Abstract
Bile salts play an important role in the digestion of lipids in vertebrates and are synthesized and conjugated to either glycine or taurine in the liver. Following secretion of bile salts into the small intestine, intestinal microbes are capable of deconjugating the glycine or taurine from the bile salts, using an enzyme called bile salt hydrolase (Bsh). Intestinal lactobacilli are regarded as major contributors to bile salt hydrolysis in vivo. Since the bile salt-hydrolyzing strain Lactobacillus plantarum WCFS1 was predicted to carry four bsh genes (bsh1, bsh2, bsh3, and bsh4), the functionality of these bsh genes was explored using Lactococcus lactis heterologous overexpression and multiple bsh deletion strains. Thus, Bsh1 was shown to be responsible for the majority of Bsh activity in L. plantarum WCFS1. In addition, bsh1 of L. plantarum WCFS1 was shown to be involved in conferring tolerance to specific bile salts (i.e., glycocholic acid). Northern blot analysis established that bsh1, bsh2, bsh3, and bsh4 are all expressed in L. plantarum WCFS1 during the exponential growth phase. Following biodiversity analysis, bsh1 appeared to be the only bsh homologue that was variable among L. plantarum strains; furthermore, the presence of bsh1 correlated with the presence of Bsh activity, suggesting that Bsh1 is commonly responsible for Bsh activity in L. plantarum strains. The fact that bsh2, bsh3, and bsh4 genes appeared to be conserved among L. plantarum strains suggests an important role of these genes in the physiology and lifestyle of the species L. plantarum. Analysis of these additional bsh-like genes in L. plantarum WCFS1 suggests that they might encode penicillin acylase rather than Bsh activity, indicating their implication in the conversion of substrates other than bile acids in the natural habitat.
Bile salts play an essential role in lipid digestion in vertebrates. They act as a detergent that emulsifies and solubilizes dietary lipids and lipid-soluble vitamins. In the liver, bile acids are synthesized and conjugated as an N-acyl amidate with the amino acid taurine or glycine before being excreted via the bile duct into the small intestine (see Fig. S1 in the supplemental material). Usually, species of the intestinal microbiota, including a number of lactobacilli, produce bile salt hydrolases that are able to deconjugate the amino acid moiety from the bile salts in the intestine (see Fig. S1 in the supplemental material). A strong correlation has been found between the habitat of a specific bacterial species or strain and Bsh activity (48), suggesting a relationship between the capability to deconjugate bile salts and survival or persistence of bacteria under gastrointestinal conditions. Furthermore, intestinal bile salt deconjugation is believed to play an important role in host physiology, as it is the gatekeeping reaction in further oxidation and dehydroxylation steps of primary bile salts (as synthesized by the host) into secondary bile salts by intestinal bacteria. Notably, the production of secondary bile acids has been linked to various intestinal diseases, such as the formation of gallstones and colon cancer (45).
According to the enzyme classification system, bile salt hydrolase (Bsh; EC 3.5.1.24) belongs to the category of enzymes that act on carbon-nitrogen bonds other than peptide bonds in linear amides. Among others, this enzyme category includes members of the β-lactam acylase family, such as penicillin and cephalosporin acylases (EC 3.5.1.11) and ceramidases (EC 3.5.1.23). Although Bsh shares significant sequence homology with some of the enzymes in the EC 3.5.1 group and the type of bond that is cleaved is identical, the types of substrates that can be converted by the various enzymes are quite heterogeneous and may vary significantly in molecule size and hydrophobicity.
Bsh activity has been found in a wide variety of mostly gram-positive species (for a review, see reference 3), including Bifidobacterium (23, 26, 49), Clostridium (22, 27, 28, 34), Enterococcus (19), Listeria (4), and Lactobacillus (2, 10-12, 18, 24, 33, 35, 43) species, with the exception of the gram-negative Bacteroides species (34, 47). Thus, Bsh activity does not appear to be limited to either pathogenic or probiotic strains. The Lactobacillus plantarum WCFS1 genome (29) was predicted to contain four related genes, annotated as bsh1 to bsh4, that are spread throughout the genome (29), and the amino acid identity levels of the corresponding putative proteins range from 21 to 39%. However, functional analysis of an L. plantarum WCFS1 bsh1 mutant suggested that Bsh1 is responsible for the majority of Bsh activity produced by this strain (33).
Here we present a functional analysis of bsh1, bsh2, bsh3, and bsh4 of L. plantarum WCFS1. To investigate the predicted functions of these genes, each of the four bsh genes was overexpressed in the Bsh-deficient species Lactococcus lactis. In addition, single, double, triple, and quadruple bsh knockout mutants of L. plantarum WCFS1 were constructed to evaluate the contributions of the individual bsh genes to hydrolysis of and/or tolerance to various substrates, including bile salts, penicillin V, and acyl-homoserine lactones. Furthermore, the evolutionary conservation of bsh homologs was investigated in several strains of the species L. plantarum, using complete genome hybridization (CGH) (38). These results indicated that bsh2, bsh3, and bsh4 appear to be conserved among L. plantarum strains, suggesting an important physiological role. In addition, the presence of bsh1 appeared to be correlated with the Bsh activity of L. plantarum strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The bacterial strains, plasmids, and primers used in this study and their relevant features are listed in Table S1 in the supplemental material. L. plantarum WCFS1 (29) and bsh mutant derivatives were grown at 37°C in MRS broth (Difco, West Molesey, United Kingdom), without aeration. The heterologous nisin-controlled expression (NICE) host L. lactis NZ9000 and its parental strain, MG1363 (21), which was used as an intermediate cloning host for NICE overexpression constructs (31, 36), were grown at 30°C in M17 broth (Oxoid, Hampshire, United Kingdom) supplemented with 0.5% glucose (wt/vol; G-M17), without aeration. Escherichia coli strains DH5α (55) and MC1061 (9, 54) were used as intermediate cloning hosts for L. plantarum mutagenesis constructs and pCR-Blunt constructs, respectively, and were grown at 37°C on TY broth (25), with aeration. When appropriate, antibiotics were added to the media. For L. plantarum, 10 μg/ml chloramphenicol and 10 μg/ml (in liquid medium) or 30 μg/ml (on solid medium) erythromycin were used. For L. lactis, 10 μg/ml chloramphenicol was used. For E. coli, 10 μg/ml chloramphenicol and 250 μg/ml erythromycin were used.
DNA and protein manipulations.
Plasmid DNA was isolated from E. coli on a small scale, using the alkaline lysis method (5). Large-scale plasmid DNA isolations were performed using Jetstar columns as recommended by the manufacturer (Genomed GmbH, Bad Oberhausen, Germany). Purification of DNA fragments from agarose gels was performed using the Wizard SV gel and PCR cleanup system (Promega, Leiden, The Netherlands). DNA isolation and transformation of L. plantarum and L. lactis were performed as described previously (16, 33). For DNA manipulations in E. coli, protein extraction, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), standard procedures were employed (46). Restriction endonucleases, Taq, Pfx, and Pwo DNA polymerases, T4 DNA ligase, and Klenow enzyme were used as prescribed by the manufacturers (Promega, Leiden, The Netherlands, and Boehringer, Mannheim, Germany). Primers were obtained from Genset Oligos (Paris, France).
RNA isolation and Northern blotting.
For RNA isolation, an overnight culture of L. plantarum WCFS1 was diluted 50-fold in 50 ml of fresh MRS medium, with or without the addition of 0.05% (wt/vol) porcine bile (Sigma, Zwijndrecht, The Netherlands), and grown to an optical density at 600 nm (OD600) of 1. Subsequently, 3 volumes of quench buffer (60% methanol, 66.7 mM HEPES, pH 6.5 [−40°C]) were added (44). The cells were immediately pelleted by centrifugation at 3,500 × g for 10 min (Megafuge 1.0R; Heraeus, Hanau, Germany), resuspended in 750 μl of ice-cold TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5), and mechanically disrupted (FastPrep FP120; Qbiogene, Illkirch, France) in the presence of 0.8 g of zirconium beads (Biospec Products, Bartlesville, OK), 0.18 g of Macaloid (Kronos Titan GmbH, Leverkusen, Germany), 50 μl of 10% SDS, and 500 μl of phenol. Subsequently, the RNA was purified from the upper, aqueous phase of the cell extract by phenol-chloroform extraction, precipitated with absolute ethanol, washed with 70% ethanol (46), and resuspended in 50 μl of MQ water. Northern blot analysis was performed as described earlier (46), using total RNA. As probes for bsh1, bsh2, bsh3, and bsh4, PCR amplification products of a large part of the genes (0.7 to 0.8 kb) were used after being amplified with Taq polymerase, using L. plantarum WCFS1 total DNA as a template in combination with the primer sets bsh1intF/bsh1R, bsh2intF/bsh2seqR, bsh3intF/bsh3R, and bsh4intF/bsh4R, respectively (see Table S2 in the supplemental material). Cross-hybridization of the individual probes with the other bsh sequences of L. plantarum WCFS1 was checked using dot blots of 0, 0.2, 0.8, 3.0, 12.5, and 50 ng of PCR amplification product encompassing bsh1, bsh2, bsh3, or bsh4 (amplified using Taq polymerase, WCFS1 total DNA, and primer set bsh1F/bshR, bsh2F/bsh2seqR, bsh3F/bsh3R, or bsh4F/bsh4R, respectively [see Table S2 in the supplemental material]).
Construction of bsh overexpression strains.
For overexpression of bsh1, a DNA fragment containing the complete bsh1 gene was amplified by PCR, using Pfx polymerase, with L. plantarum WCSF1 genomic DNA as a template and primers bsh1F and bsh1R. The resulting amplicon was cloned into pCR-Blunt (Invitrogen, Breda, The Netherlands). Subsequently, the bsh1 gene was recovered from the resulting plasmid as a 1.1-kb AflII-HindIII fragment and cloned downstream of and translationally fused to the nisA promoter in NcoI-HindIII-digested pNZ8048 (31, 36). The resulting nisin-controlled bsh1 expression plasmid was designated pNZ5306.
A DNA fragment containing bsh2 was amplified by PCR, using Pfx polymerase, with L. plantarum genomic DNA as a template and primers bsh2F2 and bsh2R. The amplicon obtained was digested with HindIII, and the resulting 1.1-kb fragment was cloned downstream of and translationally fused to the nisA promoter in pNZ8150 (36) digested with ScaI and HindIII, resulting in the bsh2 overexpression vector pNZ5307.
The overexpression plasmid for bsh3 was constructed analogously to the bsh1 plasmid pNZ5306. A bsh3-containing PCR amplicon (using Pfx polymerase, L. plantarum WCFS1 genomic DNA as a template, and primers bsh3F and bsh3R) was initially cloned into pCR-blunt (Invitrogen, Breda, The Netherlands), and a 1.1-kb fragment containing bsh3 was subcloned into pNZ8048 following the same cloning strategy as that employed with bsh1, yielding pNZ5308, which contains the bsh3 gene under control of the nisA transcription and translation signals.
Finally, a bsh4-containing DNA fragment was amplified by PCR, using Pfx polymerase, with L. plantarum WCFS1 genomic DNA as a template and primers bsh4F2 and bsh4R. The amplicon obtained was cloned into the pCR-Blunt vector. The bsh4 gene was recovered from the resulting plasmid by digestion with KpnI and ApaLI, followed by partial digestion with AflIII and cloning of the 1.1-kb, bsh4-containing fragment downstream of and translationally fused to the nisA promoter in NcoI-KpnI-digested pNZ8048, yielding the bsh4 overexpression construct pNZ5309.
For all overexpression constructs, the sequence of the cloned bsh gene was confirmed to be correct by sequencing. For overexpression studies of the bsh genes in L. lactis by use of the NICE system, pNZ5306, pNZ5307, pNZ5308, and pNZ5309 were transformed into L. lactis NZ9000.
Construction of bsh deletion mutant strains.
For the construction of deletion derivatives of L. plantarum WCFS1 that lack one or more of the bsh genes, the previously reported Cre-lox-based system for multiple gene deletions was used (33). The bsh1 deletion vector pNZ5325 and bsh1 deletion strain NZ5305 were constructed previously (33) (see Table S1 in the supplemental material). The bsh2 mutagenesis vector pNZ5329 (see Table S1 in the supplemental material) was constructed by successive cloning of the PCR-amplified 1.0-kb 5′ and 3′ chromosomal flanking regions of bsh2 (lp_0067) (using Pfx polymerase, L. plantarum WCFS1 genomic DNA as a template, and the primer sets bsh2kofr1F/bsh2kofr1R and bsh2kofr2F/bsh2kofr2R, respectively [see Table S2 in the supplemental material]) into the SwaI and Ecl136II restriction sites of pNZ5319 (see Table S1 in the supplemental material), respectively (33). Analogously, the bsh3 mutagenesis vector pNZ5332 (see Table S1 in the supplemental material) was constructed by successive cloning of the PCR-amplified 1.0-kb 5′ and 3′ chromosomal flanking regions of bsh3 (lp_3362) (using Pfx polymerase, L. plantarum WCFS1 genomic DNA as a template, and primer sets bsh3kofr1NheF/bsh3kofr1R and bsh3kofr2F/bsh3kofr2R, respectively [see Table S2 in the supplemental material]) into the PmeI and Ecl136II restriction sites of pNZ5319 (see Table S1 in the supplemental material), respectively (33). Finally, the bsh4 mutagenesis vector pNZ5336 (see Table S1 in the supplemental material) was constructed by successive cloning of the PCR-amplified 1.0- and 0.9-kb 5′ and 3′ chromosomal flanking regions of bsh4 (lp_2572) (using Pfx polymerase, L. plantarum WCFS1 genomic DNA as a template, and primer sets bsh4kofr1F/bsh4kofr1R and bsh4kofr2F2/bsh4kofr2SalR, respectively [see Table S2 in the supplemental material]) into the SwaI and Ecl136II restriction sites of pNZ5319 (see Table S1 in the supplemental material), respectively (33). For all bsh deletion constructs, the sequences of the cloned PCR-amplified regions were verified by double-strand sequence analysis (BaseClear, Roosendaal, The Netherlands).
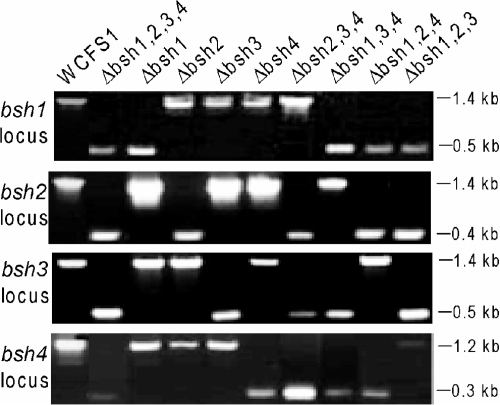
The bsh deletion strains NZ5305 (Δbsh1), NZ5307 (Δbsh2), NZ5309 (Δbsh3), NZ5311 (Δbsh4), NZ5313 (Δbsh1 Δbsh2), NZ5315 (Δbsh3 Δbsh4), NZ5324 (Δbsh1 Δbsh2 Δbsh3), NZ5326 (Δbsh1 Δbsh2 Δbsh4), NZ5328 (Δbsh1 Δbsh3 Δbsh4), NZ5330 (Δbsh2 Δbsh3 Δbsh4), and NZ5332 (Δbsh1 Δbsh2 Δbsh3 Δbsh4) were constructed as described previously (33) (see Table S1 and Fig. S1 in the supplemental material). Briefly, the desired bsh deletion vector was transformed into L. plantarum WCFS1 or one of its mutant derivatives by electroporation (33), and double-crossover gene replacement mutants in which the target gene was replaced by a lox66-P32-cat-lox71 cassette were selected based on their Cmr Ems phenotype. Correct integration of the lox66-P32-cat-lox71 cassette into the genome was confirmed by PCR amplification of the flanking regions of the integrated lox66-P32-cat-lox71 cassette, using primers annealing uniquely to genomic sequences combined with the mutagenesis vector-specific primers 85 and 87 that annealed to the P32-cat region (i.e., primers 106a/85 and 87/107a for bsh1 replacement, bsh2ko-up/85 and 87/bsh2ko-down for bsh2 replacement, bsh3ko-up/85 and 87/bsh3ko-down for bsh3 replacement, and bsh4ko-up/85 and 87/bsh4ko-down for bsh4 replacement [see Table S2 in the supplemental material]). Subsequently, the lox66-P32-cat-lox71 cassette was removed from the genome by transient expression of the Cre recombinase enzyme from a plasmid that is unstable in lactobacilli (pNZ5348), leading to stable deletion of the bsh gene, as described earlier (33). The Cre-lox-based mutagenesis system was designed specifically to allow subsequent rounds of mutagenesis within a single genetic background by using single-nucleotide mutant lox sites (lox66 and lox71) that, after recombination, lead to a double mutant lox (lox72) recombination site that is not recognized by Cre in subsequent rounds of mutagenesis (33). The L. plantarum bsh deletion mutant strains were checked by PCR, amplifying each of the four (mutated) bsh loci by using bsh1fr1F/bsh1R (yielding a 1.4-kb and a 0.5-kb product for the wild-type locus and mutated locus, respectively), bsh2fr1intF/bsh2contrR (yielding a 1.4-kb and a 0.4-kb product for the wild-type locus and mutated locus, respectively), bsh3fr1intF/bsh3R (yielding a 1.4-kb and a 0.5-kb product for the wild-type locus and mutated locus, respectively), and bsh4fr1intF/bsh4R (yielding a 1.2-kb and a 0.3-kb product for the wild-type locus and mutated locus, respectively) for bsh1, bsh2, bsh3, and bsh4, respectively (Fig. 1). Using this system, L. plantarum WCFS1 derivatives were constructed with stable deletion mutations in one, two, three, or all four of the bsh genes (see above; see Table S1 in the supplemental material).
FIG. 1.
Schematic overview of bsh mutants made in L. plantarum WCFS1 and PCR analysis encompassing the bsh locus of the mutants. Sizes of PCR products are indicated. For the wild-type locus of bsh1, bsh2, bsh3, and bsh4, the sizes of the PCR products were 1.4, 1.4, 1.4, and 1.2 kb, respectively, whereas for the mutated locus of bsh1, bsh2, bsh3, and bsh4, the sizes of the PCR products were 0.5, 0.4, 0.5, and 0.3 kb, respectively. Δbsh1,2,3,4, NZ5332; Δbsh1, NZ5305; Δbsh2, NZ5307; Δbsh3, NZ5309; Δbsh4, NZ5311; Δbsh2,3,4, NZ5330; Δbsh1,3,4, NZ5328; Δbsh1,2,4, NZ5326; Δbsh1,2,3, NZ5324.
High-performance liquid chromatography (HPLC) assay of Bsh activity.
To determine the Bsh activities of L. plantarum strains, overnight cultures were inoculated 1:10 in fresh MRS medium and cells were either grown to an OD600 of 3 or grown overnight. Cells were harvested by centrifugation for 10 min at 3,500 × g at room temperature (Megafuge 1.0R; Heraeus, Hanau, Germany) and resuspended in MRS medium to an OD600 of 100.
Overexpression of individual bsh genes in L. lactis by use of the NICE system was performed using established protocols (14). In short, overnight cultures of the L. lactis bsh overexpression strains were subcultured (1:50) in fresh G-M17 medium and grown to an OD600 of 0.5. Subsequently, 1 ng/ml nisin (Sigma, Zwijndrecht, The Netherlands) was added to these cultures, and growth was continued for 2 h. Cells were harvested by centrifugation for 10 min at 3,500 × g at room temperature (Megafuge 1.0R; Heraeus, Hanau, Germany). Cell pellets were resuspended to a final OD600 of 200 in 55 mM sodium acetate buffer, pH 6.5, containing 1 mM dithiothreitol (DTT) and 1 g of zirconium beads (52) and mechanically disrupted (FastPrep FP120; Qbiogene, Illkirch, France). Following centrifugation, cell extracts were used immediately. The protein concentration in cell extracts was determined as described previously (6).
Conversion of 1 mmol/liter of the bile salts glycocholic acid, glycodeoxycholic acid (GDC), glycochenodeoxycholic acid, taurocholic acid (TC), taurodeoxycholic acid (TDC), and taurochenodeoxycholic acid (Sigma, Zwijndrecht, The Netherlands) or of Fischer rat bile isolated by drainage from the bile duct (courtesy of Wageningen University, Wageningen, The Netherlands) by intact cells of L. plantarum or cell extracts of L. lactis bsh overexpression strains was determined at 37°C by HPLC as described previously (13). Separations were carried out with a reversed-phase resin-based column (PLRP-S; 5 μm by 300 Å by 250 mm by 4.6-mm inner diameter [Polymer Laboratories, Shropshire, United Kingdom]) and matching precolumn. Bile salts were detected using a pulsed amperometric detector (EG&G Princeton Applied Research, Princeton, NJ) equipped with a gold working electrode and a reference electrode (Ag/AgCl). Chromatograms were analyzed and integrated using the Chromeleon program (Dionex, Sunnyvale, CA), and Bsh activity was determined based on the disappearance of the conjugated bile salts used as a substrate.
Alternative acylase functionality.
Cell extracts of bsh-overexpressing L. lactis strains in physiological salt (following the procedures described above) were analyzed for alternative acylase activity (see Table S3 in the supplemental material). To determine penicillin, ampicillin, cephalosporin, acyl-homoserine lactone, and phenylacetylglycine acylase activities, 5 volumes of the cell extract was mixed with 5 volumes of 100 mM sodium acetate buffer, pH 5, containing 1 mM DTT and 1 volume of 100 mM of penicillin V, penicillin G, ampicillin, cephalosporin C, ketocaproyl-homoserine lactone, oxooctanoyl-homoserine lactone, or phenylacetylglycine and incubated overnight at 37°C. To stop the reaction, 35 volumes of 285 mM sodium acetate buffer, pH 4, was added. Free amino groups resulting from enzymatic conversion of the substrate were detected by the addition of 5 volumes of 10 mg/ml fluorescamine in acetone, centrifugation for 10 min at 3,000 × g at room temperature (TechnoSpin R; Sorval Instruments), and measurement of fluorescence in the supernatant (excitation at 360 nm and emission at 465 nm) (GENios F129004; Tecan Benelux, Giessen, The Netherlands). As a positive control, purified penicillin acylase and purified end products of the acylase reactions (6-aminopenicillanic acid, 7-aminocephalosporanic acid, homoserine lactone, and glycine) (Sigma, Zwijndrecht, The Netherlands) were used.
Furthermore, acylase activity for 6-nitro-3-(phenylacetamido) benzoic acid (NIPAB; Sigma, Zwijndrecht, The Netherlands), which is a commonly used chromogenic substrate for assaying penicillin G acylase activity, was determined as described earlier (1, 32). Briefly, 1 volume of cell extract was mixed with 9 volumes of 100 mM sodium acetate buffer containing 1 mM DTT and 2.5 mM NIPAB. Acylation of NIPAB at 25°C was determined by the increase of absorption at 405 nm, which was followed for 30 min and measured overnight. As a positive control, purified penicillin amidase of E. coli was used (Sigma, Zwijndrecht, The Netherlands).
Bile salt and penicillin V tolerance.
To evaluate the tolerance of L. plantarum WCFS1 and its bsh mutant derivatives to bile salts and penicillins, overnight cultures were inoculated 1:20 into fresh MRS medium containing 0 to 30% ox gall (wt/vol), 0 to 0.4% (wt/vol) GDC, 0 to 10% (wt/vol) TDC, and 0 to 14 μg/ml penicillin V. Growth was followed for 16 h by measurement of the OD600 at 37°C at intervals of 15 min (Spectra Max Plus 384; Molecular Devices, Sunnyvale, CA).
bsh diversity in L. plantarum strains.
The genomic diversity of L. plantarum strains (299, 299v, CIP102359, CIP104440, CIP104441, CIP104448, CIP104450, CIP104451, CIP104452, LP85-2, NCIMB12120, and SF2A35B) was previously investigated by use of strain WCFS1-derived DNA microarrays (38). This genomic genotyping database allowed the evaluation of the presence and/or absence of homologues of the four genes that were initially annotated in the L. plantarum WCFS1 genome as bsh genes (bsh1, bsh2, bsh3, and bsh4). Following statistical analysis, a positive cutoff P value of 1e−5 was used for presence calling of bsh homologs.
Detection of Bsh activity in L. plantarum strains.
The presence of Bsh activity in different L. plantarum strains (299, 299v, CIP102359, CIP104440, CIP104441, CIP104448, CIP104450, CIP104451, CIP104452, LP85-2, NCIMB12120, and SF2A35B) was detected using a bile salt plate assay, as described earlier (12). Briefly, overnight cultures of L. plantarum strains were transferred to solid MRS medium, with or without 0.5% (wt/vol) of the bile salt TDC, and incubated anaerobically for 48 h at 37°C. Bsh-active strains were recognized by the formation of opaque white colonies in the presence of TDC, which is due to the precipitation of deconjugated bile salt forms.
RESULTS
Expression of bsh genes.
L. plantarum WCFS1 contains four related bsh genes (29). The expression of these genes was studied by Northern blotting during exponential growth phase in the presence or absence of porcine bile (data not shown). The probes used for Northern blotting were specific for each of the individual bsh genes and did not show cross-hybridization. The Northern blot analysis established that bsh1, bsh2, bsh3, and bsh4 are all expressed in L. plantarum WCFS1 during the exponential growth phase. The estimated sizes of the transcripts corresponded with those predicted for monocistronic transcription of bsh1, bsh2, bsh3, and bsh4 (1.2 kb for bsh1, bsh2, and bsh3 and 1.1 kb for bsh4). Incubation of L. plantarum WCFS1 grown in liquid medium with 0.5% (wt/vol) porcine bile did not induce significant expression of any of the bsh genes.
Divergence of Bsh in L. plantarum.
In order to explore the functionality of the bsh genes of L. plantarum further, the presence of homologues of the four bsh genes of L. plantarum WCFS1 in other L. plantarum strains was determined by analysis of L. plantarum genomic DNA samples on L. plantarum WCFS1-specific microarrays, as described earlier (38), using a positive cutoff P value of 1e−5 for bsh gene presence calls. The presence and absence calls were correlated with the experimentally determined capability of these strains to hydrolyze bile salts, using a previously described Bsh plate assay (12; data not shown). Remarkably, in all L. plantarum strains, bsh2, bsh3, and bsh4 generated very good probability scores for gene presence, suggesting that these genes are highly conserved among L. plantarum strains. In contrast, the presence of a gene homologous to bsh1 appeared to vary among the strains analyzed. The bsh1 gene appeared to be absent in 4 of the 13 strains analyzed, which appeared to correlate well with the absence of Bsh activity in these four strains. The remaining nine strains appeared to contain a bsh1 homologue, as concluded from CGH analysis. Seven of these nine strains also displayed clearly detectable Bsh activity in the plate assay employed, while the other two strains did not display activity in this assay. Therefore, it is likely that Bsh activity is related to the presence of a bsh1 homologue in the species L. plantarum.
Bsh activities of individual Bsh proteins.
Since all bsh genes of L. plantarum WCFS1 appeared to be expressed during exponential growth, the contributions of the individual bsh genes to the total Bsh activity were determined. For this purpose, heterologous bsh overexpression strains of L. lactis were established. Furthermore, a set of single and multiple bsh deletion derivatives of L. plantarum WCFS1 was constructed (see Table S1 in the supplemental material).
Heterologous overexpression strains.
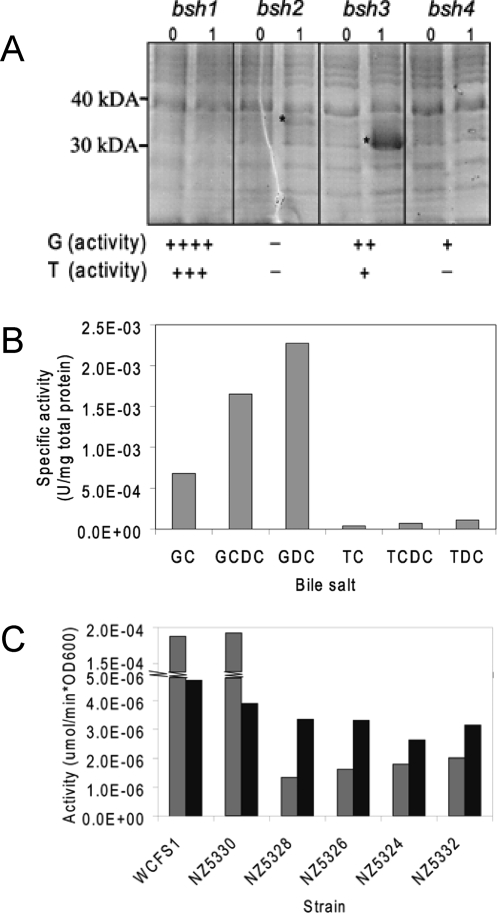
For heterologous overexpression using the NICE system (36), vectors pNZ5306, pNZ5307, pNZ5308, and pNZ5309, containing the bsh1, bsh2, bsh3, and bsh4 genes, respectively, translationally coupled to the nisin-inducible nisA promoter, were constructed and transformed into the Bsh-deficient bacterium L. lactis NZ9000. Only following nisin induction could overexpression protein products be detected by SDS-PAGE for Bsh2 and Bsh3 (Fig. 2A), at their expected molecular masses of 38 and 36 kDa, respectively. HPLC-based Bsh activity assays (13) using taurine- and glycine-conjugated bile salts of cholic acid, deoxycholic acid, and chenodeoxycholic acid as substrates (see Fig. S1 in the supplemental material) revealed that Bsh1, Bsh3, and Bsh4, but not Bsh2, are capable of bile salt deconjugation (Fig. 2A). These results confirmed the functional heterologous expression of both Bsh1 and Bsh4, despite the lack of detection of the corresponding protein products by SDS-PAGE; in heterologous expression, the amount of protein produced is not necessarily correlated to specific activity. Bsh1 and Bsh3 displayed a strong preference for glycine-conjugated bile salts compared to taurine-conjugated bile salts, while Bsh4 appeared to exclusively convert glycine-conjugated bile salts. Moreover, consistent differential substrate specificity of Bsh1, Bsh3, and Bsh4 was observed, with a substrate preference diminishing from deoxycholic acid to chenodeoxycholic acid and to cholic acid-conjugated bile salts (Fig. 2B).
FIG. 2.
(A) Protein gel and activities of heterologous overexpression products of bsh1, bsh2, bsh3, and bsh4, using the NICE system (36) in L. lactis. The predicted sizes of the Bsh1, Bsh2, Bsh3, and Bsh4 proteins were 37, 38, 36, and 36 kDa, respectively. For Bsh2 overexpression, no Bsh activity was detected. Bsh3 and Bsh4 were capable of marginal Bsh activity, with no activity found for Bsh4 for taurine-conjugated substrates. In contrast, Bsh1 overexpression yielded major Bsh activity. 0, no nisin induction; 1, induction with 1 ng/ml nisin; *, protein band differentially expressed following nisin induction. (B) Substrate preference of Bsh proteins. For all heterologous Bsh overexpression strains, substrate preference diminished from GDC to TC. As an example, the substrate preference of the heterologous Bsh1 overexpression strain is shown. (C) GDC deconjugation activities of L. plantarum WCFS1 and its bsh mutant derivatives at logarithmic and stationary growth phases. Gray bars represent the Bsh activity in the logarithmic growth phase; black bars represent the Bsh activity in stationary phase. NZ5330, Δbsh2 Δbsh3 Δbsh4 phenotype; NZ5328, Δbsh1 Δbsh3 Δbsh4 phenotype; NZ5326, Δbsh1 Δbsh2 Δbsh4 phenotype; NZ5324, Δbsh1 Δbsh2 Δbsh3 phenotype; NZ5332, Δbsh1 Δbsh2 Δbsh3 Δbsh4 phenotype.
Combinatorial bsh deletion strains.
In parallel, a set of combinatorial bsh mutant derivatives (Fig. 1; see Table S1 in the supplemental material) of L. plantarum WCFS1 was constructed using a Cre-lox-based mutation system that allows the effective deletion of multiple genes in a single genetic background (33). The bsh mutant derivatives were checked by PCR, amplifying each of the four bsh loci (Fig. 1). Growth appeared to be unaffected in these mutants compared to that of the parental strain, indicating that the deleted genes were not necessary for growth under normal laboratory conditions (data not shown).
Bsh activity was analyzed using an HPLC-based assay (13), with the purified bile salt GDC (Fig. 2C) and male Fischer rat bile (data not shown) as substrates for the triple and quadruple bsh mutant strains (NZ5324, NZ5326, NZ5328, NZ5330, and NZ5332). These experiments revealed that cells harboring an intact copy of the bsh1 gene (NZ5330) displayed Bsh activity levels that were comparable to those of the wild-type strain, confirming previous studies identifying this gene as the major Bsh-encoding gene in L. plantarum (33). Moreover, Bsh1-dependent Bsh activity declined drastically (about 40 times) in cells harvested during stationary growth phase compared with the activity in cells obtained from the logarithmic phase of growth (Fig. 2C), indicating growth phase-dependent expression of the bsh1 gene. In all triple mutant, bsh1-deficient strains (NZ5324, NZ5326, and NZ5328) and the quadruple bsh mutant strain (NZ5332), a small but detectable amount of bile salt hydrolysis was found (Fig. 2C), which appeared to be bsh2, bsh3, or bsh4 independent.
Alternative functionality of individual Bsh proteins.
Since in L. plantarum WCFS1 the bile salt hydrolase activity appeared to be independent of bsh2, bsh3, and bsh4, the functionality of these genes was investigated by determination of the activities of the Bsh proteins on a variety of putative alternative (non-bile salt) substrates (see Table S3 in the supplemental material). The Bsh proteins of L. plantarum WCFS1 share significant sequence homology with penicillin V acylase enzyme family members (including the experimentally verified penicillin V acylase of Listeria monocytogenes EGDe [4], with amino acid identity levels ranging from 30 to 26%), which are in turn related to β-lactam acylases and to acyl-homoserine lactone acylases, which play a key role in quorum sensing-dependent gene regulation in gram-negative bacteria. Bsh, penicillin acylase, β-lactam acylase, and acyl-homoserine lactone acylase all act on the same type of chemical bond, although the structures of their substrates differ considerably (see Fig. S2 in the supplemental material). Thus, the activities of the overexpression products for the individual Bsh proteins, using the NICE system (36) in L. lactis NZ9000 (using pNZ5306, pNZ5307, pNZ5308, and pNZ5309), were determined for the substrates penicillin V, penicillin G, NIPAB (which is a commonly used substrate for spectrophotometric detection of penicillin acylase activity), the β-lactams ampicillin and cephalosporin C, ketocaproyl-homoserine lactone, oxooctanoyl-homoserine lactone, and phenylacetylglycine, which is a molecule that is involved in phenylalanine metabolism and thereby readily available to L. plantarum and is cleaved by penicillin acylase of E. coli (51) (see Table S3 and Fig. S2 in the supplemental material).
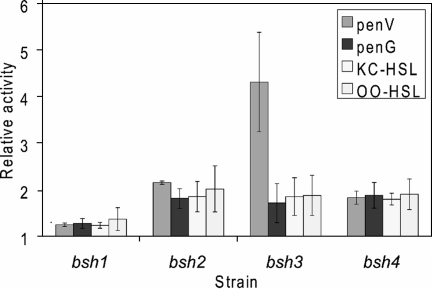
For NIPAB, ampicillin, cephalosporin C, and phenylacetylglycine, no activity could be detected in any of the Bsh overexpression strains. However, bsh3 (and, to a lesser extent, bsh2 and bsh4) overexpression resulted in an increase in acylase activity of penicillin V (on average, 4.3, 2.2, and 1.8 times, respectively) relative to that of the control strain NZ9000 in several independent experiments (Fig. 3). These findings clearly suggest a role as a penicillin acylase for Bsh3, and possibly Bsh2 and Bsh4. In addition, bsh2, bsh3, and bsh4 overexpression strains all showed increases in activities (on average, 1.9 times) toward penicillin G, ketocaproyl-homoserine lactone, and oxooctanoyl-homoserine lactone relative to those of NZ9000, suggesting a broad range of enzyme specificities. Notably, the bsh1 overexpression strain showed no significant activity toward penicillin V, penicillin G, ketocaproyl-homoserine lactone, or oxooctanoyl-homoserine lactone compared to those of NZ9000, confirming the role of bsh1 as a bona fide Bsh.
FIG. 3.
Changes in activity of heterologous overexpression products of bsh1, bsh2, bsh3, and bsh4, using the NICE system (36) in L. lactis NZ9000, relative to that in strain NZ9000 for penicillin V (penV), penicillin G (penG), ketocaproyl-homoserine lactone (KC-HSL), and oxooctanoyl-homoserine lactone (OO-HSL).
Bile salt and penicillin V tolerance.
The contributions of the individual bsh genes to the tolerance of the bile salts TDC and GDC, ox gall, and penicillin V (see Fig. S1 and S2 in the supplemental material) were determined using the triple and quadruple L. plantarum bsh mutant strains (NZ5324, NZ5326, NZ5328, NZ5330, and NZ5332). To this end, growth in the presence of a range of bile (salt) and penicillin concentrations was monitored spectrophotometrically.
L. plantarum was able to grow in the presence of up to 30% (wt/vol) of ox gall, with no significant differences in growth between the strains (data not shown). However, due to the high concentrations of ox gall used, measurements were severely hampered. In addition, L. plantarum was able to grow in the presence of >14% (wt/vol) of TDC, with no significant differences found for the strains used (data not shown). However, L. plantarum appeared to be remarkably more sensitive to GDC, with obvious differences between the strains (data not shown). The results clearly established that the presence of bsh1 in L. plantarum enhances GDC bile salt tolerance. Each of the bsh1-deficient derivatives displayed GDC-mediated growth inhibition at concentrations as low as 0.1% (wt/vol) GDC. In contrast, strains containing an intact bsh1 gene were capable of sustaining normal growth characteristics at up to 0.5 to 0.7% (wt/vol) GDC. Analogous with the limited level of hydrolytic activity toward bile salts, bsh2, bsh3, and bsh4 did not appear to contribute significantly to tolerance of GDC.
Furthermore, growth of L. plantarum was inhibited at the lowest concentration of penicillin V tested (0.3 μg/ml), with complete inhibition of growth in the presence of 8 μg/ml penicillin V. However, no difference was found between WCFS1 and its bsh mutant derivatives, indicating that none of the bsh genes appeared to influence penicillin V tolerance in L. plantarum under the conditions analyzed here.
DISCUSSION
Bile salt hydrolysis is a biologically important reaction in the intestinal tract, since it is the first step in bile salt biotransformations carried out by intestinal bacteria. The formation of secondary bile salts has a significant impact on the physiology of the host, as exemplified by suggestions regarding their implication in lowering of blood cholesterol levels (42) and in various intestinal diseases, such as the formation of gallstones and colon cancer (45). Furthermore, deconjugated bile salts were shown to induce mucin production by intestinal epithelial cells (30), possibly indicating irritation of the epithelial cells by the strong surface-active properties of deconjugated bile salts.
Remarkably, for several strains (e.g., Lactobacillus johnsonii 100-100 and NCC533, Lactobacillus acidophilus NCFM, and L. plantarum WCFS1), the presence of more than one gene encoding a Bsh homologue has been predicted. In L. plantarum WCFS1, the sequences of the bsh genes share higher levels of similarity with bsh genes from other strains or species than with each other. For example, bsh1 shares highest sequence similarity with the bsh genes of other L. plantarum strains and Enterococcus faecalis, whereas bsh2 and bsh4 share sequence similarity with the sequence annotated as bsh of Lactobacillus brevis and share no significant sequence similarity with any other organism whose sequence is publicly available; by analogy, bsh3 shares significant sequence similarity only with the sequence annotated as bsh of Lactobacillus sakei and L. brevis. The bsh genes of L. plantarum WCFS1 may have been acquired via horizontal gene transfer, as suggested earlier for L. johnsonii (18). However, the overall conservation among L. plantarum strains of the bsh2 to bsh4 genes would indicate that this acquisition occurred very early in the evolution of this species. Moreover, this high level of conservation also supports an important role of these genes in the physiology and lifestyle of the species L. plantarum.
In line with the previous finding that bsh1 is the major Bsh in L. plantarum WCFS1 (33), the presence of bsh1 appeared to correlate with the capability to hydrolyze bile salts in 11 of 13 L. plantarum strains. For 2 of 13 strains, however, a Bsh1 homolog was concluded to be present, while the capability to hydrolyze TDC was not detected in these strains. However, this apparent inconsistency can be explained by the fact that the detection of the presence of bsh1 in a particular strain does not necessarily correlate with expression of functional Bsh1 that is identical to the protein expressed by L. plantarum WCFS1. Firstly, the bsh1-like gene found by CGH analysis may contain disruptive (point) mutations, leading to detection of the gene but a lack of functionality. Secondly, the presence of a bsh1-like gene in a particular strain does not give any information on the expression of this gene under the conditions applied in the enzyme assay. Thus, the presence of a gene does not necessarily correlate with expression of this gene. Thirdly, the bsh1 homologues found may display a more stringent substrate preference than that of Bsh1 of L. plantarum WCFS1, thereby failing to convert the specific bile salt TDC used in this experiment. Thus, the Bsh1 homologues may display activity for bile salts other than TDC.
In L. plantarum WCFS1, the presence of bsh1 correlated with GDC tolerance but not with TDC tolerance. The capacity to hydrolyze bile salts has been found to be linked to bile salt tolerance in L. plantarum (15) and several other bacteria, including Lactobacillus amylovorus (24), Listeria monocytogenes (4), and Bifidobacterium (40). Thus, the preference of L. plantarum WCFS1 for deconjugation of glycine- over taurine-conjugated bile salts appears to be reflected in the differential tolerance toward GDC and TDC. This could be related to the higher toxicity of glycine-conjugated bile salts than of taurine-conjugated bile salts, leading to evolution of a preference of Bsh for glycine-conjugated bile salts. Indeed, most Bsh proteins show a preference for glycine-conjugated bile salts (3). Although the precise mechanism is unknown, bile salt hydrolysis could be of great importance for survival in vivo. Since deconjugated bile acids display reduced solubility compared to their conjugated counterparts, especially at lower pH values, bile salt hydrolysis may lead to precipitation of the bile salts and thereby relieve stress levels caused by these surface-active chemicals. Thereby, the capability to hydrolyze bile salts may contribute to the survival and persistence of bacterial strains in the intestinal tract, as previously shown for Listeria monocytogenes (17). Analogously, Bsh activity appears to be present in all lactobacilli isolated from the gastrointestinal environment (48). Notably, L. plantarum WCFS1 is capable of persisting in the mouse gastrointestinal tract for 10 days (41) and has been shown to display relatively high survival and activity during transit of the human gastrointestinal tract (53). Additional animal experiments using the set of L. plantarum WCFS1 bsh mutant derivatives constructed in this work may clarify the role of the bsh genes in persistence and survival of the organism in the gastrointestinal tract.
All four of the bsh genes of L. plantarum WCFS1 are expressed during the exponential phase of growth as monocistronic transcripts. The expression of the bsh genes did not appear to be induced as a consequence of exposure to porcine bile during growth in liquid media. In contrast, previous whole-genome transcriptome studies suggested that expression of bsh1 was induced by porcine bile when cells were grown on solid media, while the expression of bsh3 appeared to be repressed under these conditions (7). In the same study, no regulation of bsh2 or bsh4 by porcine bile was detected. Notably, these findings are in apparent agreement with the finding that bsh1 and, to a lesser extent, bsh3 are capable of bile salt hydrolysis. Nevertheless, the discrepancy in regulation of bsh expression may be due to the difference in growth conditions used (liquid versus solid medium). Explaining these different findings would require further investigation of the regulation of expression of bsh as a function of culture conditions.
Heterologous overexpression of the bsh genes by use of the NICE system in the Bsh-deficient host L. lactis NZ9000 confirmed that Bsh1 is a functional and bona fide Bsh, as described earlier (33). In addition, Bsh3 and Bsh4 displayed bile salt hydrolysis activity, although with lower enzymatic efficiencies than that of Bsh1, which suggests that these enzymes could contribute to the overall Bsh activity displayed by L. plantarum WCFS1. Analogous to what has been described for most of the Bsh enzymes studied to date (27, 37, 49, 50), the Bsh1, Bsh3, and Bsh4 enzymes displayed a clear preference for glycoconjugated over tauroconjugated bile salts. Furthermore, the substrate preference of bsh1, bsh3, and bsh4 decreased from deoxycholic to chenodeoxycholic and cholic acid as the steroid moiety of the conjugated bile salts (see Fig. S1 and S2B in the supplemental material), which has previously also been found for the Bsh activity of Lactobacillus buchneri (39). This substrate preference is probably related to the positioning and presence or absence of hydroxyl groups on the steroid moiety of the bile salts (see Fig. S1 in the supplemental material) and, concomitantly, the binding pocket properties of the enzyme.
Mutation analysis with L. plantarum WCFS1 confirmed that Bsh1 is the major Bsh. Nevertheless, differential activity of Bsh2, Bsh3, or Bsh4 under specific conditions or selective activity toward specific bile salts cannot be excluded. Screening for involvement of the bsh genes of L. plantarum WCSF1 in the conversion of various putative alternative substrates showed that Bsh3 and, to a lesser extent, Bsh2 and Bsh4 were able to hydrolyze penicillin V and penicillin G. To date, the in vivo role of penicillin acylase remains unknown. Notably, the enzyme name appears to reflect primarily its industrial application rather than its natural substrate. Therefore, Bsh2, Bsh3, and Bsh4 may play a role in acylation of compounds other than the substrates tested here, such as additional phenylacetic acid derivatives, as suggested by the capability of penicillin acylases to cleave these substrates in addition to penicillins (51). Indeed, phenylacetic acid derivatives would be available to L. plantarum in vivo, since they are formed by microbial activity on plant constituents, where L. plantarum was found to occur naturally (20). However, these substrates are not commercially available. In addition, low-level acylase activity by Bsh2, Bsh3, and Bsh4 toward two types of acyl-homoserine lactones was found, indicating the broad substrate specificity of these enzymes. Acyl-homoserine lactones could have an important function in adhesion of bacteria to the epithelium of the intestinal tract. For example, the pathogen Pseudomonas aeruginosa was found to upregulate PA-1 lectin/adhesin, an important virulence factor in this strain, in response to butanoyl-homoserine lactone in the environment (56). Therefore, bacteria that are capable of cleaving acyl-homoserine lactones could be of importance in preventing the adhesion of pathogens in the intestinal tract; however, acyl-homoserines appeared not to be the natural substrate for the Bsh proteins of L. plantarum WCFS1 due to the relatively low activity toward them.
Bsh1 showed detectable activity only toward bile salts, suggesting that Bsh1 displays a more narrow substrate specificity than that of Bsh2, Bsh3, or Bsh4.
In conclusion, Bsh1 was found to be responsible for the majority of Bsh activity in L. plantarum WCFS1, and possibly in all L. plantarum strains. Computational analyses predicted three other Bsh-encoding genes to be present in L. plantarum WCFS1, while experimental evidence showed that the functionality of these genes is unclear but possibly relates to acylase activity, with penicillin-like chemicals as substrates. Notably, the conservation of bsh2, bsh3, and bsh4 suggests an important but so far unknown role of these genes in the physiology and lifestyle of the species L. plantarum.
Supplementary Material
Footnotes
Published ahead of print on 6 June 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alkema, W. B., R. Floris, and D. B. Janssen. 1999. The use of chromogenic reference substrates for the kinetic analysis of penicillin acylases. Anal. Biochem. 275:47-53. [DOI] [PubMed] [Google Scholar]
- 2.Bateup, J. M., M. A. McConnell, H. F. Jenkinson, and G. W. Tannock. 1995. Comparison of Lactobacillus strains with respect to bile salt hydrolase activity, colonization of the gastrointestinal tract, and growth rate of the murine host. Appl. Environ. Microbiol. 61:1147-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begley, M., C. Hill, and C. G. Gahan. 2006. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72:1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begley, M., R. D. Sleator, C. G. Gahan, and C. Hill. 2005. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73:894-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bron, P. A., D. Molenaar, W. M. de Vos, and M. Kleerebezem. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100:728-738. [DOI] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 10.Christiaens, H., R. J. Leer, P. H. Pouwels, and W. Verstraete. 1992. Cloning and expression of a conjugated bile acid hydrolase gene from Lactobacillus plantarum by using a direct plate assay. Appl. Environ. Microbiol. 58:3792-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corzo, G., and S. E. Gilliland. 1999. Bile salt hydrolase activity of three strains of Lactobacillus acidophilus. J. Dairy Sci. 82:472-480. [DOI] [PubMed] [Google Scholar]
- 12.Dashkevicz, M. P., and S. D. Feighner. 1989. Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl. Environ. Microbiol. 55:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekker, R., R. van der Meer, and C. Olieman. 1991. Sensitive pulsed amperometric detection of free and conjugated bile acids in combination with gradient reversed-phase HPLC. Chromatographia 31:549-553. [Google Scholar]
- 14.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Smet, I., L. Van Hoorde, M. Vande Woestyne, H. Christiaens, and W. Verstraete. 1995. Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 79:292-301. [DOI] [PubMed] [Google Scholar]
- 16.de Vos, W. M., P. Vos, H. de Haard, and I. Boerrigter. 1989. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene 85:169-176. [DOI] [PubMed] [Google Scholar]
- 17.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchrieser, P. Glaser, and P. Cossart. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 18.Elkins, C. A., S. A. Moser, and D. C. Savage. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 147:3404-3412. [DOI] [PubMed] [Google Scholar]
- 19.Franz, C. M. A. P., I. Specht, P. Haberer, and W. H. Holzapfel. 2001. Bile salt hydrolase activity of enterococci isolated from food: screening and quantitative determination. J. Food Prot. 64:725-729. [DOI] [PubMed] [Google Scholar]
- 20.Gardner, N. J., T. Savard, P. Obermeier, G. Caldwell, and C. P. Champagne. 2001. Selection and characterization of mixed starter cultures for lactic acid fermentation of carrot, cabbage, beet and onion vegetable mixtures. Int. J. Food Microbiol. 64:261-275. [DOI] [PubMed] [Google Scholar]
- 21.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopal-Srivastava, R., and P. B. Hylemon. 1988. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J. Lipid Res. 29:1079-1085. [PubMed] [Google Scholar]
- 23.Grill, J., F. Schneider, J. Crociani, and J. Ballongue. 1995. Purification and characterization of conjugated bile salt hydrolase from Bifidobacterium longum BB536. Appl. Environ. Microbiol. 61:2577-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grill, J. P., C. Cayuela, J. M. Antoine, and F. Schneider. 2000. Isolation and characterization of a Lactobacillus amylovorus mutant depleted in conjugated bile salt hydrolase activity: relation between activity and bile salt resistance. J. Appl. Microbiol. 89:553-563. [DOI] [PubMed] [Google Scholar]
- 25.Killmann, H., C. Herrmann, A. Torun, G. Jung, and V. Braun. 2002. TonB of Escherichia coli activates FhuA through interaction with the beta-barrel. Microbiology 148:3497-3509. [DOI] [PubMed] [Google Scholar]
- 26.Kim, G. B., C. M. Miyamoto, E. A. Meighen, and B. H. Lee. 2004. Cloning and characterization of the bile salt hydrolase genes (bsh) from Bifidobacterium bifidum strains. Appl. Environ. Microbiol. 70:5603-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirby, L. C., R. A. Klein, and J. P. Coleman. 1995. Continuous spectrophotometric assay of conjugated bile acid hydrolase. Lipids 30:863-867. [DOI] [PubMed] [Google Scholar]
- 28.Kishinaka, M., A. Umeda, and S. Kuroki. 1994. High concentrations of conjugated bile acids inhibit bacterial growth of Clostridium perfringens and induce its extracellular cholylglycine hydrolase. Steroids 59:485-489. [DOI] [PubMed] [Google Scholar]
- 29.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klinkspoor, J. H., R. Kuver, C. E. Savard, D. Oda, H. Azzouz, G. N. Tytgat, A. K. Groen, and S. P. Lee. 1995. Model bile and bile salts accelerate mucin secretion by cultured dog gallbladder epithelial cells. Gastroenterology 109:264-274. [DOI] [PubMed] [Google Scholar]
- 31.Kuipers, O. P., P. G. G. A. de Ruyter, and M. Kleerebezem. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:7. [Google Scholar]
- 32.Kutzbach, C., and E. Rauenbusch. 1974. Preparation and general properties of crystalline penicillin acylase from Escherichia coli ATCC 11 105. Hoppe Seylers Z. Physiol. Chem. 355:45-53. [DOI] [PubMed] [Google Scholar]
- 33.Lambert, J. M., R. S. Bongers, and M. Kleerebezem. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73:1126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda, N. 1981. Deconjugation of bile salts by Bacteroides and Clostridium. Microbiol. Immunol. 25:1-11. [DOI] [PubMed] [Google Scholar]
- 35.McAuliffe, O., R. J. Cano, and T. R. Klaenhammer. 2005. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:4925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mierau, I., and M. Kleerebezem. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705-717. [DOI] [PubMed] [Google Scholar]
- 37.Min Kim, J., J. Young Choi, M. Sun Kim, and S. Chang Kim. 2004. In vivo excision and amplification of large human genomic segments using the Cre/loxP- and large T antigen/SV40 ori-mediated machinery. J. Biotechnol. 110:227-233. [DOI] [PubMed] [Google Scholar]
- 38.Molenaar, D., F. Bringel, F. H. Schuren, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 187:6119-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moser, S. A., and D. C. Savage. 2001. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 67:3476-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noriega, L., I. Cuevas, A. Margolies, and C. G. de los Reyes-Gavilán. 2006. Deconjugation and bile salts hydrolase activity by Bifidobacterium strains with acquired resistance to bile. Int. Dairy J. 16:6. [Google Scholar]
- 41.Pavan, S., P. Desreumaux, and A. Mercenier. 2003. Use of mouse models to evaluate the persistence, safety, and immune modulation capacities of lactic acid bacteria. Clin. Diagn. Lab. Immunol. 10:696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira, D. I., and G. R. Gibson. 2002. Effects of consumption of probiotics and prebiotics on serum lipid levels in humans. Crit. Rev. Biochem. Mol. Biol. 37:259-281. [DOI] [PubMed] [Google Scholar]
- 43.Pereira, D. I., A. L. McCartney, and G. R. Gibson. 2003. An in vitro study of the probiotic potential of a bile-salt-hydrolyzing Lactobacillus fermentum strain, and determination of its cholesterol-lowering properties. Appl. Environ. Microbiol. 69:4743-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pieterse, B., R. H. Jellema, and M. J. van der Werf. 2006. Quenching of microbial samples for increased reliability of microarray data. J. Microbiol. Methods 64:207-216. [DOI] [PubMed] [Google Scholar]
- 45.Ridlon, J. M., D. J. Kang, and P. B. Hylemon. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47:241-259. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Stellwag, E. J., and P. B. Hylemon. 1976. Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim. Biophys. Acta 452:165-176. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, H., K. Doesburg, T. Iwasaki, and I. Mierau. 1999. Screening of lactic acid bacteria for bile salt hydrolase activity. J. Dairy Sci. 82:2530-2535. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka, H., H. Hashiba, J. Kok, and I. Mierau. 2000. Bile salt hydrolase of Bifidobacterium longum: biochemical and genetic characterization. Appl. Environ. Microbiol. 66:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taranto, M. P., F. Sesma, and G. Font de Valdez. 1999. Localization and primary characterization of bile salt hydrolase from Lactobacillus reuteri. Biotechnol. Lett. 21:4. [DOI] [PubMed] [Google Scholar]
- 51.Valle, F., P. Balbas, E. Merino, and F. Bolivar. 1991. The role of penicillin amidases in nature and in industry. Trends Biochem. Sci. 16:36-40. [DOI] [PubMed] [Google Scholar]
- 52.van der Meer, J. R., J. Polman, M. M. Beerthuyzen, R. J. Siezen, O. P. Kuipers, and W. M. De Vos. 1993. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 175:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vesa, T., P. Pochart, and P. Marteau. 2000. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 14:823-828. [DOI] [PubMed] [Google Scholar]
- 54.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 55.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, L., C. Holbrook, O. Zaborina, E. Ploplys, F. Rocha, D. Pelham, E. Chang, M. Musch, and J. Alverdy. 2003. Pseudomonas aeruginosa expresses a lethal virulence determinant, the PA-I lectin/adhesin, in the intestinal tract of a stressed host: the role of epithelia cell contact and molecules of the quorum sensing signaling system. Ann. Surg. 238:754-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.