory needs in the mandatory testing of food products for multiple pathogens prior to retail distribution. To aid in multipathogen detection, a suitable selective enrichment medium is necessary to improve detection utilizing methods such as multiplex PCR (5, 20, 42, 46), DNA array hybridization (15), array-based immunosorbent assays (14), and antibody-based array biosensor techniques (35, 50).
In this study, a selective enrichment broth, SEL, was developed and evaluated for its ability to enrich a test sample with multiple pathogens concurrently if or when the pathogens were present in the same sample. SEL was formulated by modifying the recipe for BLEB and contains four different antimicrobial agents, acriflavine, cycloheximide, fosfomycin, and nalidixic acid (Table 2), along with tryptic soy broth, yeast extract, sodium pyruvate, and sodium phosphate, which are proven to support the growth of healthy and injured food-borne pathogens (9, 11). Sodium pyruvate and sodium phosphates protect bacteria from pH changes and reactive oxygen atoms (32), and the selective antibiotics inhibit the growth of background resident microorganisms (31).
Overall, the individual growth patterns of the three target pathogens in SEL were satisfactory, and the performance of SEL as a selective enrichment broth was superior to those of mEC+n for E. coli and FB for Listeria and equivalent to that of RV broth for Salmonella. For E. coli O157:H7, SEL was able to support growth after inoculation with 10 and 1,000 CFU/ml, while mEC+n failed to support growth after inoculation with 10 CFU/ml. The inability of mEC+n to support growth at this inoculation level was in agreement with the findings in a previous report (25). The lack of growth may be due to the strain used or the incubation temperature or agitation conditions employed in this study. Moreover, bile salts and novobiocin present in mEC+n most likely exerted inhibitory effects culminating in reduced or no growth at lower cell numbers (28, 49). The inability of mEC+n to support growth at 10 CFU/ml or lower is unacceptable, since an infectious dose of E. coli O157:H7 is in the range of 10 to 100 CFU (25). The growth rate of L. monocytogenes in SEL was superior to that in FB (47). In addition, other bacteria, Bacillus, Enterococcus, and Streptococcus spp., which showed poor or no growth in SEL (Table 1) can grow in FB (12).
In a mixed-culture experiment, SEL allowed the concurrent growth of the three target pathogens and competition among the pathogens and their initial levels were determinants of their growth kinetics. When the pathogens were mixed in equal proportions, E. coli O157:H7 showed the shortest LPD and the highest maximum cell density, while L. monocytogenes showed the lowest maximum cell density and Salmonella growth was intermediate (Table 4). Lower cell numbers for L. monocytogenes were expected because this pathogen is a slow-growing and poor competitor (2). When the bacteria were mixed at various ratios, the growth pattern of each pathogen was proportional to the initial cell number. This detailed growth kinetics profile of each pathogen in a mixture in SEL would aid in the selection of a suitable method for the accurate detection of these three pathogens if present in the same sample.
The application of SEL as an enrichment medium for the detection of three target pathogens in inoculated meat samples by antibody-based lateral-flow immunoassays and nucleic acid-based PCR assays was investigated. As expected, individual-pathogen-specific ICLFA strips gave positive reactions when bacteria were grown in SEL, suggesting that all three pathogens can be detected using antibody-based methods. Moreover, the ICLFA reaction intensities in SEL were stronger than those in the respective individual selective enrichment broths (Fig. 3). This result suggests that SEL promoted increased expression of antibody-reactive antigens compared to the expression of these antigens in its counterparts. A selective- or nonselective-medium-mediated reduction in the expression of antigen or in an antigen-antibody reaction has been demonstrated previously for Salmonella, E. coli O157:H7, and L. monocytogenes (7, 21, 22, 24). Pathogen-specific multiplex PCR assays were also successful in detecting each pathogen from the mixture without producing any nonspecific amplification (Fig. 4 and 5). Furthermore, the growth of two nontarget bacteria, Enterobacter aerogenes and Streptococcus mutans, in SEL (Table 1) did not interfere with the PCR amplification of the specific target genes of the three pathogens (data not shown).
In the inoculated-meat experiments, both ICLFA and PCR assays were performed with SEL-enriched samples at various time intervals. In most cases, positive ICLFA reactions were obtained after 12 h of enrichment, with approximate cell populations of 6 to 8.2 log CFU/ml, while positive PCR results were obtained after 8 h of growth, with cell counts of 4.48 to 5.7 log CFU/ml, irrespective of the type of meat sample (see Fig. S2 in the supplemental material), confirming that PCR is more sensitive than the ICLFA. (Note that 8 h is the earliest time point at which we tested.) In general, these limits of detection for ICLFA and PCR are in agreement with the data in previous reports (8, 19, 44).
The ability of selective enrichment broth to resuscitate temperature-, preservative-, salt-, and acid-stressed cells (1, 3, 33, 36) encountered during food processing, storage, or sanitization is critical for detection. It is also well known that injured cells can cause an improper estimation of the decimal reduction time (D value) and z value (temperature required to change the D value to transverse by 1 log) during thermal processing (41) and, most importantly, that the injured pathogens can revive and grow under favorable conditions (9). We have demonstrated that SEL allowed the recovery of acid (pH 5.5)- and temperature (4°C)-stressed cells and that, overall, the recovery rates were comparable to those in nonselective TSBYE broth (Table 5). However, the recovery rates for pH 4.5-stressed cells were variable; SEL successfully resuscitated Listeria cells but failed to resuscitate E. coli O157:H7 and Salmonella cells.
In this study, the performance of SEL was compared with that of UPB (2), a currently known universal enrichment broth for Salmonella, Listeria, E. coli O157:H7, and Yersinia spp. (43, 51, 54). Overall, SEL supported improved growth of all three target pathogens compared to that in UPB and the individual selective enrichment broths RV, mEC+n, and FB. In addition, SEL inhibited greater numbers of natural food-borne bacteria, including some nascent food isolates, than UPB (Table 1; see Table S2 in the supplemental material). Of the two ETEC strains tested, the O78:H11 strain did not grow in SEL, while the O25:K98:NM strain did. The lack of growth of O78:H11 was determined to be due to the presence of nalidixic acid and acriflavine (data not shown). Further research on the types and concentrations of antibiotics needed for all ETEC strains to grow in SEL is warranted.
When both SEL and UPB were used as enrichment broths with spiked meat samples, the overall bacterial cell counts in UPB were slightly better than those in SEL, but the differences were not statistically significant. Though PCR and ICLFA results for the two media were comparable, the PCR with UPB-enriched samples yielded slightly improved detection of L. monocytogenes at 8 h compared to that with SEL-enriched samples (see Fig. S2 in the supplemental material).
In summary, SEL, a selective enrichment broth, promoted the concurrent growth of three major food-borne pathogens, S. enterica, E. coli O157:H7, and L. monocytogenes, present at various cell numbers in cultures and meat samples. The performance of SEL was superior to that of UPB and the respective individual selective enrichment broths. Based on the data presented in this study, SEL can be considered a selective enrichment broth for the detection of three major food-borne pathogens by antibody- or nucleic acid-based assays. Currently, SEL is being evaluated for the detection of pathogens in naturally or artificially contaminated meat samples by biosensor-based assays, including light scattering and the use of fiber optic sensors.
Supplementary Material
Acknowledgments
This research was supported through a cooperative agreement with the Agricultural Research Service of the U.S. Department of Agriculture, project number 1935-42000-035, and the Center for Food Safety Engineering at Purdue University.
Sincere thanks to B. K. Hahm for technical assistance and Heather Day, Seung Ohk, and Amornrat Aroonnual for critical review of the manuscript.
Footnotes
Published ahead of print on 6 June 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdul-Raouf, U. M., L. R. Beuchat, and M. S. Ammar. 1993. Survival and growth of Escherichia coli O157:H7 in ground, roasted beef as affected by pH, acidulants, and temperature. Appl. Environ. Microbiol. 59:2364-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, J. S., and N. A. Cox. 1992. Universal preenrichment broth for the simultaneous detection of Salmonella and Listeria in foods. J. Food Prot. 55:256-259. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin, M. M., and A. R. Datta. 1995. Acid tolerance of enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 61:1669-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhagwat, A. A. 2006. Microbiological safety of fresh-cut produce: where are we now?, p. 121-165. In K. R. Mathews (ed.), Microbiology of fresh produce. ASM Press, Washington, DC.
- 5.Bhagwat, A. A. 2003. Simultaneous detection of Escherichia coli O157:H7, Listeria monocytogenes and Salmonella strains by real-time PCR. Int. J. Food Microbiol. 84:217-224. [DOI] [PubMed] [Google Scholar]
- 6.Bhunia, A. K. 2008. Biosensors and bio-based methods for the separation and detection of foodborne pathogens, p. 1-44. In S. Taylor (ed.), Advances in food and nutrition research, vol. 54. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 7.Bird, C. B., R. J. Hoerner, and L. Restaino. 2001. Comparison of Reveal 20-hours method and BAM culture method for detection of Escherichia coli O157:H7 in selected food and environment swabs: collaborative study. J. AOAC Int. 84:737-751. [PubMed] [Google Scholar]
- 8.Bird, C. B., R. L. Miller, M. M. Miller, K. Schniederm, and G. Rodvich. 1999. Reveal for Salmonella test system. J. AOAC Int. 82:625-633. [PubMed] [Google Scholar]
- 9.Buduamoako, E., S. Toora, R. F. Ablett, and J. Smith. 1992. Evaluation of the ability of primary selective enrichment to resuscitate heat-injured and freeze-injured Listeria monocytogenes cells. Appl. Environ. Microbiol. 58:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnett, S. L., and L. R. Beuchat. 2000. Human pathogens associated with raw produce and unpasteurized juices, and difficulties in decontamination. J. Ind. Microbiol. Biotechnol. 25:281-287. [DOI] [PubMed] [Google Scholar]
- 11.Busch, S. V., and C. W. Donnelly. 1992. Development of a repair-enrichment broth for resuscitation of heat-injured Listeria monocytogenes and Listeria innocua. Appl. Environ. Microbiol. 58:14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capita, R., C. Alonso-Calleja, B. Moreno, and M. C. Garcia-Fernandez. 2001. Occurrence of Listeria species in retail poultry meat and comparison of a cultural/immunoassay for their detection. Int. J. Food Microbiol. 65:75-82. [DOI] [PubMed] [Google Scholar]
- 13.Cebula, T. A., W. L. Payne, and P. Feng. 1995. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J. Clin. Microbiol. 33:248-250. (Erratum, 33:1048.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, C. S., and R. A. Durst. 2006. Simultaneous detection of Escherichia coli O157:H7, Salmonella spp. and Listeria monocytogenes with an array-based immunosorbent assay using universal protein G-liposomal nanovesicles. Talanta 69:232-238. [DOI] [PubMed] [Google Scholar]
- 15.Chiang, Y.-C., C.-Y. Yang, C. Li, Y.-C. Ho, C.-K. Lin, and H.-Y. Tsen. 2006. Identification of Bacillus spp., Escherichia coli, Salmonella spp., Staphylococcus spp. and Vibrio spp. with 16S ribosomal DNA-based oligonucleotide array hybridization. Int. J. Food Microbiol. 107:131-137. [DOI] [PubMed] [Google Scholar]
- 16.Doyle, M. P., and J. L. Schoeni. 1987. Isolation of Escherichia coli O157:H7 from retail fresh meats and poultry. Appl. Environ. Microbiol. 53:2394-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ericsson, H., H. Unnerstad, J. G. Mattsson, M. L. Danielsson-Tham, and W. Tham. 2000. Molecular grouping of Listeria monocytogenes based on the sequence of the inlB gene. J. Med. Microbiol. 49:73-80. [DOI] [PubMed] [Google Scholar]
- 18.FDA. 5 March 2001, revision date. Bacteriological analytical manual, 8th ed. AOAC International, Arlington, VA. http://www.cfsan.fda.gov/∼ebam/bam-mm.html.
- 19.Fratamico, P. M., L. K. Bagi, and T. Pepe. 2000. A multiplex polymerase chain reaction assay for detection and identification of Escherichia coli O157:H7 in food and bovine feed. J. Food Prot. 63:1032-1037. [DOI] [PubMed] [Google Scholar]
- 20.Fratamico, P. M., and T. P. Strobaugh. 1998. Simultaneous detection of Salmonella spp. and Escherichia coli O157:H7 by multiplex PCR. J. Ind. Microbiol. Biotechnol. 21:92-98. [Google Scholar]
- 21.Geng, T., B. K. Hahm, and A. K. Bhunia. 2006. Selective enrichment media affect the antibody-based detection of stress-exposed Listeria monocytogenes due to differential expression of antibody-reactive antigens identified by protein sequencing. J. Food Prot. 69:1879-1886. [DOI] [PubMed] [Google Scholar]
- 22.Geng, T., K. P. Kim, R. Gomez, D. M. Sherman, R. Bashir, M. R. Ladisch, and A. K. Bhunia. 2003. Expression of cellular antigens of Listeria monocytogenes that react with monoclonal antibodies C11E9 and EM-7G1 under acid-, salt- or temperature-induced stress environments. J. Appl. Microbiol. 95:762-772. [DOI] [PubMed] [Google Scholar]
- 23.Gracias, K. S., and J. L. McKillip. 2004. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can. J. Microbiol. 50:883-890. [DOI] [PubMed] [Google Scholar]
- 24.Hahm, B. K., and A. K. Bhunia. 2006. Effect of environmental stresses on antibody-based detection of Escherichia coli O157:H7, Salmonella enterica serotype Enteritidis and Listeria monocytogenes. J. Appl. Microbiol. 100:1017-1027. [DOI] [PubMed] [Google Scholar]
- 25.Hepburn, N. F., M. MacRae, M. Johnston, J. Mooney, and I. D. Ogden. 2002. Optimizing enrichment conditions for the isolation of Escherichia coli O157 in soils by immunomagnetic separation. Lett. Appl. Microbiol. 34:365-369. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen, C. N. 1999. The influence of commonly used selective agents on the growth of Listeria monocytogenes. Int. J. Food Microbiol. 50:221-226. [Google Scholar]
- 27.Jaradat, Z. W., G. E. Schutze, and A. K. Bhunia. 2002. Genetic homogeneity among Listeria monocytogenes strains from infected patients and meat products from two geographic locations determined by phenotyping, ribotyping and PCR analysis of virulence genes. Int. J. Food Microbiol. 76:1-10. [DOI] [PubMed] [Google Scholar]
- 28.Jeffries, L. 1959. Novobiocin-tetrathionate broth: a medium of improved selectivity for the isolation of salmonellae from faeces. J. Clin. Pathol. 12:568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, J., E. Larkin, M. Steele, C. Poppe, and J. A. Odumeru. 1998. Evaluation of universal preenrichment broth for the recovery of foodborne pathogens from milk and cheese. J. Dairy Sci. 81:2798-2803. [DOI] [PubMed] [Google Scholar]
- 30.Kawasaki, S., N. Horikoshi, Y. Okada, K. Takeshita, T. Sameshima, and S. Kawamoto. 2005. Multiplex PCR for simultaneous detection of Salmonella spp., Listeria monocytogenes, and Escherichia coli O157:H7 in meat samples. J. Food Prot. 68:551-556. [DOI] [PubMed] [Google Scholar]
- 31.Kim, H. 2006. A selective enrichment medium for simultaneous growth and detection of Escherichia coli O157:H7, Listeria monocytogenes and Salmonella Enteritidis from food. Purdue University, West Lafayette, IN.
- 32.Knabel, S. J., and S. A Thielen. 1994. Enhanced recovery of severely heat-injured, thermotolerant Listeria monocytogenes from USDA and FDA primary enrichment media using a novel, simple, strictly anaerobic method. J. Food Prot. 58:29-34. [DOI] [PubMed] [Google Scholar]
- 33.Koutsoumanis, K. P., and J. N. Sofos. 2004. Comparative acid stress response of Listeria monocytogenes, Escherichia coli O157:H7 and Salmonella Typhimurium after habituation at different pH conditions. Lett. Appl. Microbiol. 38:321-326. [DOI] [PubMed] [Google Scholar]
- 34.Lathrop, A. A., P. P. Banada, and A. K. Bhunia. 2008. Differential expression of InlB and ActA in Listeria monocytogenes in selective and nonselective enrichment broths. J. Appl. Microbiol. 104:627-639. [DOI] [PubMed] [Google Scholar]
- 35.Ligler, F. S., C. R. Taitt, L. C. Shriver-Lake, K. E. Sapsford, Y. S. Shubin, and J. P. Golden. 2003. Array biosensor for detection of toxins. Anal. Bioanal. Chem. 377:469-477. [DOI] [PubMed] [Google Scholar]
- 36.Lioa, C. H., and W. F. Fett. 2005. Resuscitation of acid-injured Salmonella in enrichment broth in apple juice and on the surface of fresh-cut cucumber and apple. Lett. Appl. Microbiol. 41:487-492. [DOI] [PubMed] [Google Scholar]
- 37.Lynch, M., J. Painter, R. Woodruff, and C. Braden. 2006. Surveillance for foodborne-disease outbreaks—United States, 1998-2002. MMWR Surveill. Summ. 55:1-42. [PubMed] [Google Scholar]
- 38.Maciorowski, K. G., P. Herrera, F. T. Jones, S. D. Pillai, and S. C. Ricke. 2006. Cultural and immunological detection methods for Salmonella spp. in animal feeds: a review. Vet. Res. Commun. 30:127-137. [DOI] [PubMed] [Google Scholar]
- 39.Maldonado, Y., J. C. Fiser, C. H. Nakatsu, and A. K. Bhunia. 2005. Cytotoxicity potential and genotypic characterization of Escherichia coli isolates from environmental and food sources. Appl. Environ. Microbiol. 71:1890-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, F. A., T. R. S. Brandao, P. Teixeira, and C. L. M. Silva. 2006. Recovery of heat-injured Listeria innocua. Int. J. Food Microbiol. 112:261-265. [DOI] [PubMed] [Google Scholar]
- 42.Mukhopadhyay, A., and U. K. Mukhopadhyay. 2007. Novel multiplex PCR approaches for the simultaneous detection of human pathogens: Escherichia coli O157:H7 and Listeria monocytogenes. J. Microbiol. Methods 68:193-200. [DOI] [PubMed] [Google Scholar]
- 43.Nam, H. M., S. E. Murinda, L. T. Nguyen, and S. P. Oliver. 2004. Evaluation of universal pre-enrichment broth for isolation of Salmonella spp., Escherichia coli O157:H7 and Listeria monocytogenes from dairy farm environmental samples. Foodborne Pathog. Dis. 1:37-44. [DOI] [PubMed] [Google Scholar]
- 44.Oberst, R. D., M. P. Hays, L. K. Bohra, R. K. Phebus, C. T. Yamashiro, C. Paszko-Kolva, S. J. Flood, J. M. Sargeant, and J. R. Gillespie. 1998. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5′ nuclease (TaqMan) assay. Appl. Environ. Microbiol. 64:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oktay, H. I., and D. Heperkan. 2006. Evaluation of ISO method and Vidas automated system for identifying Listeria and Salmonella in selected food. J. Rapid Methods Autom. Microbiol. 14:133-135. [Google Scholar]
- 46.Settanni, L., and A. Corsetti. 2007. The use of multiplex PCR to detect and differentiate food- and beverage-associated microorganisms: a review. J. Microbiol. Methods 69:1-22. [DOI] [PubMed] [Google Scholar]
- 47.Silk, T. M., T. M. T. Roth, and C. W. Donnelly. 2002. Comparison of growth kinetics for healthy and heat-injured Listeria monocytogenes in eight enrichment broths. J. Food Prot. 65:1333-1337. [DOI] [PubMed] [Google Scholar]
- 48.Soumet, C., G. Ermel, N. Rose, V. Rose, P. Drouin, G. Salvalt, and P. Coulin. 1999. Evaluation of multiplex PCR assay for simultaneous identification of Salmonella spp., Salmonella Enteritidis and Salmonella Typhimurium from environmental swabs of poultry houses. Lett. Appl. Microbiol. 28:113-117. [DOI] [PubMed] [Google Scholar]
- 49.Stephens, P. J., and J. A. Joynson. 1998. Direct inoculation into media containing bile salts and antibiotics is unsuitable for the detection of acid/salt stressed Escherichia coli O157:H7. Lett. Appl. Microbiol. 27:147-151. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, A. D., J. Ladd, Q. Yu, S. Chen, J. Homola, and S. Jiang. 2006. Quantitative and simultaneous detection of four foodborne bacterial pathogens with a multi-channel SPR sensor. Biosens. Bioelectron. 22:752-758. [DOI] [PubMed] [Google Scholar]
- 51.Thippareddi, H., R. K. Phebus, D. Y. C. Fung, and C. L. Kastner. 1995. Use of universal pro-enrichment medium supplemented with oxyrase for the simultaneous recovery of Escherichia coli O157:H7 and Yersinia enterocolitica. J. Rapid Methods Autom. Microbiol. 4:37-50. [Google Scholar]
- 52.Vidal, R., M. Vidal, R. Lagos, M. Levine, and V. Prado. 2004. Multiplex PCR for diagnosis of enteric infections associated with diarrheagenic Escherichia coli. J. Clin. Microbiol. 42:1787-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodward, M. J., and S. E. Kirwan. 1996. Detection of Salmonella Enteritidis in eggs by the polymerase chain reaction. Vet. Rec. 138:411-413. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, T., and M. P. Doyle. 2001. Evaluation of universal preenrichment broth for growth of heat-injured pathogens. J. Food Prot. 64:1751-1755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.