Abstract
Lactococcin 972 (Lcn972) is a nonlantibiotic bacteriocin that inhibits septum biosynthesis in Lactococcus lactis rather than forming pores in the cytoplasmic membrane. In this study, a deeper analysis of the molecular basis of the mode of action of Lcn972 was performed. Of several lipid cell wall precursors, only lipid II antagonized Lcn972 inhibitory activity in vivo. Likewise, Lcn972 only coprecipitated with lipid II micelles. This bacteriocin inhibited the in vitro polymerization of lipid II by the recombinant S. aureus PBP2 and the addition to lipid II of the first glycine catalyzed by FemX. These experiments demonstrate that Lcn972 specifically interacts with lipid II, the substrate of both enzymes. In the presence of Lcn972, nisin pore formation was partially hindered in whole cells. However, binding of Lcn972 to lipid II could not compete with nisin in lipid II-doped 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) liposomes, possibly indicating a distinct binding site. The existence of a putative cotarget for Lcn972 activity is discussed in the context of its narrow inhibitory spectrum and the localized action at the division septum. To our knowledge, this is the first unmodified bacteriocin that binds to the cell wall precursor lipid II.
As the most outer macrostructure in the cell, the cell wall is the major sensory interface between the cell and the environment. It is needed to maintain the cell shape and counteract the inner osmotic pressure and is the anchoring structure for many enzymes (1). The cell wall is, therefore, crucial for survival. The major component of the cell wall in Gram positives is the peptidoglycan (PG), a large polymer made of covalently linked units of N-acetylglucosamine-N-acetylmuramic acid disaccharide pentapeptide (GlcNAc-MurNAc-pentapeptide). PG synthesis proceeds stepwise by linking MurNAc-pentapeptide from the UDP-activated form to the lipid carrier (C55-P), yielding lipid I, which is next converted into lipid II by addition of GlcNAc. Lipid II is subsequently translocated to the outside and polymerized by transglycosylation and further cross-linked via the pentapeptides by transpeptidases (see references 22 and 23 and references therein).
The cell wall is the target of many antibiotics, e.g., penicillin, the first discovered antibiotic that blocks the transpeptidation step carried out by the penicillin-binding proteins (PBPs). It has been lately recognized that several bacteriocins also interfere with cell wall biosynthesis. Bacteriocins are ribosomally synthesized antimicrobial peptides that usually kill bacteria by making pores in the cytoplasmic membrane (9). They are highly active and often combine two modes of action in the same molecule for potent antimicrobial activity. The class I or lantibiotic (lantionine-containing) nisin was the first pore-forming bacteriocin shown to directly interact with lipid II prior to pore formation (5, 8). Other members of the lantibiotic family, such as plantaricin C, gallidermin, and lacticin 3147, also bind to lipid II (4, 25, 26). Therefore, lipid II appears to be a prime target for lantibiotics. Unmodified bacteriocins (or class II) make use of other “receptors” or docking molecules such as components of the mannose phosphotransferase system to facilitate pore formation (10).
Lactococcin 972 (Lcn972) is the only nonlantibiotic bacteriocin described thus far whose primary target is not the cytoplasmic membrane. Instead, Lcn972 blocks the incorporation of cell wall precursors at the septum area, thereby inhibiting cell division (14, 16). We have performed global transcriptional analysis of cells treated with Lcn972 and demonstrated that the cell response is orchestrated by the two-component regulatory system CesSR (15). CesSR is homologous to the LiaSR two-component system in Bacillus subtilis (17) and to VraSR in Staphylococcus aureus involved in vancomycin resistance (13). The histidine kinases of these two-component system belong to a new family of the intramembrane-sensing histidine kinases, which are postulated to sense cell envelope stress at the membrane interface (18). Accordingly, they are highly induced by lipid II interacting antibiotics such as bacitracin, nisin, and vancomycin, among others.
Based on these observations, we have postulated that Lcn972 might also induce CesSR through interaction with or binding to lipid II. In the present study, we show that Lcn972 efficiently inhibits PG polymerization by PBP2 and the addition of Gly to lipid II by FemX, both enzymes that use lipid II as a substrate. However, in spite of the lipid II binding properties, Lcn972 cannot fully compete with nisin. To our knowledge, this is the first example of a nonlantibiotic bacteriocin that specifically binds to lipid II.
MATERIALS AND METHODS
Chemicals.
All chemicals were of analytical grade or better. 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) was purchased from Avanti-Polar Lipids, Inc., and stored at −20°C in chloroform. Lipid I and lipid II were synthesized and purified as previously described (21). C55-P was purchased from Larodan Fine Chemicals (Sweden), and C55-PP was obtained from Sigma (Germany). Protein concentration was determined by using the bicinchoninic acid protein assay reagent (Pierce Chemical Corp.) with bovine serum albumin as standard.
Bacterial strains and culture conditions.
The Lcn972 producer Lactococcus lactis IPLA972 and the indicator strain L. lactis MG1614 were routinely grown in M17 (Scharlab, Barcelona, Spain) plus glucose at 0.5% (GM17) at 30°C. Glucose was increased up to 2% to purify Lcn972 to achieve higher yields.
Purification of lactococcin 972.
Lcn972 was purified as described elsewhere (15). Very briefly, late exponential supernatants were subjected to cation-exchange chromatography and subsequently to hydrophobic interaction. All of the chromatographic steps were run in phosphate buffer 50 mM (pH 6.8). The specific activity of the Lcn972 preparations used in the present study was 50 arbitrary units (AU)/μg. AU were defined as the inverse of the highest dilution that gives a clear inhibition halo on L. lactis MG1614 by the agar diffusion test and were expressed per ml. When needed, a more concentrated Lcn972 solution was obtained by ultracentrifugation in a Centriprep filter device with an Ultracell-YM3 (3,000 cutoff) membrane (Millipore).
MIC and antagonization tests.
MICs were determined in microtiter plates by the broth microdilution method. Serial twofold dilutions of Lcn972 were made in GM17, and the plates were inoculated with 105 CFU of L. lactis MG1614/ml in a volume of 0.2 ml. Plates were incubated at 30°C for 18 h. The MIC was read as the lowest peptide concentration causing inhibition of visible growth. Determinations were carried out in triplicate. The antagonization test was performed in a similar fashion, mixing in each well Lcn972 (8× the MIC; 0.16 μM) and the lipid cell wall precursors (lipid II, lipid I, C55-PP, and C55-P) in a 2:1 molar ratio (precursor-peptide). The lipids were predissolved in 5 μl of methanol.
Coprecipitation of Lcn972 with lipid micelles.
The lipids lipid II or C55-PP (0.7 nmol) were emulsified in 50 mM phosphate buffer (pH 6.8) and mixed in a molar ratio 1:1 with Lcn972 diluted in the same buffer. The mixtures were incubated on ice for 10 min. The samples were centrifuged at 20,000 × g at 4°C for 60 min. The supernatant was transferred to a clean tube, and residual Lcn972 activity and protein content were determined as indicated above. Controls in the absence of lipids were also carried out. The experiment was done once.
Potassium release from whole cells.
Cells in the exponential phase were harvested at an optical density at 600 nm (OD600) of 1.5 (3,300 × g, 5°C, 3 min), washed with 50 ml of cold choline-buffer (300 mM choline chloride, 30 mM MES [4-morpholineethanesulfonic acid], 20 mM Tris [pH 6.5]), and resuspended in the same buffer to an OD600 of 30. The concentrated cell suspension was kept on ice and used within 30 min. For each measurement, cells were diluted in choline-buffer (25°C) to an OD600 of about 3. Nisin-induced potassium efflux was monitored by using a microprocessor pH meter (pH213; Hanna Instruments) with a MI-442 potassium electrode and MI-409F reference electrode, and leakage was expressed relative to the total amount of potassium release induced by the addition of 0.1 μM nisin. Before each experiment, the electrodes were calibrated with standard solutions containing 0.01, 0.1, or 1 mM KCl in buffer, and calculations of the percent potassium efflux were performed as described previously (19). For the competition assays, cells were preincubated for 3 min with Lcn972 prior to nisin addition. All of the experiments were performed in duplicate.
CF efflux experiments.
Large unilamellar vesicles were prepared for carboxyfluorescein (CF) experiments by the extrusion technique essentially as described by Wiedemann et al. (24). Vesicles were made of DOPC supplemented with 0.1 mol% lipid II (referring to the total amount of phospholipids). CF-loaded vesicles were prepared with 50 mM CF and then diluted in 1.5 ml of K+ buffer (50 mM MES-KOH, 100 mM K2SO4 [pH 6.0]) in a final concentration of 25 μM phospholipid on a phosphorous base. After addition of the peptide(s), the increase of fluorescence intensity was measured at 520 nm (excitation at 492 nm) on a RF-5301 spectrophotometer (Shimadzu) at room temperature. The leakage was documented relative to the total amount of marker release after solubilization of the vesicles by addition of 10 μl of 20% Triton X-100. In competition experiments, liposomes were incubated with Lcn972, vancomycin, or gallidermin A12L for 1 min prior to nisin addition. All of the experiments were performed twice.
Inhibition of the in vitro PBP2 activity.
Recombinant PBP2 from S. aureus NCTC 8325 was produced in E. coli as a C-terminal His6-tagged fusion and purified with Ni-NTA resin (Qiagen) according to the manufacturer's instructions (T. Schneider, unpublished data). The reaction was carried out in 50 μl of 10 mM MES (Sigma) buffer (pH 5.5) containing the substrate lipid II (2 nmol) and 7 μg of recombinant PBP2 at 37°C for 60 min. When indicated, moenomycin (1.3 nmol) or Lcn972 (1.5 nmol) was added. The reaction products were extracted with 1 volume of n-butanol-pyridine acetate (2:1 [vol/vol]; pH 4.2) and analyzed by thin-layer chromatography (TLC; silica plates, 60F254; Merck) using chloroform-methanol-water-ammonia (88:48:10:1) as the solvent (20).
Inhibition of the in vitro FemX activity.
The FemX assay was performed as described by Schneider et al. (21) with slight modifications. A total of 10 nmol of peptide was incubated with 5 nmol of lipid II for 15 min at room temperature. Afterward, the reaction assay in 100 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 0.8% Triton X-100, 2 mM ATP, 50 nmol of glycine (1:50 [U-14C]glycine, 3.7 GBq mmol−1; Amersham Pharmacia Biosciences), 25 μg of tRNA, 10 μg of glycyl-tRNA-synthetase, and 2.7 μg of FemX was completed in a final volume of 100 μl. After incubation for 1 h at 30°C, the lipid intermediates were extracted and analyzed by TLC as described above. Radiolabeled spots or lanes were visualized and quantified by using a phosphor storage screen and a Storm 820 optical scanner (GE Healthcare, Munich, Germany).
RESULTS
Inhibition of Lcn972 activity by cell wall precursors.
An antagonization test was initially carried out to determine whether any of the lipid cell wall precursors (lipid II, lipid I, C55-P, and C55-PP) could inhibit the biological activity of Lcn972. The assay was performed in microtiter plate wells containing Lcn972 at 8× the MIC for L. lactis MG1614 (MIC = 0.02 μM) and the cell wall precursors. In these conditions only lipid II, in a lipid/peptide molar ratio of 1:1 or higher, was able to antagonize Lcn972 activity, and growth of the indicator strain was observed (Table 1). None of the other precursors interfered with Lcn972 inhibitory activity.
TABLE 1.
Lcn972 inhibitory activity in the presence of cell wall precursors
| Cell wall precursor:Lcn972 and molar concn or ratioa | L. lactis MG1614 growth |
|---|---|
| Control (no Lcn972) | + |
| Lcn972 (0.16 μM) | - |
| Lipid II | |
| 2:1 | + |
| 1:1 | + |
| 0.5:1 | - |
| Lipid I (2:1) | - |
| C55-P (2:1) | - |
| C55-PP (2:1) | - |
Lcn972 was added at 0.16 μM (8× the MIC).
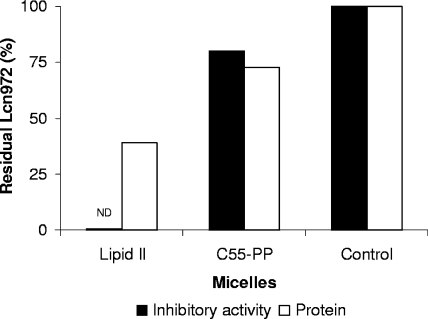
Furthermore, when Lcn972 was mixed with micelles made of either C55-PP or lipid II, coprecipitation of Lcn972 was only observed with the later (Fig. 1). C55-PP micelles removed only 25% of the peptide, as well as of the inhibitory activity. Recruitment of Lcn972 by C55-PP micelles could be due to electrostatic interactions between the cationic Lcn972 and the negatively charged C55-PP. On the contrary, removal of lipid II micelles occurred alongside the removal of most of the peptide, and no residual inhibitory activity was detected in the samples (Fig. 1). These experiments represented the first indication that Lcn972 could specifically bind to lipid II.
FIG. 1.
Coprecipitation of Lcn972 with lipid micelles. Lcn972 was mixed in a 1:1 molar ratio with lipid II or C55-PP micelles. The residual inhibitory activity and protein concentration were determined in the supernatants obtained after centrifugation. Values in the control sample (without lipids) were taken as 100%. ND, not detected.
Lcn972 inhibits PBP2 activity in vitro.
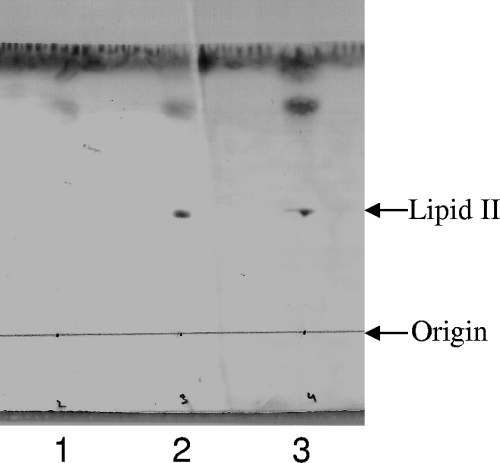
PBP2 is the only class A PBP of S. aureus with glycosyltransferase and transpeptidase activity. This enzyme catalyzes polymerization of the PG units outside the cell using lipid II as a substrate (2). Lcn972 inhibited PBP2 activity in vitro, as shown in Fig. 2. In the presence of either Lcn972 or the transglycosylase inhibitor moenomycin, the substrate lipid II could be extracted from the reaction mixture after incubation and was revealed by TLC. On the contrary, no lipid II was recovered in the control reaction (without inhibitors), as it is presumably polymerized by PBP2.
FIG. 2.
Inhibition of PBP2 activity by Lcn972. Lipid II was used as the substrate of the in vitro PBP2 assay. The products of the reaction mixtures were extracted with butanol-pyridine acetate 2:1 and analyzed by TLC. Lanes: 1, PBP2 control reaction without inhibitors; 2, PBP2 reaction plus moenomycin (transglycosylase inhibitor); 3, PBP2 reaction plus Lcn972. The arrows indicate the application spot (origin) and the position of lipid II. The results from a representative experiment are shown.
Lcn972 inhibits Gly incorporation to lipid II.
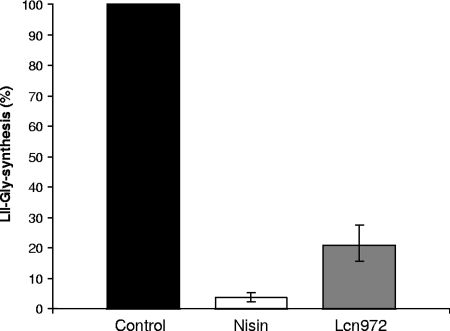
Incorporation of Gly to lipid II is catalyzed by the recombinant FemX that turns lipid II into lipid II-Gly1 (21). This in vitro assay has the advantage over the PBP2 assay described above in that it enables us to quantify the extent of the inhibition caused by Lcn972. We have performed this assay using lipid II or lipid II which had been preincubated with nisin or Lcn972 in a 1:2 lipid/peptide molar ratio (Fig. 3). The reaction was strongly inhibited by Lcn972, and only ca. 20% of the lipid II molecules were radioactively labeled. Likewise, preincubation of lipid II with nisin also inhibited Gly incorporation (Fig. 3). Comparison of both inhibiting activities demonstrated that Lcn972, like nisin, is a strong inhibitor of FemX. Therefore, Lcn972 could bind to lipid II tightly enough to hamper the accessibility of FemX to its substrate.
FIG. 3.
Inhibition of glycine incorporation to lipid II by Lcn972. Lipid II was preincubated with nisin (□) or Lcn972 (░⃞). The in vitro FemX reaction was carried out in the presence of [U-14C]glycine, recombinant tRNA-synthetase and purified tRNA. The reaction products were analyzed by TLC, and inhibition was scored measuring the radioactive labeling of lipid II. The control reaction without any peptide (▪) was taken as 100% [U-14C]glycine incorporation.
Nisin pore formation is partially hindered by Lcn972 in whole cells but not in lipid II-doped DOPC liposomes.
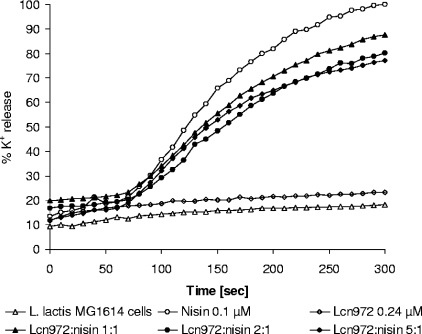
Lipid II acts as a docking molecule that facilitates pore formation by nisin and other pore-forming lantibiotics (5, 8). We hypothesized that, if Lcn972 binds to lipid II, the presence of this peptide might interfere with this process as nisin and Lcn972 would share the same target. To check this, we proceeded to determine K+ efflux from whole cells to score nisin pore formation in the presence or absence of Lcn972 (Fig. 4). When the cells were preincubated with Lcn972, K+ efflux was partially inhibited in a concentration-dependent fashion. However, Lcn972 was not able to entirely prevent nisin pore formation. Even when used in a 10:1 (Lcn972/nisin) molar ratio, inhibition of K+ release was never higher than 20 to 25% (data not show). Controls carried out without nisin confirmed that Lcn972 was unable to form pores that allowed K+ release (Fig. 4).
FIG. 4.
Lcn972 interference with nisin pore formation in L. lactis MG1614 whole cells. Peptides were added after 30 s, and the potassium release was monitored with a potassium sensitive electrode. When indicated, cells were preincubated for 3 min with several Lcn972 concentrations (0.12, 0.24, and 0.56 μM) prior to nisin addition (0.1 μM). Potassium leakage is expressed relative to the total amount of potassium (100% value) released after addition of 0.1 μM nisin. Controls without peptide addition and with each peptide alone were also performed. The results from a representative experiment are shown.
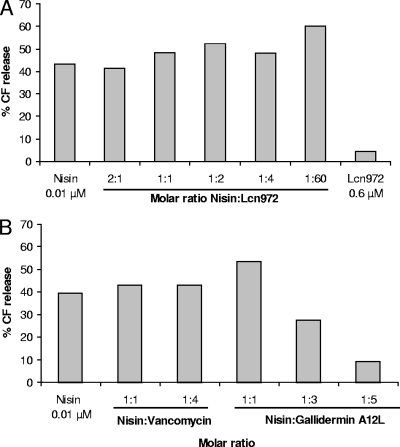
Nisin pore formation in the presence of Lcn972 was also studied in liposomes, a more defined system than whole cells. It had been shown that in the presence of lipid II, nisin induces marker release from liposomes at very low concentrations (24). We performed similar experiments with lipid II-doped DOPC liposomes that were preincubated with Lcn972. In contrast to the results obtained with whole cells, Lcn972 could not prevent nisin pore formation (Fig. 5A). Vancomycin was tested as well. This well-known lipid II-binding molecule also failed to block nisin pore formation (Fig. 5B). However, gallidermin A12L clearly antagonized nisin activity in a concentration-dependent fashion (Fig. 5B). Gallidermin A12L is a mutant of the lantibiotic gallidermin unable to form pores but that retains the ability to bind lipid II (4). Gallidermin and nisin share the same lipid II binding motif, which is missing in Lcn972 and vancomycin, and this could tentatively explain the antagonistic behavior in the liposome assays. On the other hand, despite the ability of Lcn972 to interact with lipid II, Lcn972 did not disturb the liposomes (Fig. 5A), even when used at very high concentrations.
FIG. 5.
Interference of Lcn972 (A) and the lipid II-binding peptides vancomycin and gallidermin A12L (B) with nisin pore formation in liposomes made of DOPC supplemented with 0.1 mol% lipid II. Liposomes were preincubated for 1 min with Lcn972, vancomycin, or gallidermin A12L prior to nisin addition. Nisin induced CF release was determined 2.5 min after addition. The 100% leakage level was determined by addition of Triton X-100. The results from a representative experiment are shown.
DISCUSSION
The results of the present study have proved Lcn972 as the first nonlantibiotic bacteriocin that specifically interacts with the cell wall precursor lipid II. Several lines of evidence support this conclusion. First, previous studies showed that Lcn972 inhibits the incorporation of cell wall precursors and the synthesis of the septum between daughter cells (14). Moreover, Lcn972 activates the two-component regulatory system CesSR, which is strongly induced by molecules that interfere with lipid II and its biosynthetic cycle (15). In the present study, we have shown that (i) lipid II, but not other lipid cell wall precursors, antagonizes the bactericidal activity of Lcn972 in vivo, (ii) Lcn972 coprecipitates with lipid II micelles, and (iii) Lcn972 inhibits the activity of two enzymes that use lipid II as substrate (PBP2 and FemX). The data obtained from the FemX assay clearly reveal Lcn972 as a potent inhibitor since the reaction was inhibited by at least 80%.
Lipid II has been shown to be the target or docking molecule of several lantibiotics, including non-pore-forming lantibiotics (mersacidin), pore-forming lantibiotics (nisin, plantaricin C, gallidermin, etc), and two-peptide systems such as lacticin 3147 (4, 5, 8, 25, 26). Two different structural motifs have been proposed to be involved in lipid II binding by lantibiotics (3). These motifs are based on the structural studies of the two leading lipid II binding lantibiotics nisin and mersacidin. In both cases the intramolecular rings imposed by the lanthionine residues are involved in lipid II binding (revised by Breukink and de Kruijff [6]). In the case of nisin, the N-terminal rings A and B were found to form a binding cage for the pyrophosphate linkage group of lipid II (11). Lcn972 is composed of nonmodified amino acids and lacks any lanthionine ring or even cysteines that may form intramolecular bridges. Hence, Lcn972 might display a novel lipid II binding motif that would warrant structural studies of the putative complex Lcn972-lipid II.
In spite of the fact that nisin and Lcn972 bind to the same molecule, Lcn972 could not fully compete with nisin. Targeted pore formation by nisin was only partially inhibited by Lcn972 in whole cells. Moreover, nisin pore formation in lipid II-doped liposomes took place regardless the presence of Lcn972. The high affinity of nisin for lipid II with a binding constant 2 × 107 M−1 (24) could explain these results. Alternatively, Lcn972 might recognize another motif in lipid II rather than the pyrophosphate cage. Actually, vancomycin, which binds to the d-Ala-d-Ala terminus of the pentapeptide of lipid II, was not able to hinder nisin pore formation. On the contrary, gallidermin A12L, which bears the same binding motif as nisin, clearly inhibited nisin-induced marker release from the liposomes.
Other as-yet-unidentified cellular factors have been postulated to be involved in the specific action of several lipid II binding peptides. It has been shown that low lipid II levels do not determine resistance to nisin (12). Moreover, the contribution to killing of cell wall synthesis inhibition and pore formation seems to vary on a species-specific basis for each antimicrobial peptide (25, 26). In the light of the new results that involve lipid II, a universal molecule in the prokaryotic world, as a target for Lcn972, it is reasonable to think of the existence of a putative cotarget that facilitates or promote access to lipid II. Lcn972 displays a very narrow inhibitory spectrum, and only certain species of Lactococcus are susceptible. On the other hand, Lcn972 is not active against nondividing cells, and electron micrographs showed that Lcn972 specifically inhibits septum formation (14) rather than promoting an overall drastic reduction of the cell wall thickness, as described for other lipid II binding lantibiotics such as mersacidin (7). These observations tentatively point to one (or more) components of the divisome as putative cotargets. Further studies to prove this and to define the molecular basis of Lcn972 mode of action are currently in progress. The results could be instrumental in the design of novel and highly targeted antibiotics.
Acknowledgments
I.W. and B.M. were awarded a personnel exchange grant funded by the Deutscher Akademischer Austauschdienst of Germany (D/04/39997) and Ministerio de Educación y Ciencia of Spain (HA2004-0086). This study has been funded by grant BIO-4312 (Ministerio de Educación y Ciencia of Spain) and the German Research Foundation (Sa292/11-1).
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC.
- 2.Barrett, D., C. Leimkuhler, L. Chen, D. Walker, D. Kahne, and S. Walker. 2005. Kinetic characterization of the glycosyltransferase module of Staphylococcus aureus PBP2. J. Bacteriol. 187:2215-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonelli, R. R., I. Wiedemann, and H. G. Sahl. 2006. Lantibiotics, p. 97-105. In A. Kastin (ed.), Handbook of biologically active peptides. Elsevier, New York, NY.
- 4.Bonelli, R. R., T. Schneider, H. G. Sahl, and I. Wiedemann. 2006. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob. Agents Chemother. 50:1449-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. G. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 6.Breukink, E., and B. de Kruijff. 2006. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5:321-323. [DOI] [PubMed] [Google Scholar]
- 7.Brötz, H., G. Bierbaum, A. Markus, E. Mollitor, and H. G. Sahl. 1995. Mode of action of the lantibiotic mersacidin: inhibition of peptidoglycan biosynthesis via a novel mechanism? Appl. Environ. Microbiol. 39:714-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brötz, H., M. Josten, I. Wiedemann, U. Schneider, F. Gotz, G. Bierbaum, and H. G. Sahl. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin, and other lantibiotics. Mol. Microbiol. 30:317-327. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 10.Diep, D. B., M. Skaugen, Z. Salehian, H. Holo, and I. F. Nes. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. USA 104:2384-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu, S. T., E. Breukink, E. Tischenko, M. A. Lutters, B. de Kruijff, R. Kaptein, A. M. Bonvin, and N. A. van Nuland. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11:963-967. [DOI] [PubMed] [Google Scholar]
- 12.Kramer, N. E., E. J. Smid, J. Kok, B. de Kruijff, O. P. Kuipers, and E. Breukink. 2004. Resistance of gram-positive bacteria to nisin is not determined by lipid II levels. FEMS Microbiol. Lett. 239:157-161. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 14.Martínez, B., A. Rodríguez, and J. E. Suárez. 2000. Lactococcin 972, a bacteriocin that inhibits septum formation in lactococci. Microbiology 146:949-955. [DOI] [PubMed] [Google Scholar]
- 15.Martínez, B., A. L. Zomer, A. Rodríguez, J. Kok, and O. P. Kuipers. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64:473-486. [DOI] [PubMed] [Google Scholar]
- 16.Martínez, B., J. E. Suárez, and A. Rodríguez. 1996. Lactococcin 972, a homodimeric lactococcal bacteriocin whose primary target is not the plasma membrane. Microbiology 142:2393-2398. [DOI] [PubMed] [Google Scholar]
- 17.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 18.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133-144. [DOI] [PubMed] [Google Scholar]
- 19.Orlov, D. S., T. Nguyen, and R. I. Lehrer. 2002. Potassium release, a useful tool for studying antimicrobial peptides. J. Microbiol. Methods 49:325-328. [DOI] [PubMed] [Google Scholar]
- 20.Rick, P. D., G. L. Hubbard, M. Kitaoka, H. Nagaki, T. Kinoshita, S. Dowd, V. Simplaceanu, and C. Ho. 1998. Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella typhimurium defective in ECA synthesis. Glycobiology 8:557-567. [DOI] [PubMed] [Google Scholar]
- 21.Schneider, T., M. M. Senn, B. Berger-Bachi, A. Tossi, H. G. Sahl, and I. Wiedemann. 2004. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol. Microbiol. 53:675-685. [DOI] [PubMed] [Google Scholar]
- 22.van Heijenoort, J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R-36R. [DOI] [PubMed] [Google Scholar]
- 23.van Heijenoort, J. 2007. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev. 71:620-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiedemann, I., E. Breukink, C. van Kraaij, O. P. Kuipers, G. Bierbaum, B. de Kruijff, and H. G. Sahl. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772-1779. [DOI] [PubMed] [Google Scholar]
- 25.Wiedemann, I., T. Bottiger, R. R. Bonelli, A. Wiese, S. O. Hagge, T. Gutsmann, U. Seydel, L. Deegan, C. Hill, P. Ross, and H. G. Sahl. 2006. The mode of action of the lantibiotic lacticin 3147: a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 61:285-296. [DOI] [PubMed] [Google Scholar]
- 26.Wiedemann, I., T. Bottiger, R. R. Bonelli, T. Schneider, H. G. Sahl, and B. Martínez. 2006. Lipid II-based antimicrobial activity of the lantibiotic plantaricin C. Appl. Environ. Microbiol. 72:2809-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]