Abstract
We collected Mycobacterium avium isolates from clinical and drinking-water sources and compared isolates among themselves and to each other using molecular methods. Four clinical isolates were related to water isolates. Groups of indistinguishable clinical isolates were identified. The groups of identical clinical isolates suggest a common source of exposure.
Mycobacterium avium has been associated with infections among individuals of all ages and has been found in drinking water (2, 3, 6, 9). We isolated M. avium from point-of-use (POU) water taps delivering municipal drinking water within a single county (2, 4). We used molecular methods to compare the genetic relatedness of M. avium isolates from water and isolates from residents receiving the water to determine if we could identify drinking water as a source of exposure.
We collaborated with personnel from laboratories that isolate and identify mycobacteria from human specimens collected during patient care activities. We obtained clinical isolates of M. avium complex that were collected from county residents during the study period. We recorded each anatomic site of collection and each patient's residential zip code. The study was approved by and conducted in adherence with the human subjects' requirements of the Institutional Review Board of the University of North Carolina at Chapel Hill.
A subset of viable M. avium complex clinical isolates was selected for molecular analysis using specific selection criteria. Isolates were more likely to be analyzed when they were isolated from a sterile site and the patient's zip code was included in the municipal water distribution area. M. avium complex isolates were identified as either M. avium or Mycobacterium intracellulare using multilocus enzyme electrophoresis (MEE) (11, 12). Isolates identified as M. avium which shared an electrophoretic type (ET) were further subtyped by pulsed-field gel electrophoresis (PFGE) using 40 U of XbaI for each restriction reaction (7). The Tenover criteria were used to evaluate relatedness of PFGE fragment patterns (10).
Groups of related isolates were evaluated to confirm that isolates were collected from unique individuals. If isolates were collected from the same individual or if we could not confirm that the isolates were collected from unique individuals, we retained only the isolate with the earliest collection date from the group.
Twenty-seven M. avium isolates from four drinking-water POU sites were typed using MEE. Among 23 isolates and 4 identical ETs, PFGE was used for greater genetic discrimination, yielding 4 identical or closely related PFGE groups.
We received 253 M. avium complex clinical isolates: 200 (79%) from respiratory sites, 37 (15%) from normally sterile sites, and 16 (6%) from other sites, such as wounds and stool. We analyzed 126 of these using MEE; of these, 88 (70%) were identified as M. avium. Fifty-six of the 88 (64%) M. avium isolates were from respiratory sites, 25 (28%) from normally sterile sites, and 7 (8%) from other sites, such as wounds and stool.
Seventy-four isolates were from patients with a home zip code served by the municipal water system. Among these clinical isolates, we found one that was closely related to an environmental M. avium isolate (Fig. 1); the zip code of the patient's residence was adjacent to the drinking-water sample site, providing an additional geographic association between these isolates. Three other clinical M. avium isolates shared an ET with another M. avium environmental isolate but when compared by PFGE analysis were only possibly related (Fig. 2); there was no apparent geographic relationship among these isolates.
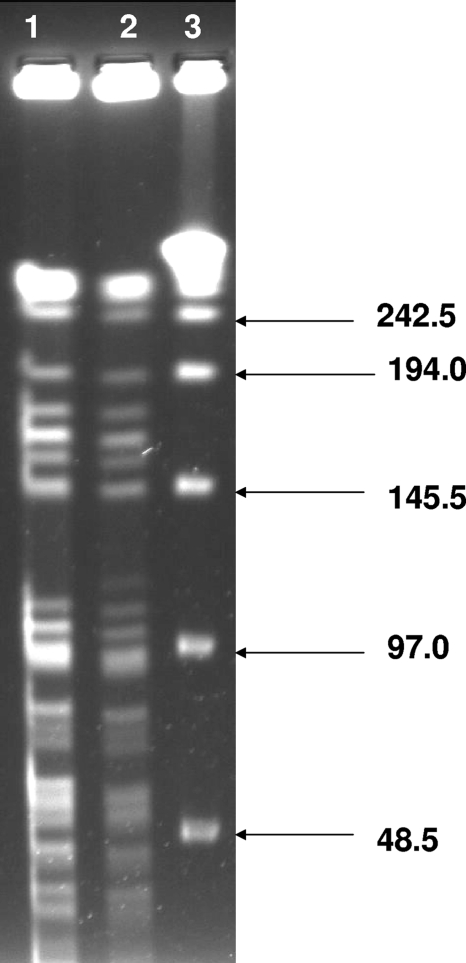
FIG. 1.
Closely related clinical (lane 1) and drinking-water (lane 2) M. avium isolates. Lane 3 contains a 48.5-kb ladder.
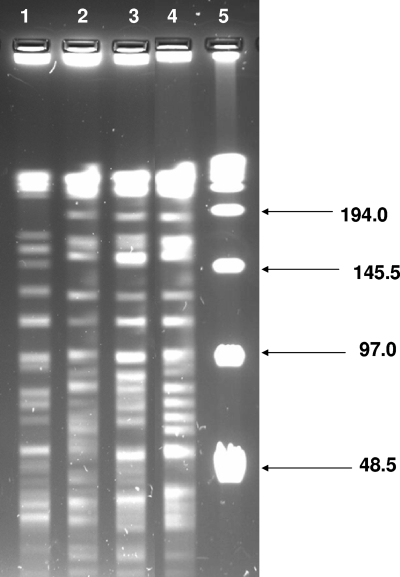
FIG. 2.
Possibly related M. avium isolates from drinking water (lane 1) and patients (lanes 2 to 4) all shared an electrophoretic group. Lane 5 contains a 48.5-kb ladder.
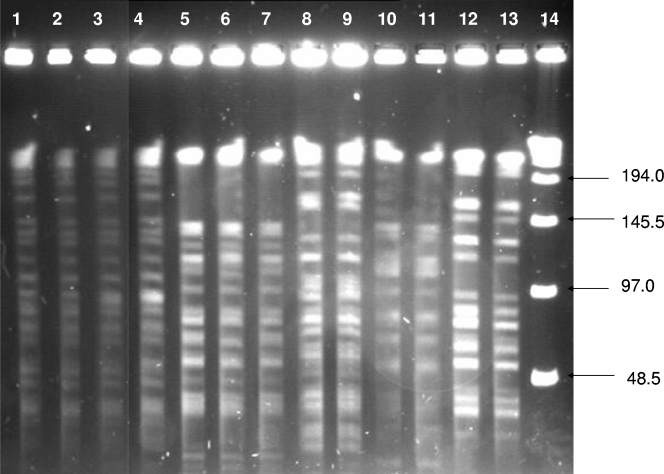
A subset of 15 clinical isolates shared five PFGE groups (A to E). Each group was comprised of indistinguishable isolates collected from two to four individuals. We display 13 of these clinical isolates (Fig. 3). Indistinguishable isolates were found to be collected from different patients on different dates over a 2- to 41-month period. Pairs of patients within two groups shared a home zip code. The dates of clinical sample collection between these geographically related pairs varied; one pair was collected 6 months apart, the other pair 4 months apart.
FIG. 3.
Groups of indistinguishable M. avium isolates from clinical samples: PFGE groups A to E. Group A, lanes 1 to 4; group B, lanes 5 to 7; group D, lanes 8 and 9; group E, lanes 10 and 11; group C, lanes 12 and 13. Lane 14 contains a 48.5-kb ladder.
We found evidence of a drinking-water source of exposure for one patient, whose M. avium isolate was closely related to an isolate from a drinking-water sample collected adjacent to the patient's home zip code. We found an additional three patients whose isolates were possibly related to an M. avium isolate from drinking water. These results are surprising given the multiple potential sources of exposure in the study area, the limited number of drinking-water sample sites, and the large amount of genetic heterogeneity reported to occur among M. avium isolates (7).
We identified five PFGE groups of indistinguishable clinical isolates that were collected over a 2- to 41-month period. These groups likely represent patients with common environmental sources of M. avium exposure, although specific sources of exposure are unknown. Clusters of M. avium infections sharing a PFGE group have previously been reported among individuals (8), sometimes among those with no identified epidemiologic links (5). Clusters such as these may be attributed to either persistence of a common source of exposure or delayed detection of infection or colonization among patients exposed to a transitory common source of exposure. Either exposure scenario is plausible; M. avium strains are reported to persist at drinking-water POU taps for up to 26 months (4). Diagnosis of M. avium complex infections is frequently missed or delayed, particularly among patients with no commonly recognized risk factors for infection (1). Limitations of the study include the lack of patient-specific information, including exposure history and disease status. We analyzed a convenience sample of clinical isolates; unknown biases associated with identification, collection, and viability may have occurred. Environmental isolates were collected from a limited number of drinking-water sites. It is probable that many more strains of M. avium could be isolated from the millions of POU sites located within the municipal utility's distribution area.
Molecular techniques provide the strongest support to link human M. avium infections and environmental exposures. However, progression of infection may be indolent, isolation of M. avium from environmental samples is difficult, and molecular methods are not widely used. Control of this environmentally transmitted infectious disease will continue to be a challenge until detection of new cases of illness increases, methods to identify and isolate M. avium from environmental samples improve, and methods to link human and environmental strains of M. avium are more widely accessible.
Acknowledgments
We thank Marie Coyle and LaDonna Carlson of Harborview Medical Center, Seattle, WA; Paul Swenson of the Seattle & King County Public Health Laboratory; Craig Colombel of the Washington State Public Health Laboratory; and Margaret Floyd, Centers for Disease Control and Prevention, Atlanta, GA, for technical support. We gratefully acknowledge the contributions of Stacy L. Pfaller of the U.S. Environmental Protection Agency, Office of Research and Development, National Exposure Research Laboratory, Cincinnati, OH, and the technical assistance and expertise of Sandy Dunkel, Brian Hoyt, and Moya Joubert from the participating utility.
Isolates were purchased through contracts 1D-5128-NAEX, 2D-5783-NAEX, and 2D-6115-NAEX.
The views expressed in this report are those of the individual authors and do not necessarily reflect the views and policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Choudhri, S., J. Manfreda, J. Wolfe, S. Parker, and R. Long. 1995. Clinical significance of nontuberculous mycobacteria isolates in a Canadian tertiary care center. Clin. Infect. Dis. 21:128-133. [DOI] [PubMed] [Google Scholar]
- 2.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma, Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falkinham, J. O., III, C. D. Norton, and M. W. LeChevallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilborn, E. D., T. C. Covert, M. A. Yakrus, S. I. Harris, S. F. Donnelly, E. W. Rice, S. Toney, S. A. Bailey, and G. N. Stelma, Jr. 2006. Persistence of nontuberculous mycobacteria in a drinking water system after addition of filtration treatment. Appl. Environ. Microbiol. 72:5864-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunimoto, D. Y., M. S. Peppler, J. Talbot, P. Phillips, S. D. Shafran, and Canadian HIV Trials Network Protocol 010 Study Group. 2003. Analysis of Mycobacterium avium complex isolates from blood samples of AIDS patients by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:498-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Dantec, C., J. P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl. Environ. Microbiol. 68:5318-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazurek, G. H., S. Hartman, Y. Zhang, B. A. Brown, J. S. Hector, D. Murphy, and R. J. Wallace, Jr. 1993. Large DNA restriction fragment polymorphism in the Mycobacterium avium-M. intracellulare complex: a potential epidemiologic tool. J. Clin. Microbiol. 31:390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pestel-Caron, M., G. Graff, G. Berthelot, J. L. Pons, J. F. Lemeland, and Groupe de Recherche sur les Antimicrobiens et les Micro-organismes. 1999. Molecular analysis of Mycobacterium avium isolates by using pulsed-field gel electrophoresis and PCR. J. Clin. Microbiol. 8:2450-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos, R., F. Oliveira, J. Fernandes, S. Goncalves, F. Macieira, and M. Cadete. 2005. Detection and identification of mycobacteria in the Lisbon water distribution system. Water Sci. Technol. 52:177-180. [PubMed] [Google Scholar]
- 10.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasem, C. F., C. M. McCarthy, and L. W. Murray. 1991. Multilocus enzyme electrophoresis analysis of the Mycobacterium avium-M. intracellulare-M. scrofulaceum complex. J. Clin. Microbiol. 29:264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yakrus, M. A., M. W. Reeves, and S. B. Hunter. 1992. Characterization of isolates of Mycobacterium avium serotypes 4 and 8 from patients with AIDS by multilocus enzyme electrophoresis. J. Clin. Microbiol. 30:1474-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]