Abstract
Monitoring recreational waters for fecal contamination by standard methodologies involves culturing indicator bacteria, such as fecal coliforms and enterococci. Delayed reporting of microbial water quality parameters increases the likelihood of public exposure to pathogens of fecal origin, making the development of rapid methods important for public health protection. A rapid assay for enterococci was developed using a combined ultrafiltration-biosensor procedure. Twelve 100-liter water samples were collected from upper Tampa Bay over a 9-month period. The samples were collected on site by dead-end hollow-fiber ultrafiltration. Postfiltration processing of the initial retentates included sonication and micrometer-level sieve passage to remove interfering particles. Centrifugation was utilized for secondary concentration. Grab samples were collected simultaneously with the ultrafiltered samples. Concentrations of enterococci in all grab and ultrafiltration samples were determined by the standard method (EPA method 1600) for calculation of recovery efficiencies and concentration factors. Levels of enterococci increased twofold in initial retentates and by 4 orders of magnitude in final retentates over ambient concentrations. An aliquot of each final retentate was adsorbed onto polystyrene waveguides for immunoassay analysis of enterococci with a microfluidic fiber optic biosensor, the Raptor. Enterococci were detected when concentrations in the ambient water exceeded the regulatory standard for a single sample (≥105 CFU/100 ml). The combined ultrafiltration-biosensor procedure required 2.5 h for detection compared to 24 for the standard method. This study demonstrated that enterococci can be detected rapidly using on-site ultrafiltration, secondary concentration, and biosensor analysis.
Standard water-monitoring technologies for fecal contamination currently involve culturing indicator microorganisms (e.g., Escherichia coli or Enterococcus spp.) (48). High concentrations of these microbes in water signal possible contamination by fecal material that could harbor microbial pathogens. While the indicator paradigm and its application are meant to ensure water safety and protect persons from fecal pathogens, it has become apparent within the past 2 decades that the current application of the indicator concept is flawed as it pertains to the assessment of recreational-water quality.
Enterococci are currently used as indicators of fecal contamination in recreational waters at Florida beaches (13). Levels of enterococci are assessed by membrane filtration followed by incubation on membrane-Enterococcus indoxyl-β-d-glucoside (mEI) agar for 24 h (EPA method 1600) (49). During the incubation period, recreational-water users may be exposed to unsafe conditions before beach advisories are issued. The corollary to this is that once an advisory is issued, the elevated concentrations of enterococci may have dispersed, so the swimming advisory is posted needlessly, causing economic harm to the businesses near the posted beach(es). Another limitation of the standard regulatory method is the possibility that shoreline sands act as a reservoir for enterococci. Shibata et al. found decreasing levels of indicator bacteria with increasing distance from the shoreline (39). Typically, water quality samples are collected near shore, which can artificially increase concentrations of enterococci and lead to further inaccuracies in the posting of advisories. It is imperative that new, rapid approaches be devised for the identification of fecal pollution events in order to protect public health and the designated recreational uses of coastal waters.
One strategy for rapid detection of microbial targets couples the specific binding of antibodies to their targets to the rapid response of sensor systems, known as biosensors (27). The ideal microbial sensor assay would be sensitive, specific, and able to detect low concentrations of target microbes. Biosensor immunoassays offer several advantages over rapid nucleic-acid-based detection systems. Although PCR assays can detect approximately 30 cells in a short time (14, 17, 20), the assays require a relatively “clean” sample that is free of inhibitors. Antibody-based biosensor assays have been performed on complex matrices, such as ground beef, apple juice, sprout irrigation waters, talc, chlorinated and chloraminated potable water, river water, and throat specimens (8, 9, 23, 24, 27, 31, 46). The following microorganisms have been detected within one or more of these matrices: Escherichia coli O157:H7, Salmonella enterica serovar Typhimurium, Bacillus anthracis, Bacillus atrophaeus, and vaccinia virus. Biosensor-based techniques can accurately detect the presence of these organisms in hours, as opposed to days or weeks with the standard methods (4, 10, 11, 44). Based on these data, recreational waters could also be monitored by biosensor methods without interference from background materials.
Rapid detection of biological targets in water is complicated by their low levels in ambient samples, which make concentration necessary in order to obtain the quantity of microorganisms needed for biosensor detection (32, 36, 43). Detection limits of biological targets in complex matrices average 104 to 105 CFU/ml (8, 9, 23, 24, 46). Hollow-fiber ultrafiltration provides significant concentration of cells from water with little increase in assay time and is currently the most promising technology for processing large volumes of water. Several recently developed methods have utilized hollow-fiber ultrafiltration for the concentration of microbes from water (19, 22, 25, 26, 30, 40). Water and particles smaller than the filter's molecular weight cutoff are forced through the filter pores. Parasites, bacteria, and viruses are retained within the fibers (19, 30). The material trapped within the fiber cores can be recovered by backflushing with water or a buffer after sample concentration and can subsequently be subjected to further analysis (23).
Ultrafiltration concentration of microorganisms from recreational water coupled to biosensor detection of the concentrated targets has the potential to greatly advance the field of microbial water quality. Concentration of microorganisms, such as enterococci, in recreational waters by hollow-fiber ultrafiltration reduces the time from collection to detection, thereby simultaneously reducing the potential exposure of swimmers to pathogens and the unnecessary closing of beaches. The goal of this research was to develop a more rapid method of measuring enterococci in recreational coastal waters. The system features dead-end hollow-fiber ultrafiltration, adapted from a previous study (47), coupled to immunoassay biosensor detection. The process reduces the time required for the detection of enterococci by an order of magnitude (from 24 h to 2.5 h). Rapid detection of other fecal indicator organisms and enteric pathogens is possible with this system due to the filter's ability to concomitantly retain parasites, bacteria, and viruses (19, 22).
MATERIALS AND METHODS
Antibodies.
A polyclonal rabbit antiserum against group D streptococci was purchased from American Research Products, Belmont, MA. The immunoglobulin G was affinity purified from the serum with a HiTrap protein A HP column (GE Healthcare Life Sciences, Piscataway, NJ), followed by three buffer exchanges with 0.1 M carbonate buffer (pH 8.5; Sigma-Aldrich) through an Amicon Ultra 4 centrifugal filter (Millipore Corp., Bedford, MA). An affinity-purified rabbit polyclonal antibody to Mycobacterium tuberculosis was purchased from Meridian Life Science, Inc. (Cincinnati, OH) for use in baseline measurements in Raptor assays prior to application of the anti-group D Streptococcus antibody. A lyophilized, affinity-purified, Cy5-labeled goat anti-rabbit secondary antibody was obtained from Jackson ImmunoResearch, West Grove, PA. A goat anti-rabbit antibody conjugated to horseradish peroxidase was purchased from Kirkegaard and Perry Laboratories, Inc., Gaithersburg, MD. The lyophilized antibodies were rehydrated as described by DeMarco et al. (9).
Bacteria.
The sensitivity of the anti-Streptococcus group D antibody for the detection of enterococci was determined by using the following bacteria: Enterococcus faecalis ATCC 19433, Enterococcus faecium ATCC 19434, Enterococcus faecium ATCC 35667, Enterococcus faecium C68, Enterococcus durans ATCC 6056, Enterococcus saccharolyticus ATCC 43076, Enterococcus avium ATCC 14025, Enterococcus casseliflavus ATCC 700327, Enterococcus gallinarum ATCC 49573, and Escherichia coli ATCC 15597. Enterococcus faecalis was utilized for the development of the indirect Raptor assay.
ELISA.
The anti-group D Streptococcus antibody was evaluated for sensitivity by an enzyme-linked immunosorbent assay (ELISA) prior to use in the indirect Raptor assay by its ability to bind to each of the bacteria listed in the preceding section. Bacteria were incubated in 5 ml of tryptic soy broth (BBL, Sparks, MD) with shaking (150 rpm) at 37°C for 5 h. Cells were harvested by centrifugation at 14,636 × g for 7 min at 4°C on an Eppendorf microcentrifuge, model 5415 R (Eppendorf North America, Westbury, NY). The supernatant fluids were decanted and pellets resuspended in 1 ml of 0.1 M carbonate bicarbonate (CBC) buffer (10.59 g Na2CO3 and 8.40 g NaHCO3 per liter [pH 9.3]). Direct counts were performed using a Cellometer (Nexcelom Bioscience, Lawrence, MA), and the concentration of each cell suspension was adjusted to 1.0 × 109 cells/ml. The suspensions were serially diluted (1:10) in CBC buffer to final concentrations of 1.0 × 104 to 1.0 × 108 cells/ml. Each cell concentration of each bacterium in duplicate or 0.1 M CBC (blanks) (8 wells per plate) was adsorbed to three wells of a 96-well microtiter plate (100 μl in each well) (Maxisorp; Nalgene Nunc International, Rochester, NY) for 18 h at 4°C.
Each well of the plates was washed three times with 100 μl of 0.01 M Dulbecco's phosphate-buffered saline (PBS) with 0.1% Tween 20 (PBST) by using a BioTek ELx50 Auto Strip washer (BioTek Instruments, Inc., Winooski, VT). One hundred fifty microliters of blocking buffer (1% bovine serum albumin and casein [wt/vol] in 0.01 M Dulbecco's PBS) was added to each well. The suspension was incubated for 30 min at 25°C, followed by three additional washes of each well with 100 μl PBST. The anti-group D Streptococcus antibody (100 μl at 10 μg/ml in blocking buffer) was applied to each well for 30 min at 25°C. The horseradish peroxidase-labeled goat anti-rabbit antibody (100 μl at 10 μg/ml in blocking buffer) was added to each well after three washes with 100 μl PBST. The antibody was incubated for 30 min at 25°C, followed by three additional washes with PBST (100 μl in each well). Bound peroxidase-labeled antibody was detected by the QuantaBlu fluorogenic peroxidase substrate kit (Pierce, Rockford, IL).
Each plate was analyzed on the SpectraMax Gemini XS microplate spectrofluorometer (Molecular Devices, Sunnyvale, CA) with the following parameters: excitation, 340 nm; emission, 470 nm; cutoff, 455 nm; photomultiplier tube setting, auto. The relative fluorescence for each cell concentration of each bacterium was divided by the average relative fluorescence for the 16 blank wells to generate a signal-to-noise (S/N) ratio for each bacterium at each cell concentration. Triplicate S/N values for each cell concentration were averaged and standard deviations calculated. Average S/N ratios above 2.0 were considered positive.
Raptor instrument and waveguide preparation.
The indirect biosensor assay for enterococci was developed with a portable microfluidic evanescent wave fiber optic biosensor (Raptor; Research International, Monroe, WA). The Raptor was developed by the Naval Research Laboratory (Washington, DC) for lateral-flow detection of biological targets from small sample volumes (i.e., ≤2 ml per sample port) (1, 2). Target analytes on polystyrene waveguides are bound by fluorophore-labeled reporter antibodies, and fluorescent molecules within 100 to 1,000 nm of the waveguide surface are excited by the evanescent field of the laser. A portion of the emission energy recouples into the waveguide and is quantified by the photodiode of the Raptor in picoamperes (pA).
Fiber optic waveguides were cleaned in a sonicating bath containing isopropanol and then rinsed with deionized water, and the distal tip was masked with black paint to provide a light dump for the biosensor's laser (24). Four waveguides were fixed into each coupon with optical adhesive for each final retentate sample to be analyzed. Each coupon containing four waveguides was then inserted into the Raptor for sample interrogation.
Indirect Raptor assay with cells in buffer.
Enterococcus faecalis was grown for 18 h at 37°C on tryptic soy agar (BBL, Sparks, MD). Cells were removed from the agar surface with sterile swabs and were suspended in CBC buffer. Cell concentrations were determined with a Cellometer, and the suspension was adjusted to a final concentration of 5.0 × 108 cells/ml. The stock suspension was used to prepare serial dilutions in CBC buffer (1:10), resulting in final cell concentrations ranging from 5.0 × 104 to 5.0 × 108/ml for use in the biosensor immunoassays. Cells of the same concentration (125 μl) were directly adsorbed to three waveguides in a coupon, and sterile CBC buffer (125 μl) was adsorbed to the fourth waveguide (negative control) for each assay. Cells and buffer were incubated on the waveguides for 10 min at 37°C. Each waveguide was rinsed twice with 125 μl of PBST to remove any unbound cells after sample incubation. The coupons were sealed and inserted into the biosensor. Each cell concentration was assayed in triplicate (three coupons per cell concentration).
The Raptor was programmed to run the following program for baseline readings: an antibody directed against a nontarget bacterium, M. tuberculosis (1 ml of antibody at 10 μg/ml in blocking buffer in each of the four sample ports), was applied to each of the four waveguides within the coupon for 3 min. The antibody was chosen for the baseline readings because of the inability of M. tuberculosis to grow in marine waters (21). Concentrated samples from marine water would eventually be tested by the Raptor immunoassay in the combined procedure. Nonspecific binding of the nontarget antibody to the concentrated samples would be limited, and stable baseline readings would be generated. The reagent (detection antibody) consisted of a Cy5-labeled antibody at 5 μg/ml in blocking buffer. The detection antibody was incubated on each waveguide within the coupon for 3 min. Four baseline readings (pA) were taken of each assay coupon for each cell concentration. Each E. faecalis concentration was detected by addition of the anti-group D Streptococcus antibody (1 ml at 10 μg/ml in blocking buffer) to each sample port of the Raptor. The program described above was run to generate the sample readings (pA). The target antibody was incubated on each waveguide a second time to confirm any positive sample readings from the first application.
A waveguide normalization factor was generated by dividing the emission value for the baseline readings (pA) from the four waveguides within a coupon by the lowest baseline reading within a coupon after the fourth baseline reading was obtained (41). This factor was used to normalize the emission values in each immunoassay so as to remove variability between the waveguides. Signal normalizations were necessary to account for the inherent variability of the fiber optic waveguides, which are individually molded and can differ widely in their baseline readings (41). Limits of detection (LOD) were calculated for each waveguide after normalization by adding the average of its baseline readings plus three times the standard deviations of the baseline for each waveguide in a coupon (24, 41). The LOD was subtracted from the emission value for each sample reading (pA) to achieve the signal above the LOD (SALOD). If the SALOD was greater than zero and larger than the SALOD from the negative-control waveguide (CBC buffer), the sample was considered positive for enterococci. If the SALOD was less than zero or less than or equal to the SALOD of the negative control, the sample was considered negative. The SALOD values calculated from the replicates for each cell concentration were averaged, and the standard deviation was calculated in order to determine the SALOD ranges.
Sampling location and dates.
Ben T. Davis Municipal Beach is a narrow beach located on Rocky Point Island in upper Tampa Bay (latitude, 27°57′58″N; longitude, 82°34′50″W). Twelve 100-liter samples were collected from this location at slack tide between August 2006 and April 2007.
Hollow-fiber concentration system.
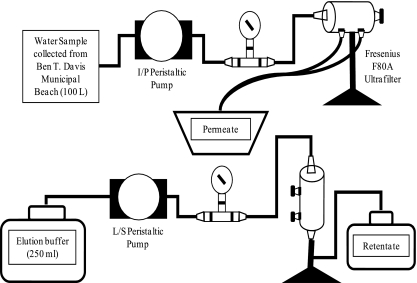
The recreational dead-end concentrator (Rec DEC) (Fig. 1) utilized a new F80A Hemoflow polysulfone high-flux capillary dialyzer (Fresenius Medical Care North America, Lexington, MA) for each of the 12 samples. Each filter consists of an array of hollow fibers with a fiber length of 25 cm, a membrane thickness of 40 μm, an inner fiber radius of 100 μm, a total surface area of 1.8 m2, and a molecular weight cutoff of approximately 15,000 to 20,000 (19).
FIG. 1.
Schematic of the Rec DEC in sample collection (top) and sample elution (bottom) modes.
Beach water was fed into each filter by a Masterflex I/P Precision brushless peristaltic pump drive with a Masterflex I/P Easy-Load pump head (Cole Parmer Instrument Co., Vernon Hills, IL). The pump was powered in the field by a portable Xantrex XPower Powerpack 600 HD battery (Xantrex Technology Inc., Burnaby, British Columbia, Canada). Fifty feet of Masterflex I/P BioPharm platinum-cured silicone I/P 70 tubing, with a nylon mesh screen attached to the uptake end, transported the water from the sampling location to the Rec DEC onshore. A pressure gauge was mounted in-line with the collection tubing to monitor the internal pressure of the system during filtration. The tubing was connected to the filter with a rigid polyvinyl chloride dialysis connector (Qosina Corp., Edgewood, NY) through one of the two inlet ports on the filter cartridge. The dead end was created by inserting a soft polyvinyl chloride dialysis connector (Qosina) into the other inlet port and closing it with a Scienceware acetal screw clamp (Thermo Fisher Scientific, Waltham, MA).
Concentration procedure.
Microbes within the water (2 m from shore at a depth of 0.5 m) were concentrated with the filter in the horizontal position. The pump was operated initially at a speed of 5 U to purge air from the collection line and to allow the filter to fill with water with the dead-end port open. The dead-end clamp was closed after water began to flow from the permeate ports. The pump speed was increased 1 U every 30 s until the final speed of 35 U was reached. Filtration continued until 100 liters was processed, as determined by the flow from the permeate ports. Filtration occurred in approximately 50 min while an internal system pressure of 25 to 30 lb/in2 was maintained. A permeate flow rate of at least 2.5 liters/min was achieved at top speed. After filtration was completed, the filter ports were closed for storage at 4°C during transport. All Rec DEC components were cleaned between sampling events with 10% bleach followed by 10% sodium thiosulfate (19), or with 0.8% Lysol followed by deionized water for metal components.
Sample elution from the filter.
The residual volume (70 to 100 ml) within the filter cartridge was removed by vacuum filtration prior to application of the elution buffer. Elution was performed with the filter in the vertical position using 250 ml of elution buffer (4 M urea-50 mM lysine [pH 9.0]) (7). The buffer was fed into the filter through one of the inlet ports by a Masterflex L/S peristaltic pump and Masterflex L/S 36 gauge Tygon tubing. The other inlet port was closed by clamping the Tygon tubing, while the permeate ports were capped (Fig. 1). The pump was operated at a speed of 2 U to feed elution buffer into the filter. The upper permeate port was opened briefly to purge air from the buffer line. The buffer was allowed to interact with the filter fibers for 2 min. The initial retentate was collected by opening the clamp on the lower inlet port tubing and operating the pump at a speed of 4 U until the flow from the filter ceased. The residual initial retentate remaining in the filter was removed by vacuum filtration and combined with the pumped retentate to produce the total initial retentate.
Postfiltration sample processing.
The initial retentate was sonicated for 60 s at 14 W with the Sonic Dismembrator, model 100 (Fisher Scientific, Pittsburgh, PA). Sonication of the sample disrupts the attachment of microbial cells to sediment particles and has been shown to increase the recovery of indicator organisms adsorbed to sediment particles (3). The retentate was poured through stacked stainless steel sieves (pore sizes, 75 μm, 53 μm, and 38 μm) to remove particles that could obstruct the microfluidics of the Raptor (33). The flowthrough volume was collected from the sieve reservoir and subjected to secondary concentration by centrifugation. Centrifugation was performed at 15,180 × g for 5 min at 4°C on a Sorvall Evolution RC Superspeed refrigerated centrifuge (Thermo Electron Corporation, Asheville, NC). The supernatant fluid was decanted and the pellet resuspended in 4 ml of 0.1 M CBC buffer to yield the final retentate.
Membrane filtration.
A small subsample (5 ml) from each stage in the procedure (elution, sonication and sieve passage, and centrifugation) was reserved for membrane filtration. A 1-liter ambient grab sample from Ben T. Davis Municipal Beach was collected for each sampling date and was stored on ice until the time of processing. The grab samples and ultrafiltration subsamples were filtered through Gelman GN-6 Meticel membrane filters (diameter, 47 mm; pore size, 0.45 μm; Pall Corporation, East Hills, NY). Membranes were placed on mEI agar and incubated at 41°C for 24 h according to EPA method 1600 (49). All resultant colonies with blue halos were counted as enterococci. Counts were used to compare the EPA method with cell concentrations during the ultrafiltration procedure and to calculate recovery efficiencies and concentration factors (19).
Indirect Raptor assay with concentrated samples.
Final retentates were analyzed for the presence of enterococci by the indirect Raptor immunoassay as described in “Indirect Raptor assay with cells in buffer” above, with a few modifications. Each cell suspension was incubated on two waveguides (125 μl each) in a coupon. Positive- and negative-control waveguides, consisting of an aliquot of the final retentate (62.5 μl) mixed with an equal volume of E. faecalis at 1 × 108 cells/ml in CBC buffer and sterile CBC buffer, respectively, were incubated on the other two waveguides in each coupon. The waveguides were incubated with the respective samples for 10 min at 37°C. The indirect assay and all SALOD calculations were performed as described above.
Statistical analysis.
Differences in log10-transformed ambient cell concentrations and those resulting from the ultrafiltration procedure were analyzed using two-tailed paired t tests (GraphPad InStat, version 3.0; GraphPad Software Inc., San Diego, CA). Concentration factors for the final retentates were calculated to quantify the increase in cell concentration compared to ambient grab samples. Similar ratios have been calculated to quantify changes in cell counts following concentration (6). The concentration factor for hollow-fiber filtration and postfiltration processing is defined as the ratio of the concentration of enterococci in the final retentate to the concentration of enterococci in the ambient water. The recovery efficiency (percent recovery) of the ultrafiltration procedure was calculated by determining the number of enterococci in the ambient water and that in the final retentate (19). After the values were determined, the recovery efficiency was calculated by dividing the total number in the final retentate by the total number in the ambient water. The percentage of recovery was determined by multiplying the fraction by 100.
Linear regression was used to compare the E. faecalis concentrations in buffer to the Raptor SALOD values generated by each concentration, as well as the SALOD values from concentrated samples and ambient enterococcal levels (GraphPad InStat, version 3.0). Binary logistic regression was used to compare the Raptor SALOD values to ambient concentrations relevant to regulatory thresholds in Florida marine waters (≥105 CFU/100 ml) (SPSS 15.0 for Windows; SPSS Inc., Chicago, IL) (16). The Nagelkerke R square, which can range from 0.0 to 1.0, measures the effect size (the strength of the relationship); stronger associations have values closer to 1.0. Relationships were considered significant when the P value for the model chi-square was <0.05 and the confidence interval for the odds ratio did not include 1.0. Greater odds ratios indicate a higher probability of change in the dependent variable with a change in the independent variable. Statistical significance for all data was accepted at the 95% confidence level (α ≤ 0.05).
RESULTS
ELISA.
The sensitivity of the anti-group D Streptococcus antibody was assessed by ELISA prior to its use in the Raptor immunoassay. Positive detection (S/N ratio, ≥2.0) with each of the species of Enterococcus occurred from 1.0 × 105 to 1.0 × 108 cells/ml. S/N ratios ranging from 14.8 to 18.1 were obtained at 1.0 × 108 cells/ml. As the concentration of enterococci decreased, the S/N ratio decreased to a minimum of 2.6 at 1.0 × 105 cells/ml.
Indirect biosensor detection of E. faecalis in buffer.
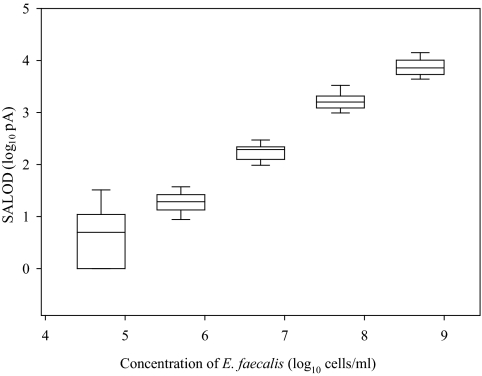
The anti-group D Streptococcus antibody was tested for its usefulness as a detection antibody in an indirect Raptor assay for enterococci. Sensitivity assays were performed with samples of E. faecalis in buffer (CBC). Each replicate sample was interrogated by laser excitation for the presence of E. faecalis. Positive detection occurred at concentrations equal to or greater than 5.0 × 105 cells/ml, with an average SALOD of 20 (Fig. 2). The average SALOD values for different cell concentrations do not overlap, while a slight convergence was observed for SALOD standard deviations at the lower cell concentrations. A linear decrease in the signal with decreasing cell concentration was observed (r2 = 0.94; P < 0.001). The average SALOD values for the negative-control waveguides (CBC buffer) were higher than those for 5.0 × 104 cells/ml.
FIG. 2.
Comparison of the concentrations of E. faecalis and the SALOD values generated during the indirect Raptor assays. Each box displays the range of SALODs for each cell concentration. Each box includes the median (center line) SALOD and the 25th and 75th percentiles (lower and upper box limits, respectively). Error bars represent the 10th and 90th percentiles of the SALOD values for each cell concentration.
Ambient enterococcal concentrations versus concentrations of cells recovered from ultrafilters and postfiltration processing.
Twelve grab samples and 12 100-liter concentrated samples from the Ben T. Davis Municipal Beach were analyzed for enterococci by membrane filtration. Ambient enterococcal concentrations ranged from −0.4 to 2.4 log10 CFU/100 ml based on method 1600. Hollow-fiber filtration and backflushing increased the concentrations of enterococci by 2 orders of magnitude (P = 0.018). Sonication and sieve passage increased the concentration of enterococci by 1 log unit while removing small particles from the initial retentate that could interfere with the Raptor's microfluidics (P = 0.032). Secondary concentration increased the levels of enterococci by another 2 orders of magnitude (P = 0.0002). The concentration of enterococci in the final retentates ranged from 0.6 to 5.3 log10 CFU/100 ml. The levels of enterococci in the final retentate were at least 4 orders higher than those in the ambient samples (P < 0.0001).
The calculated total number of enterococci present in 100 liters of ambient water ranged from 2.6 to 5.4 log10 CFU (Table 1). The total number in 4 ml of final retentate ranged from 1.2 to 5.9 log10 CFU. Concentration factors were lowest on 11 January 2007 (1,000-fold) and highest on 22 August 2006 (approximately 177,000-fold). Recovery efficiencies ranged from 4% to 708%, with an average of 251%.
TABLE 1.
Ultrafiltration recovery efficiencies and concentration factors of enterococci from ambient samples
| Date (mo/day/yr) | Total CFU (log10)a in:
|
Recovery (%)b | Concn factorc | |
|---|---|---|---|---|
| Ambient sample (100 liters) | Final retentate (4 ml) | |||
| 8/15/06 | 4.7 | 4.6 | 80 | 20,000 |
| 8/22/06 | 5.1 | 5.9 | 708 | 176,991 |
| 9/5/06 | 5.4 | 5.6 | 160 | 40,000 |
| 10/17/06 | 5.0 | 5.6 | 370 | 92,593 |
| 12/5/06 | 3.0 | 3.6 | 400 | 100,000 |
| 1/11/07 | 2.6 | 1.2 | 4 | 1,000 |
| 1/16/07 | 4.0 | 3.9 | 88 | 21,978 |
| 2/20/07 | 3.8 | 3.8 | 105 | 26,154 |
| 3/20/07 | 3.6 | 4.1 | 300 | 75,000 |
| 3/27/07 | 3.3 | 3.3 | 100 | 25,000 |
| 3/30/07 | 3.4 | 3.9 | 320 | 80,000 |
| 4/3/07 | 3.0 | 3.6 | 376 | 94,000 |
Based on method 1600.
Calculated as (total CFU in the final retentate)/(total CFU in the ambient sample) × 100.
Calculated as (CFU/ml of the final retentate)/(CFU/ml of the ambient sample).
Recoveries over 100% have been observed by others using hollow-fiber ultrafiltration, but those experiments were performed by spiking with known amounts of the target microbes (18, 19, 34). The values for the total CFU in 100 liters of ambient water are extrapolations of concentrations obtained by membrane filtration of 100 ml according to method 1600. The total CFU may underestimate or overestimate the number of enterococci present in each 100-liter sample. The distribution of enterococci in the larger volume of water should not be assumed to be the same as that in the smaller volume; however, use of the extrapolated values was necessary to evaluate the concentration procedure. Variability of viable counts on growth media has also been documented (5, 29, 37) and has been observed following dead-end ultrafiltration and elution (19, 23). Reduced recoveries of viable cells following ultrafiltration may be due to stresses encountered by cells following removal from the aquatic environment and subsequent hollow-fiber filtration. Immunoassays can detect viable and nonviable cells, so any variability in viable counts could be negated and enterococci could be detected in poor-quality water regardless of the range of calculated recoveries (42, 52).
Indirect detection of enterococci in the final retentates.
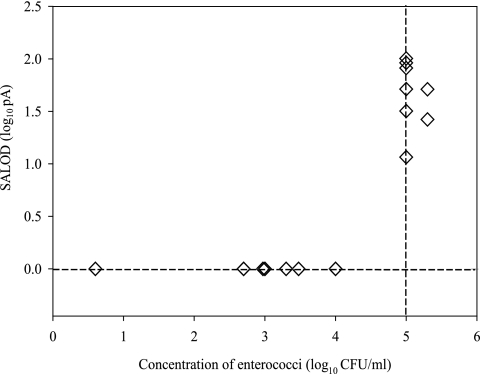
An aliquot of the final retentate was tested by the indirect Raptor immunoassay to determine its ability to detect enterococci. SALOD values were below zero when the ambient concentrations of enterococci were below the regulatory threshold for a single-sample maximum for Florida recreational marine waters (≥105 CFU/100 ml) (Fig. 3). SALOD values ranged from 11 to 100 when the single-sample maximum was exceeded. A correlation was established between log10-transformed SALOD values from final retentates and log10-tranformed concentrations of enterococci in ambient samples (R2 = 0.59; P < 0.0001). Binary logistic regression was used to assess the predictive relationship between the occurrence of ambient enterococcal concentrations equal to or greater than the single-sample maximum and the occurrence of a positive SALOD from the Raptor immunoassay. The Nagelkerke R square measures the predictive power of the model. The strength of the relationship ranges from 0.0 to 1.0, with stronger associations having values closer to 1.0. The odds ratio compares the probability that a change in the independent variable will lead to a change in the dependent variable. Larger odds ratios indicate stronger relationships between the variables. A correlation was established between the detection of enterococci in final retentates with the Raptor and the occurrence of ambient concentrations relevant to regulatory thresholds (Nagelkerke R2 = 0.61; P < 0.0001).
FIG. 3.
SALOD values generated by enterococci in final retentates. Each retentate was adsorbed to waveguides for biosensor detection (n = 26). The minimum concentration of enterococci that can be detected with the indirect Raptor assay is 1.0 × 105 CFU/ml.
DISCUSSION
There is a need for rapid methods for identification of fecal pollution events in order to minimize the exposure of the public to microbial pathogens, for the safety of those using the nation's recreational waters. Various procedures that would permit “same-day” warning systems have recently been developed in an attempt to meet this need. The combined procedure described in this report couples on-site dead-end hollow-fiber ultrafiltration to Raptor detection of enterococci. The portable format of the filtration system should prove useful for rapid monitoring of recreational-water quality. Detection with the combined procedure occurred when concentrations of enterococci in the ambient water exceeded the regulatory standard for a single sample (≥105 CFU/100 ml). The main advantage of this technology over existing methods is the speed with which results are provided: 2.5 h compared with 24 h for traditional membrane filtration methods and 3 to 4 h for quantitative PCR (QPCR)-based methods (32, 49).
Other recently developed rapid techniques for the detection of enterococci include QPCR and transcription-mediated amplification (a genetic method that targets RNA rather than DNA) (32). Raptor analysis following hollow-fiber ultrafiltration is unique with respect to the monitoring of recreational waters for indicator bacteria. The other technologies rely on amplifying captured targets (i.e., replication of DNA). The biosensor technology measures the captured organisms directly after the processing of a large volume of water to increase their concentration to the levels necessary for biosensor detection. The targets are removed directly from the environment, and detection is not a result of multiplying the few initially captured targets, thus making possible the decrease in analysis time. This demonstrates the potential of the Rec DEC-Raptor procedure for rapid monitoring of water quality.
A more thorough assessment of fecal pollution events at recreational coastal waters is possible with the combined ultrafiltration-biosensor methodology. Such an assessment would better protect public health and the designated uses of these waters. The ability to monitor 100 liters of recreational water is an advantage of the procedure over the standard methodology and the rapid molecular techniques. The larger volume of water processed with the Rec DEC-Raptor procedure provides a more complete picture of the true water quality at the beach than does a smaller volume (e.g., 100 ml for the standard method and for membrane filtration followed by QPCR). The concentration-biosensor system would be more expensive to implement initially due to equipment costs but would provide a more rapid, representative determination of water quality than method 1600. Similar filtration techniques have recently been validated by the EPA for the monitoring of potable and source waters (50).
A further advantage of the system is the possibility of directly measuring microbial pathogens that can be simultaneously concentrated with the hollow-fiber filter. Direct pathogen detection in recreational waters would alleviate the issues associated with indicator organisms, such as false positives from possible regrowth in sands and sediments (39). Detection of the pathogens following ultrafiltration concentration has been accomplished. Kearns et al. detected B. atrophaeus, the surrogate organism for B. anthracis, through biosensor analysis following hollow-fiber concentration from chlorinated and chloraminated potable water (23). Direct assessment of recreational waters for the presence of pathogens, not simply indicators, could be accomplished through the combined procedure of dead-end ultrafiltration and biosensor detection. Furthermore, new immunoassay-based biosensor technologies have the ability to detect the presence of multiple target microbes within a single sample simultaneously (38, 45).
Many coastal waters, particularly those near densely populated urban areas, have decreased water quality (51). Urbanization exacerbates the problem by increasing the area of impervious surfaces, such as roads and parking lots, in affected watersheds, resulting in greater stormwater runoff and microbial contamination during rain events (28). Transport of enteric microorganisms to coastal waters can occur if stormwaters are not treated or contained (12, 15, 35). The Rec DEC-Raptor procedure confers the ability to detect these and other microbial inputs into coastal waters rapidly Not only can indicator bacteria be detected within 2.5 h, but the possibility exists for direct assessment of pathogens within the body of recreational water. Public health would be enhanced by the ability to detect waterborne pathogens more rapidly and to post advisories prior to the current minimum of 24 h postsampling.
Acknowledgments
This research was supported by a grant from the U.S. Army Research, Development, and Engineering Command.
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Anderson, G. P., J. P. Golden, L. K. Cao, D. Wijesuriya, L. C. Shriver-Lake, and F. S. Ligler. 1994. Development of an evanescent wave fiber optic biosensor. IEEE Eng. Med. Biol. Mag. 13:358-363. [Google Scholar]
- 2.Anderson, G. P., K. D. King, K. L. Gaffney, and L. H. Johnson. 2000. Multi-analyte interrogation using the fiber optic biosensor. Biosens. Bioelectron. 14:771-777. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, W. H., and T. S. Hammack. 2003. Chapter 5, Salmonella. In FDA bacteriological analytical manual online, 8th ed. U.S. Food and Drug Administration, Washington, DC.
- 5.Bissonnette, G. K., J. J. Jezeski, A. McFeters, and D. G. Stuart. 1975. Influence of environmental stress on enumeration of indicator bacteria from natural waters. Appl. Microbiol. 29:186-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlucci, A. F., and P. M. Williams. 1965. Concentration of bacteria from sea water by bubble scavenging. ICES J. Mar. Sci. 30:28-33. [Google Scholar]
- 7.Chang, L. T., S. R. Farrah, and G. Bitton. 1981. Positively charged filters for virus recovery from wastewater treatment plant effluents. Appl. Environ. Microbiol. 42:921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeMarco, D. R., and D. V. Lim. 2002. Detection of Escherichia coli O157:H7 in 10- and 25-gram ground beef samples with an evanescent-wave biosensor with silica and polystyrene waveguides. J. Food Prot. 65:596-602. [DOI] [PubMed] [Google Scholar]
- 9.DeMarco, D. R., E. W. Saaski, D. A. McCrae, and D. V. Lim. 1999. Rapid detection of Escherichia coli O157:H7 in ground beef using a fiber-optic biosensor. J. Food Prot. 62:711-716. [DOI] [PubMed] [Google Scholar]
- 10.Espy, M. J., F. R. Cockerill III, R. F. Meyer, M. D. Bowen, G. A. Poland, T. L. Hadfield, and T. F. Smith. 2002. Detection of smallpox virus DNA by LightCycler PCR. J. Clin. Microbiol. 40:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, P., and S. D. Weagant. 2002. Chapter 4a, Diarrheagenic Escherichia coli. In FDA bacteriological analytical manual online, 8th ed. U.S. Food and Drug Administration, Washington, DC.
- 12.Ferguson, C., A. M. de Roda Husman, N. Altavilla, D. Deere, and N. Ashbolt. 2003. Fate and transport of surface water pathogens in watersheds. Crit. Rev. Environ. Sci. Technol. 33:299-361. [Google Scholar]
- 13.Florida Administrative Code. 1998. Surface water quality standards. http://www.flrules.org/gateway/ruleNo.asp?id=62-302.530.
- 14.Fode-Vaughan, K. A., J. S. Maki, J. A. Benson, and M. L. P. Collins. 2003. Direct PCR detection of Escherichia coli O157:H7. Lett. Appl. Microbiol. 37:239-243. [DOI] [PubMed] [Google Scholar]
- 15.Griffin, D. W., C. J. Gibson III, E. K. Lipp, K. Riley, J. H. Paul III, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harwood, V. J., A. D. Levine, T. M. Scott, V. Chivukula, J. Lukasik, S. R. Farrah, and J. B. Rose. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71:3163-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haugland, R. A., S. C. Siefring, L. J. Wymer, K. P. Brenner, and A. P. Dufour. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559-568. [DOI] [PubMed] [Google Scholar]
- 18.Hill, V. R., A. M. Kahler, N. Jothikumar, T. B. Johnson, D. Hahn, and T. L. Cromeans. 2007. Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl. Environ. Microbiol. 73:4218-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, V. R., A. L. Polaczyk, D. Hahn, J. Narayanan, T. L. Cromeans, J. M. Roberts, and J. E. Amburgey. 2005. Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl. Environ. Microbiol. 71:6878-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibekwe, A. M., and C. M. Grieve. 2003. Detection and quantification of Escherichia coli O157:H7 in environmental samples by real-time PCR. J. Appl. Microbiol. 94:421-431. [DOI] [PubMed] [Google Scholar]
- 21.Jamieson, W., P. Madri, and G. Claus. 1976. Survival of certain pathogenic microorganisms in sea water. Hydrobiologia 50:117-121. [Google Scholar]
- 22.Juliano, J., and M. D. Sobsey. 1998. Simultaneous concentration of Cryptosporidium, bacteria, and viruses from water by hollow-fiber ultrafiltration. In Proceedings of the Water Quality Technology Conference 1998. American Water Works Association, Denver, CO.
- 23.Kearns, E. A., S. Magana, and D. V. Lim. 19 February 2008. Automated concentration and recovery of microorganisms from drinking water using dead-end filtration. J. Appl. Microbiol. doi: 10.1111/j. 1365-2672.2008.03757.x. [DOI] [PubMed]
- 24.Kramer, M. F., and D. V. Lim. 2004. A rapid and automated fiber optic-based biosensor assay for the detection of Salmonella in spent irrigation water used in the sprouting of sprout seeds. J. Food Prot. 67:46-52. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn, R. C., and K. H. Oshima. 2001. Evaluation and optimization of a reusable hollow fiber ultrafilter as a first step in concentrating Cryptosporidium parvum oocysts from water. Water Res. 35:2779-2783. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn, R. C., and K. H. Oshima. 2002. Hollow-fiber ultrafiltration of Cryptosporidium parvum oocysts from a wide variety of 10-L surface water samples. Can. J. Microbiol. 48:542-549. [DOI] [PubMed] [Google Scholar]
- 27.Lim, D. V., J. M. Simpson, E. A. Kearns, and M. F. Kramer. 2005. Current and developing technologies for monitoring agents of bioterrorism and biowarfare. Clin. Microbiol. Rev. 18:583-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallin, M. A., K. E. Williams, E. C. Esham, and R. P. Lowe. 2000. Effect of human development on bacteriological water quality in coastal watersheds. Ecol. Appl. 10:1047-1056. [Google Scholar]
- 29.McKay, A. M. 1992. Viable but non-culturable forms of potentially pathogenic bacteria in water. Lett. Appl. Microbiol. 14:129-135. [Google Scholar]
- 30.Morales-Morales, H. A., G. Vidal, J. Olszewski, C. M. Rock, D. Dasgupta, K. H. Oshima, and G. B. Smith. 2003. Optimization of a reusable hollow-fiber ultrafilter for simultaneous concentration of enteric bacteria, protozoa, and viruses from water. Appl. Environ. Microbiol. 69:4098-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narang, U., G. P. Anderson, F. S. Ligler, and J. Burans. 1997. Fiber optic-based biosensor for ricin. Biosens. Bioelectron. 12:937-945. [DOI] [PubMed] [Google Scholar]
- 32.Noble, R. T., and S. B. Weisberg. 2005. A review of technologies for rapid detection of bacteria in recreational waters. J. Water Health 3:381-392. [DOI] [PubMed] [Google Scholar]
- 33.Olszewski, J., L. Winona, and K. H. Oshima. 2005. Comparison of 2 ultrafiltration systems for the concentration of seeded viruses from environmental waters. Can. J. Microbiol. 51:295-303. [DOI] [PubMed] [Google Scholar]
- 34.Polaczyk, A. L., J. Narayanan, T. L. Cromeans, D. Hahn, J. M. Roberts, J. E. Amburgey, and V. R. Hill. 2008. Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-L tap water samples. J. Microbiol. Methods 73:92-99. [DOI] [PubMed] [Google Scholar]
- 35.Reddy, K. R., R. Khaleel, and M. R. Overcash. 1981. Behavior and transport of microbial pathogens and indicator organisms in soils treated with organic wastes. J. Environ. Qual. 10:255-266. [Google Scholar]
- 36.Reynolds, K. A. 2003. On tap: detecting waterborne pathogens—a look at past, present and future approaches. Water Conditioning Purif. Mag. 45:86-89. http://www.wcponline.com/NewsView.cfm?ID=2384. [Google Scholar]
- 37.Roszak, D. B., and R. R. Colwell. 1987. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51:365-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sapsford, K. E., M. M. Ngundi, M. H. Moore, M. E. Lassman, L. C. Shriver-Lake, C. R. Taitt, and F. S. Ligler. 2006. Rapid detection of foodborne contaminants using an Array Biosensor. Sensors Actuators B 113:599-607. [Google Scholar]
- 39.Shibata, T., H. M. Solo-Gabriele, L. E. Fleming, and S. Elmir. 2004. Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Water Res. 38:3119-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmons, O. D., III, M. D. Sobsey, C. D. Heaney, F. W. Schaefer III, and D. S. Francy. 2001. Concentration and detection of Cryptosporidium oocysts in surface water samples by method 1622 using ultrafiltration and capsule filtration. Appl. Environ. Microbiol. 67:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson-Stroot, J. M., E. A. Kearns, P. G. Stroot, S. Magaña, and D. V. Lim. 2008. Monitoring biosensor capture efficiencies: development of a model using GFP-expressing Escherichia coli O157:H7. J. Microbiol. Methods 72:29-37. [DOI] [PubMed] [Google Scholar]
- 42.Stopa, P. J. 2000. The flow cytometry of Bacillus anthracis spores revisited. Cytometry 41:237-244. [DOI] [PubMed] [Google Scholar]
- 43.Straub, T. M., and D. P. Chandler. 2003. Towards a unified system for detecting waterborne pathogens. J. Microbiol. Methods 53:185-197. [DOI] [PubMed] [Google Scholar]
- 44.Szabo, J. G., E. W. Rice, and P. L. Bishop. 2007. Persistence and decontamination of Bacillus atrophaeus subsp. globigii spores on corroded iron in a model drinking water system. Appl. Environ. Microbiol. 73:2451-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taitt, C. R., J. P. Golden, Y. S. Shubin, L. C. Shriver-Lake, K. E. Sapsford, A. Rasooly, and F. S. Ligler. 2004. A portable array biosensor for detecting multiple analytes in complex samples. Microb. Ecol. 47:175-185. [DOI] [PubMed] [Google Scholar]
- 46.Tims, T. B., and D. V. Lim. 2004. Rapid detection of Bacillus anthracis spores directly from powders with an evanescent wave fiber-optic biosensor. J. Microbiol. Methods 59:127-130. [DOI] [PubMed] [Google Scholar]
- 47.Trindade, M., M. Kramer, and D. Lim. 2005. An evanescent wave fiber optic biosensor for the detection of fecal enterococci in beach water, abstr. Q-314. Abstr. 105th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 48.U.S. Environmental Protection Agency. 2000. Improved enumeration methods for the recreational water quality indicators: enterococci and Escherichia coli. EPA 821-R-97-004. U.S. Environmental Protection Agency, Washington, DC.
- 49.U.S. Environmental Protection Agency. 2002. Method 1600: enterococci in water by membrane filtration using membrane-Enterococcus indoxyl-β-d-glucoside agar (mEI). EPA 821-R-02-022. U.S. Environmental Protection Agency, Washington, DC.
- 50.U.S. Environmental Protection Agency. 1999. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA 821-R-99-006. U.S. Environmental Protection Agency, Washington, DC.
- 51.Vitousek, P. M., H. A. Mooney, J. Lubchenco, and J. M. Melillo. 1997. Human domination of Earth's ecosystems. Science 277:494-499. [Google Scholar]
- 52.Zhang, Q., L. Zhu, H. Feng, S. Ang, F. S. Chau, and W.-T. Liu. 2006. Microbial detection in microfluidic devices through dual staining of quantum dots-labeled immunoassay and RNA hybridization. Anal. Chim. Acta 556:171-177. [DOI] [PubMed] [Google Scholar]