Abstract
In previous studies, we have shown that direct protein-protein interaction between the two regulators ArgR and AhrC in Lactococcus lactis is required for arginine-dependent repression of the biosynthetic argC promoter and the activation of the catabolic arcA promoter. Here, we establish the global ArgR and AhrC regulons by transcriptome analyses and show that both regulators are dedicated to the control of arginine metabolism in L. lactis.
In many bacteria, arginine metabolism is controlled by transcriptional regulators of the ArgR family, including the well-characterized ArgR of Escherichia coli (13), AhrC of Bacillus subtilis (5), and ArgR of Bacillus stearothermophilus (7). Regulation is mediated through the interaction of the regulators with operators designated ARG boxes, which consist of 18-bp palindromic structures overlapping the core promoters of target genes (4, 5, 17). Low-G+C-content gram-positive bacteria often harbor more than one ArgR-type homologue (see reference 2 for an overview), and separate studies recently demonstrated that two arginine regulators are necessary for the regulation of arginine metabolism in the lactic acid bacteria Lactococcus lactis and Lactobacillus plantarum (11, 15). The deletion or mutation of any single arginine regulator in these organisms results in the complete disruption of arginine-mediated regulation. L. lactis AhrC is known to facilitate the binding of ArgR to ARG boxes in the biosynthetic argC promoter and to sequester and prevent ArgR from binding to ARC boxes (ARG box half sites) in the catabolic arcA promoter (12). In this way, arginine leads to the repression of arginine biosynthesis and the activation of arginine catabolism.
Here, we establish the complete regulons of ArgR and AhrC in L. lactis and show that these regulators are dedicated to the control of arginine metabolism.
ArgR and AhrC are dedicated regulators of arginine metabolism.
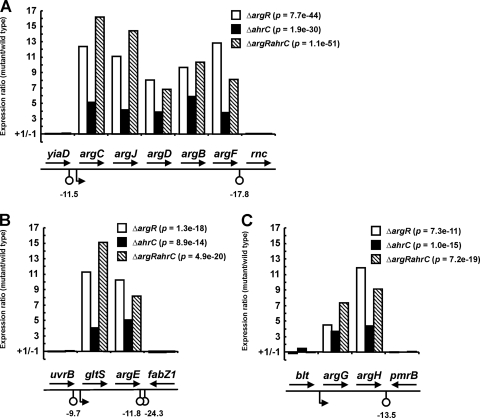
The global regulatory effects of ArgR and AhrC of L. lactis were studied by DNA microarray-based transcriptome analyses as described previously, by using amplicon-based L. lactis IL-1403 slides (6, 10). IL-1403 amplicon sequences and the microarray data used are available online at http://molgen.biol.rug.nl/publication/arg_data/. RNA was isolated from cells grown to mid-exponential phase in chemically defined medium (CDM) containing 10 mM arginine (11). The transcription profile of L. lactis MG1363 was compared to those of its two single regulator mutants MGΔargR and MGΔahrC and the double regulator mutant MGΔargRahrC (11). Significant derepression of all the genes in the argCJDBF, gltS-argE, and argGH operons of the arginine biosynthesis pathway in all three mutants was observed (Fig. 1). These results extend the known ArgR/AhrC regulons considerably and also establish the operon structures of the regulated genes, in agreement with the predicted promoter and putative terminator structures (Fig. 1). Whereas the levels of derepression of all biosynthesis genes in MGΔargR, as well as those in MGΔargRahrC, varied between 5- and 15-fold, the levels of derepression of these genes in MGΔahrC never exceeded 6-fold (Fig. 1). The lower mutant-versus-wild-type ratios obtained for MGΔahrC than for MGΔargR and MGΔargRahrC can be explained by the previous observation that ArgR, independently of AhrC, can weakly bind to the argC promoter in the presence of arginine (12). Still, both arginine regulators are essential for the complete repression of arginine bio-synthesis, as the loss of either of the two regulators leads to an increase in the expression of all involved genes.
FIG. 1.
Derepression of the arginine biosynthesis operons of L. lactis, as measured by DNA microarray analyses. Cells were grown to mid-exponential phase in CDM with 10 mM arginine (11). The argCJDBF (A), gltS-argE (B), and argGH (C) operons and their neighboring genes, predicted promoters, and putative stem-loop (terminator) structures (calculated as ΔG° in kilocalories per mole) are drawn schematically, not to scale. The bars in the graphs show the ratios of expression in L. lactis strains MGΔargR, MGΔahrC, and MGΔargRahrC relative to that in L. lactis MG1363. The change in expression (n-fold) is shown on the y axis, with positive and negative values indicating up- and downregulation, respectively; if the expression of a gene in the mutant was equal to that in the wild-type strain, this resulted in a value near the x axis (+1/−1). The combined P values for the respective operons, thus excluding the values for surrounding genes, are given in parentheses next to the symbols.
Regulation of the arginine catabolic gene cluster.
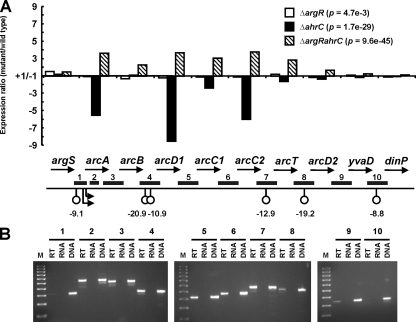
The transcriptome analyses demonstrated a complex pattern of regulation of the arc gene cluster encoding the catabolic arginine deiminase (ADI) pathway (Fig. 2). The deletion of argR had no significant effect on arc expression, while the deletion of ahrC resulted in general downregulation, and the expression of arc genes in MGΔargRahrC was increased relative to that in the wild-type strain (Fig. 2A). The arcB gene did not show significant changes in expression in any of the regulator mutants (Fig. 2A), which seems surprising considering that the changes in the expression of the other arc genes were highly significant. L. lactis arc is recognized as one of the largest and most complex of bacterial ADI pathway operons (19). The ADI operon arcDABC of Pseudomonas aeruginosa is subject to posttranscriptional regulation via mRNA processing and transcription termination, resulting in differential gene expression according to the requirement of the gene products in the cell (8, 9). A similar process may be in effect in L. lactis, although the role therein, if any, of the putative stem-loop structures downstream of arcB (Fig. 2A) remains to be investigated.
FIG. 2.
(A) Deregulation of the L. lactis arginine catabolic arcABD1C1C2TD2-yvaD gene cluster (not drawn to scale). Refer to the legend to Fig. 1 for details. Numbered horizontal bars refer to regions examined by RT-PCR. (B) Results from RT-PCR investigation of the arc operon structure. Primer pairs were designed to amplify the intergenic regions indicated by numbered bars in panel A. Regions 1 and 2 correspond to RT-PCR probes of the arcA promoter region (negative control) and an intragenic arcA region (positive control), respectively. “DNA” indicates a positive PCR control with chromosomal DNA as the template. “RT” and “RNA” indicate PCRs performed with total RNA with and without reverse transcriptase treatment, respectively, the latter acting as a negative control. M, molecular size markers.
RT-PCR analysis of the arc gene cluster.
To verify the arc operon structure as determined by transcriptome analyses, reverse transcription (RT)-PCR analyses of total RNA from cells grown in CDM with 10 mM arginine were carried out using primer pairs spanning all intergenic and neighboring regions. Only a little RT-PCR product corresponding to the arcT-arcD2 and arcD2-yvaD regions was formed, while no product was obtained by probing between argS and arcA and between yvaD and dinP, defining the arginine catabolic operon to comprise arcABD1C1C2TD2-yvaD (Fig. 2B). The lower level of transcription of promoter-distal genes than of genes further upstream in the operon is likely caused by the possible terminator (ΔG = −19.2 kcal/mol) in the arcT-arcD2 intergenic region, which would allow only partial transcription elongation (Fig. 2). The putative yvaD product does not show homology to any known protein, and its function was not investigated further.
ARG box operators in L. lactis.
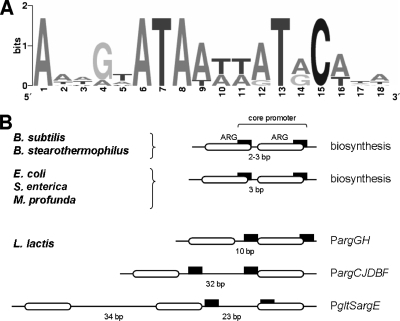
Overrepresented sequence motifs in the regions upstream of the ArgR and AhrC target genes were identified using the MotifSampler online tool (18). Considering the regulatory patterns and the resulting operon structures (Fig. 1 and 2), only the regions upstream of the first genes of the operons were subjected to the motif search. By employing the Genome2D software tool (1), an 18-bp motif was found to be present in all three arginine biosynthetic promoters (Fig. 3). A weight matrix based on the lactococcal ArgR motif was applied to all the promoter regions of L. lactis by using Genome2D (1). The obtained motif sequences were ranked according to their levels of similarity to the consensus sequence. By using the matrix, 8 of the 10 targets with the highest levels of similarity to the consensus sequence were determined to be present in the promoters of the arginine biosynthetic genes of L. lactis MG1363 (data not shown). The identified motif has a high degree of similarity to the 5′-WnTGnATWWWWATnCAnW-3′ consensus ARG box motif of E. coli (uppercase letters represent highly conserved residues) (13) (Fig. 3A). However, the typical 2- to 3-bp spacing between double ARG boxes in E. coli and other organisms was not observed in L. lactis, where the spacing (in multitudes of 10 to 11 bp) suggests the ARG boxes to be present on the same face of the helix but separated by one, two, or three helical turns (Fig. 3B). Note that no clear ARG box was obtained for the arcA promoter region, which is in agreement with results obtained previously by DNase I footprinting (12).
FIG. 3.
L. lactis ARG operators. (A) Consensus logo of ARG box operators in the arginine biosynthetic promoters of L. lactis, created using the WebLogo tool (http://weblogo.berkeley.edu/) (3, 16). (B) Schematic comparison of ARG box organization patterns in different organisms (S. enterica, Salmonella enterica serovar Typhimurium; M. profunda, Moritella profunda). Core promoter −35 (left) and −10 (right) sequences are indicated as black boxes. White boxes represent 18-bp ARG boxes. The numbers below the horizontal lines (indicating chromosomal DNA) refer to the lengths of the spacer regions between ARG boxes.
Indirect effect on the pyrimidine genes of arginine regulator mutants.
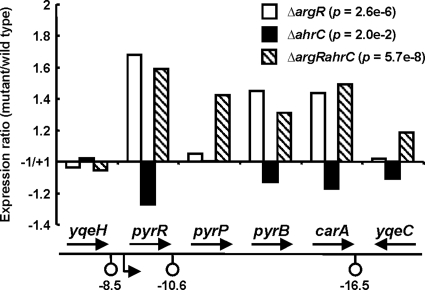
The levels of expression of pyrimidine de novo biosynthesis pathway genes (pyr) in MGΔargR and MGΔargRahrC were between 1.3- and 1.7-fold higher than those in strain MG1363, whereas no significant effects in MGΔahrC were observed (Fig. 4; data are shown only for the pyrR-carA operon). To assess the validity of these DNA microarray results, the activities of orotate phosphoribosyltransferase (PyrE, the gene for which was upregulated 1.7- and 1.5-fold in MGΔargR and MGΔargRahrC, respectively) and orotidine-phosphate decarboxylase (PyrF, the gene for which was upregulated 1.5-fold in both MGΔargR and MGΔargRahrC) were determined (Table 1). The activities of these two enzymes of the pyrimidine de novo pathway confirmed the results of the transcriptome analyses, showing that the impaired arginine regulation in the two mutants lacking ArgR did have a significant albeit weak effect on downstream pyrimidine metabolism (Table 1). The pyr gene expression profiles correlated well with the expression profiles of the arginine biosynthesis genes and not with those of the catabolic genes. Thus, in MGΔargR and MGΔargRahrC, the flux toward arginine was most likely increased due to the high-level expression of the arginine biosynthetic genes. As a consequence, carbamoylphosphate, which is required for the synthesis of both arginine and pyrimidine, was directed toward arginine, resulting in carbamoylphosphate and pyrimidine depletion. To counteract this effect, the pyrimidine biosynthetic genes, including both carA and carB (the latter is upregulated 1.9- and 1.5-fold in MGΔargR and MGΔargRahrC, respectively), were induced, possibly via the known transcriptional regulator PyrR (14).
FIG. 4.
Deregulation of pyrimidine de novo biosynthesis genes in the pyrRPB-carA operon of L. lactis. See the legend to Fig. 1 for details.
TABLE 1.
Specific activities of orotate phosphoribosyltransferase (PyrE) and orotidine-phosphate decarboxylase (PyrF) in MG1363 and its isogenic regulator deletion derivativesa
| Strain | Specific enzymatic activity (U/mg of protein) (upregulation [n-fold])b of:
|
|
|---|---|---|
| PyrE | PyrF | |
| MG1363 | 88 ± 17 (1) | 197 ± 3 (1) |
| MGΔargR | 200 ± 37 (2.3) | 256 ± 15 (1.3) |
| MGΔahrC | 110 ± 31 (1.3) | 146 ± 24 (−1.3) |
| MGΔargRahrC | 221 ± 76 (2.5) | 251 ± 26 (1.3) |
Cells were grown to mid-exponential phase in CDM with 10 mM arginine. Values shown are the averages and standard deviations determined from results obtained in three independent experiments.
Degree of upregulation in the indicated strain compared to the activity in the wild-type strain (MG1363).
Concluding remarks.
The presence of two to four ArgR-type regulators in many low-G+C-content gram-positive bacteria led us to speculate as to possible global target genes of the two homologues in L. lactis. In support of the findings of our previous mechanistic studies (11, 12), we here show that both regulators are dedicated to the functional control of arginine metabolism. Additionally, the data extend the known ArgR/AhrC regulons of L. lactis, define the biosynthetic (arg) and catabolic (arc) operon structures, and show that even weak indirect effects can be significantly measured for the de novo pyrimidine genes in response to disrupted arginine regulation.
Acknowledgments
We thank Anne de Jong and Aldert Zomer for advice on and assistance with the DNA microarray studies.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Baerends, R. J. S., W. K. Smits, A. de Jong, L. W. Hamoen, J. Kok, and O. P. Kuipers. 2004. Genome2D: a visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol. 5:R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky, B. R. 2002. Biosynthesis of amino acids of the glutamate and aspartate families, alanine and polyamines, p. 203-231. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 3.Crooks, G. E., G. Hon, J.-M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunin, R., T. Eckhardt, J. Piette, A. Boyen, A. Pierard, and N. Glansdorff. 1983. Molecular basis for modulated regulation of gene expression in the arginine regulon of Escherichia coli K-12. Nucleic Acids Res. 11:5007-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czaplewski, L. G., A. K. North, M. C. Smith, S. Baumberg, and P. G. Stockley. 1992. Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. Mol. Microbiol. 6:267-275. [DOI] [PubMed] [Google Scholar]
- 6.den Hengst, C. D., S. A. F. T. van Hijum, J. M. W. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332-34342. [DOI] [PubMed] [Google Scholar]
- 7.Dion, M., D. Charlier, H. Wang, D. Gigot, A. Savchenko, J. N. Hallet, N. Glansdorff, and V. Sakanyan. 1997. The highly thermostable arginine repressor of Bacillus stearothermophilus: gene cloning and repressor-operator interactions. Mol. Microbiol. 25:385-398. [DOI] [PubMed] [Google Scholar]
- 8.Gamper, M., B. Ganter, M. R. Polito, and D. Haas. 1992. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J. Mol. Biol. 226:943-957. [DOI] [PubMed] [Google Scholar]
- 9.Gamper, M., and D. Haas. 1993. Processing of the Pseudomonas arcDABC mRNA requires functional RNase E in Escherichia coli. Gene 129:119-122. [DOI] [PubMed] [Google Scholar]
- 10.Kuipers, O. P., A. de Jong, R. J. S. Baerends, S. A. F. T. van Hijum, A. L. Zomer, H. A. Karsens, C. D. den Hengst, N. E. Kramer, G. Buist, and J. Kok. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie van Leeuwenhoek 82:113-122. [PubMed] [Google Scholar]
- 11.Larsen, R., G. Buist, O. P. Kuipers, and J. Kok. 2004. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. J. Bacteriol. 186:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen, R., J. Kok, and O. P. Kuipers. 2005. Interaction between ArgR and AhrC controls regulation of arginine metabolism in Lactococcus lactis. J. Biol. Chem. 280:19319-19330. [DOI] [PubMed] [Google Scholar]
- 13.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinussen, J., J. Schallert, B. Andersen, and K. Hammer. 2001. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol. 183:2785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicoloff, H., F. Arsene-Ploetze, C. Malandain, M. Kleerebezem, and F. Bringel. 2004. Two arginine repressors regulate arginine biosynthesis in Lactobacillus plantarum. J. Bacteriol. 186:6059-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song, H., H. Wang, D. Gigot, D. Dimova, V. Sakanyan, N. Glansdorff, and D. Charlier. 2002. Transcription regulation in thermophilic bacteria: high resolution contact probing of Bacillus stearothermophilus and Thermotoga neapolitana arginine repressor-operator interactions. J. Mol. Biol. 315:255-274. [DOI] [PubMed] [Google Scholar]
- 18.Thijs, G., K. Marchal, M. Lescot, S. Rombauts, B. de Moor, P. Rouze, and Y. Moreau. 2002. A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 9:447-464. [DOI] [PubMed] [Google Scholar]
- 19.Zuñiga, M., G. Perez, and F. Gonzalez-Candelas. 2002. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 25:429-444. [DOI] [PubMed] [Google Scholar]