Abstract
Legionella pneumophila is the causative agent of Legionnaires' disease. This bacterium is ubiquitous in aqueous environments and uses amoebae as an intracellular replicative niche. Real-time PCR has been developed for rapid detection of Legionella DNA in water samples. In addition to culturable bacteria, this method may also detect dead and viable but noncultivable (VBNC) legionellae. In order to understand the significance of positive PCR results in this setting, we prepared water samples containing known concentrations of L. pneumophila and analyzed them comparatively by means of conventional culture, real-time PCR, viability labeling, and immunodetection (solid-phase cytometry). We also examined the influence of chlorination on the results of the four methods. The different techniques yielded similar results for nonchlorinated water samples but not for chlorinated samples. After treatment for 24 h with 0.5 and 1 ppm chlorine, all cultures were negative, PCR and immunodetection showed about 106 genome units and bacteria/ml, and total-viable-count (TVC) labeling detected 105 and 102 metabolically active bacteria/ml, respectively. Thus, PCR also detected bacteria that were VBNC. The recoverability of VBNC forms was confirmed by 5 days of coculture with Acanthamoeba polyphaga. Therefore, some TVC-positive bacteria were potentially infective. These data show that L. pneumophila PCR detects not only culturable bacteria but also VBNC forms and dead bacterial DNA at low chlorine concentrations.
Legionella pneumophila, the bacterium responsible for Legionnaires' disease and Pontiac fever, is ubiquitous in natural and man-made aqueous environments and requires free-living amoebae for its intracellular replication (1, 15, 31). Under appropriate conditions, L. pneumophila can also survive for long periods as a free organism in low-nutrient environments (4, 30). Regular monitoring of potentially contaminated water sources is essential to prevent legionellosis outbreaks (21, 27). Culture with selective media is the standard method for the detection, isolation, and identification of L. pneumophila in clinical and environmental samples (18, 19), but it can take more than 7 days. Cost-effective and reliable real-time quantitative PCR methods have been developed for rapid detection/quantification of Legionella DNA in water samples and are often used as a routine monitoring tool (14, 36). The results are expressed as the number of genome units (GU) per liter, but the precise equivalence with the number of CFU has not been established. Culture and PCR agree well on samples from hot water systems but not from cooling towers. Culture is always less sensitive than PCR (2, 23, 36).
Discrepancies between PCR and culture results can be explained by several factors. Legionella growth can be inhibited or masked by overgrowth of contaminating microorganisms (18). Furthermore, L. pneumophila can enter a viable but noncultivable (VBNC) state, from which it can recover after passage in amoebae (12, 30). These VBNC legionellae may be detected by PCR, along with dead bacteria, possibly explaining, at least in part, why PCR values are usually higher than those obtained by culture.
In order to understand the significance of positive L. pneumophila PCR results for water samples, we prepared L. pneumophila-containing water samples, with and without chlorination, and tested them comparatively by (i) conventional culture, (ii) a real-time PCR assay for total L. pneumophila DNA, (iii) a fluorescence assay for viable bacteria (VBNC and cultivable bacteria), and (iv) immunodetection of all living and dead intact L. pneumophila cells. Potential VBNC bacteria detected by the fluorescence assay were tested for infectivity and recultivability in a coculture technique with amoebae.
MATERIALS AND METHODS
Bacterial culture.
The strain used in this study was Legionella pneumophila Paris (CIP 107629). It was stored at −80°C. After thawing, the cells were streaked onto BCYEα agar {buffered activated charcoal and yeast extract in 2-[(2-amino-2-oxoethyl)amino]ethanesulfonic acid; Oxoid, Dardilly, France} for 48 h at 37°C. They were then restreaked onto the same medium for another 48 h at 37°C. A patch of L. pneumophila culture was then used to inoculate 3 ml of sterile tryptone-salt solution (peptone at 1 g/liter, NaCl at 8.5 g/liter) in Culligan water (pH 7.5; Culligan, Champagne aux Monts d'Or, France). After 2 min of sonication at 104 W, the optical density at 600 nm was adjusted to 1 (assumed to correspond to 109 CFU/ml), and the cells were immediately used for chlorination experiments. GVPC medium (BCYE agar containing a glycine, vancomycin, polymyxin B, and cycloheximide supplement; Biomerieux, France) was also used.
Chlorination.
Free chlorine solution was freshly prepared by diluting bleach containing 9.6% active chlorine (Lacroix, Bois-Colombes, France) with sterile ultrapure water (Culligan, Champagne aux Monts d'Or, France) and phosphate-buffered saline (10× Dulbecco's phosphate-buffered saline without CaCl2 and MgCl2; GIBCO, Cergy-Pontoise, France) to obtain a 10,000-ppm stock solution in 1× phosphate-buffered saline. The concentration of free chlorine was determined with a pocket colorimeter analysis system (Chlore CHEMets kit; CHEMetrics Inc., Calverton, VA). For chlorination experiments, bacteria were first diluted to 107 CFU/ml in water and then treated with the different chlorine concentrations for the times indicated. The chlorinated samples were then neutralized with sterile sodium thiosulfate (final concentration, 20 mg/liter), and serial dilutions theoretically containing 107 to 102 Legionella organisms/ml were prepared for analysis by culture, viability assay, immunodetection, and real-time PCR.
Culturability.
After chlorine exposure and thiosulfate neutralization, the samples and controls were serially diluted to contain 107 to 102 Legionella organisms/ml before being plated onto BCYE agar and incubated at 37°C for 7 days. The results are expressed in log CFU/ml.
Sample preparation and quantitative PCR.
After exposure to chlorine, the samples and controls were serially diluted to concentrations of 107 to 102 Legionella organisms/ml, and 1 ml of the 107-cell/ml dilution was added to 100 ml of fresh water before vacuum filtration. One hundred microliters of the 106-cells/ml dilution was used directly for DNA extraction, without prior filtration, in order to determine whether DNA was retained on the filter and amplified. Samples were prepared as previously described (36) with the GeneExtract instrument (GeneSystems, France), which includes a DNA extraction system that is able to handle five water samples and one negative control simultaneously. After purification (GeneSystems, France), DNA was eluted in a final volume of 300 μl of elution buffer. Quantitative PCR was performed with the GeneDisc-Cycler apparatus (GeneSystems, France) and the GeneDisc Legionella pneumophila kit, a ready-to-use molecular biology device. GeneDisc software uses the threshold cycle and the positive-control fluorescence value to detect PCR inhibitors. For each sample, the GeneDisc-Cycler indicates the final result as the number of genome units per liter. For each batch of GeneDiscs, linearity was confirmed by constructing an external five-point standard curve ranging from 25 to 2.5 × 105 GU of L. pneumophila serogroup 1 (ATCC 33152) per well.
Quantification of viable bacteria by solid-phase cytometry.
After exposure to chlorine, the samples and untreated controls were serially diluted to concentrations from 107 to 102 Legionella organisms/ml, and 100 μl of the 104- to 102-cells/ml dilutions was filtered under a maximum vacuum of 100 mm Hg through labeling membranes (ChemFilter CB 04; AES-Chemunex, Ivry-sur-Seine, France). The viability of L. pneumophila was analyzed as previously described (8), using the ChemChrome V6 probe and the TVC (total viable cell) Bioburden kit (AES-Chemunex, Ivry-sur-Seine, France). Briefly, this method is based on a nonfluorescent precursor, which is internalized and cleaved into a green fluorescent product (emission, 520 nm) by esterases present in viable bacteria. The cells remain fluorescent only if their membranes are intact and the probe is unable to diffuse out (8). All fluorescent events detected, including background signals, were counted by the cytometer and were then visually discriminated under a microscope (Olympus BX41) equipped with a motorized stage.
Quantification of living and intact dead L. pneumophila organisms by solid-phase cytometry.
Living and intact dead L. pneumophila organisms were counted with the “Detection of Legionella pneumophila” kit (AES-Chemunex, Ivry-sur-Seine, France) according to the manufacturer's instructions. Briefly, samples and controls were serially diluted from 107 to 102 Legionella organisms/ml, and 100 μl of the 104- to 102-cells/ml dilutions was filtered under a maximum vacuum of 100 mm Hg through labeling membranes (ChemFilter CB 04; AES-Chemunex, Ivry-sur-Seine, France). After filtration, the membranes were incubated in a petri dish for 1 h at 30°C in the dark, on a 100-μl drop of the primary antibody solution (ChemId LpA; Chemunex). The membranes were then washed on a support pad soaked with 500 μl of B32 solution. The membranes were then saturated with 500 μl of CSE/2 solution, vacuum filtered, and incubated in a petri dish for 30 min at 30°C in the dark on a 100-μl drop of secondary-antibody solution (ChemId LpB; Chemunex). The membranes were placed on the holder (support pad; Chemunex) presoaked with 100 μl of mounting medium, and scanned with a ChemScan RDI according to the manufacturer's instructions. All fluorescent events were manually discriminated under a microscope (Olympus BX41) with a motorized stage.
Direct resuscitation of chlorinated VBNC legionellae.
One million L. pneumophila organisms in chlorinated and untreated control samples were inoculated directly into 3 ml of Legionella growth medium (LGM) in order to determine if treated and nongrowing VBNC legionellae would be able to replicate in liquid medium. After 5 days in an orbital-shaker incubator, 100 μl of each culture was plated onto BCYE agar and blood agar plates (negative control).
Infection of protozoan cells.
Axenic cultures of Acanthamoeba polyphaga strain Linc AP-1 were prepared as adherent cells in 10 ml of PYG broth [2% proteose peptone no. 3, 0.1% yeast extract, 0.1 M glucose, 4 mM MgSO4, 0.4 M CaCl2, 0.1% sodium citrate, 0.05 M Fe(NH4)2-6H2O, 2.5 mM NaH2PO3, 2.5 mM K2HPO3 (pH 6.5)] at 30°C for 10 days. Monolayers were prepared in 24-well tissue culture plates (Corning Costar Corporation, Cambridge, MA) using Pages' amoebal saline (PAS) buffer as previously described (16). L. pneumophila was grown for 72 h at 37°C on BCYE plates and was subsequently resuspended in PAS buffer for amoebal infection, as described below. The concentration of viable L. pneumophila bacteria was determined (nearly 106 CFU/ml for untreated bacteria and 107 CFU/ml for treated bacteria). Control wells containing PAS buffer without A. polyphaga were included. A. polyphaga, at 105 CFU per well in 24-well plates, was infected with 106 CFU (multiplicity of infection, 10) of viable L. pneumophila (as shown by TVC measurements). The plates were centrifuged at 500 × g for 5 min to improve cell-cell contact and were incubated for 1 h at 37°C. At the end of the infection period, the monolayers were washed three times with PAS buffer to remove nonadherent cells and then were incubated for 5 days. Supernatants and amoebal cell lysates prepared with 0.05% Triton X-100 (10, 11) were combined, and aliquots were plated onto BCYE agar plates for L. pneumophila colony counting. Other aliquots were used for real-time PCR and immunodetection. A Nikon TMS inverted microscope was used to count the cells for the infection and to observe the state of the amoebae.
RESULTS
Quantification of L. pneumophila cells and DNA.
Water samples spiked with 106 Legionella CFU/ml yielded concentrations of 1.8 × 106 ± 0.86 × 105 CFU/ml after growth on BCYE medium, 0.91 × 106 ± 1.37 × 105 GU/ml by GeneSystems PCR, 2.7 × 106 ± 2.6 × 105 viable L. pneumophila organisms/ml with the TVC Bioburden kit, and 3.39 × 106 ± 1.6 × 105 intact L. pneumophila cells by immunodetection. If we consider that each unit used represents an entity of Legionella/ml, the results of the four techniques show similar concentrations of these Legionella entities in untreated water and are therefore comparable (data not shown).
Quantification of bacteria and DNA after addition of chlorine.
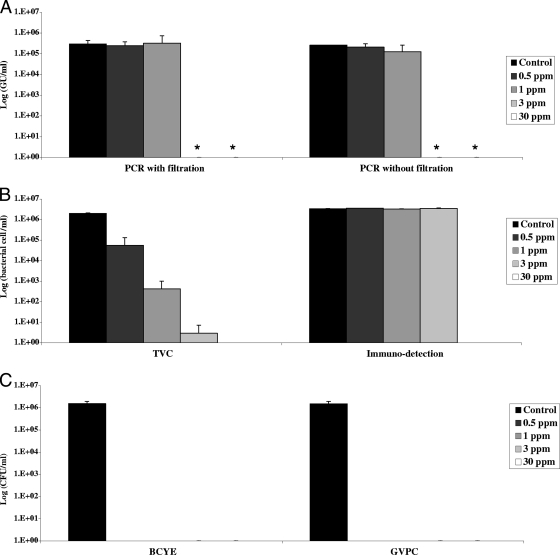
After 24 h of incubation with 0.5, 1, 3, or 30 ppm chlorine, all L. pneumophila cultures were negative (Fig. 1). PCR results were not modified by 0.5 or 1 ppm chlorine, but the signals were below the quantification limit with 3 or 30 ppm chlorine. PCR results were similar before and after filtration. TVCs fell proportionally with the chlorine concentration. The immunodetection results were not affected by 0.5, 1, or 3 ppm chlorine, but the signal was totally abolished by 30 ppm chlorine. Chlorine thus affected bacterial viability and, more importantly, cultivability, while it had a lesser effect on PCR detection of bacterial DNA and immunodetection of dead and viable bacterial cells.
FIG. 1.
Effects of increasing chlorine concentrations on bacterial quantification. For the control sample, water containing 106 legionellae was serially diluted and analyzed by the four methods described in Materials and Methods. Water containing 107 legionellae/ml was treated for 24 h with various concentrations of chorine. After neutralization, serial dilutions were quantified by the techniques described in Materials and Methods. Results are expressed in GU/ml for real-time PCR (A), in cells/ml for the solid-phase cytometry techniques (B), and in CFU/ml for culture on BCYE agar (C). Values are means ± standard deviations from two independent experiments. *, value below the quantification limit.
Kinetics of the effect of 0.5 ppm chlorine on the quantification of bacteria and DNA.
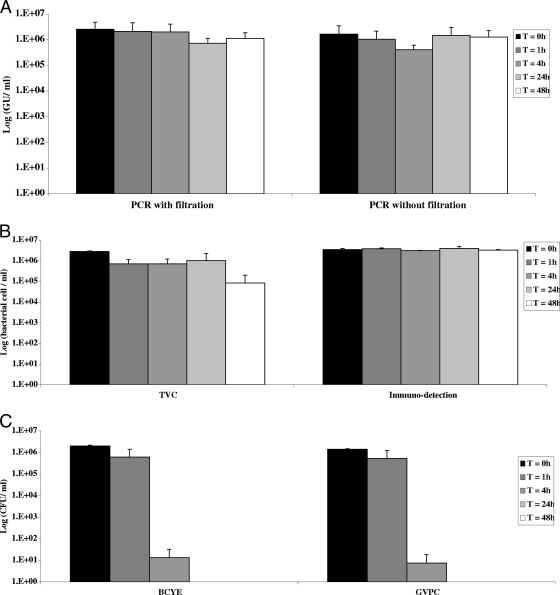
The effect of 0.5 ppm chlorine was measured after 1, 4, 24, and 48 h of incubation (Fig. 2). BCYE and GVPC culture results fell fivefold after 4 h of treatment, and no growth was observed after longer treatment periods. The PCR signals remained constant over time, while the TVC signals fell only after 48 h of treatment, by 1 log unit. Immunodetection results remained stable over time. Thus, the time interval between the addition of chlorine and the detection of bacteria in water samples did not affect quantification by PCR or immunodetection, but it did affect bacterial viability and culturability after 48 h and a few hours of treatment, respectively.
FIG. 2.
Kinetics of the effect of 0.5 ppm chlorine. Water containing 107 legionellae/ml was treated for 1 to 48 h with 0.5 ppm chlorine. After neutralization, serial dilutions were quantified by the techniques described in Materials and Methods. Results are expressed as described in the legend to Fig. 1. Values are means ± standard deviations from two independent experiments. T, time.
Production and detection of VBNC bacteria.
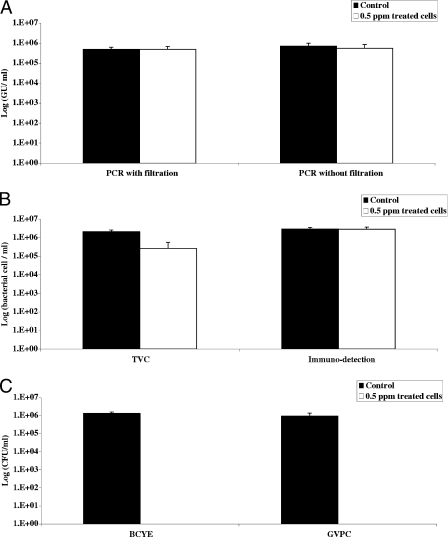
Since these results suggested that PCR and immunodetection detected total bacteria, including viable bacteria (as measured by TVC) and noncultivable bacteria (quantified by growth on media), we wondered whether similar results would be obtained using VBNC bacteria artificially prepared with L. pneumophila Paris. We first determined the best conditions under which to obtain VBNC bacteria, which correspond to a positive TVC signal and no growth on media. These conditions were obtained with 0.5 ppm chlorine after 24 h of incubation (Fig. 3). Under these conditions, a 1-log-unit decrease in the TVC signal was observed, whereas the PCR and immunodetection results did not change. No growth on BCYE or GVPC medium was detected. The experiment was repeated three times for the control and treated samples and was reproducible. It was therefore possible to obtain VBNC bacteria from L. pneumophila Paris strain after 24 h of incubation in the presence of 0.5 ppm chlorine.
FIG. 3.
Preparation of VBNC. The graph shows the reproducibility of the chlorination (0.5 ppm) procedure. Legionellae in water were quantified by the techniques described in Materials and Methods. The results are expressed as described in the legend to Fig. 1.
Restoration of the culturability of VBNC legionellae.
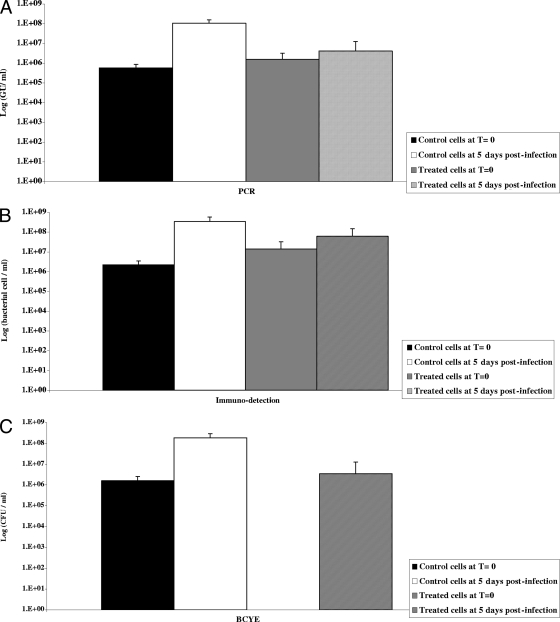
VBNC legionellae obtained as described above were tested for their capacity to infect A. polyphaga. For this purpose, 106 viable L. pneumophila organisms were inoculated onto 105 A. polyphaga amoebae per well in 24-well plates. After 5 days of incubation at 30°C, microscopic examination showed numerous amoebae containing motile and spinning L. pneumophila bacteria. The detection signal of non-chlorine-treated bacteria increased by 2 log units as determined by PCR, immunodetection, and BCYE culture after 5 days of coculture with A. polyphaga. However, for chlorine-treated samples, the PCR and immunodetection signals increased by 0.5 log unit during the incubation period (Fig. 4), and the bacteria finally regained their culturability, yielding 106 L. pneumophila CFU/ml after 5 days of incubation. This recovery of culturability was not detected when amoebae were omitted from the wells (data not shown). No cultivable legionellae were detected when chlorinated bacteria were inoculated directly into LGM for 5 days. Together, these results showed that L. pneumophila PCR results include VBNC forms.
FIG. 4.
Restoration of the culturability of VBNC bacteria. Legionella suspensions were analyzed for living and metabolically active cells by using the TVC labeling kit as described in Materials and Methods. The volume of each cell suspension containing 106 CFU/ml viable L. pneumophila was determined. A. polyphaga (105 CFU per well in 24-well plates) was infected with 106 CFU/ml (multiplicity of infection, 10) of viable L. pneumophila (measured by TVC). After 1 h of incubation at 37°C, the monolayers were washed, incubated for 5 days, and then lysed with 0.05% Triton X-100. Aliquots were plated on BCYE agar for colony counting and were quantified by real-time PCR and immunodetection. Legionella viability could not be measured on the samples, because viable amoebae could be labeled at the same time, making the reading difficult. Control chlorinated bacteria (treated cells) incubated in PAS buffer without A. polyphaga remained culture negative. The results are expressed as described in the legend to Fig. 1. T, time.
DISCUSSION
To our knowledge, this is the first study to compare the results of PCR, solid-phase cytometry, and standard methods for quantifying L. pneumophila in graduated water samples. We confirm the large discrepancies between PCR and culture for L. pneumophila quantification in treated water samples (2, 3, 7, 14, 23-25, 34-36). We show that L. pneumophila DNA detected by PCR in water samples treated with chlorine for 24 h includes VBNC forms and dead bacteria. Indeed, culture was negative after the addition of 0.5 ppm chlorine for 24 h, whereas the PCR signal was preserved and the number of metabolically active bacteria (detected by TVC) was barely affected. With 1 ppm chlorine, PCR and immunodetection signals did not change but TVC values were halved. In the presence of 3 ppm chlorine, fewer than 10 bacteria were labeled by TVC, whereas the PCR signal was below the quantification limit and the immunodetection results were unaffected, suggesting that dead bacteria were detected by antibodies. All signals were below the quantification limit at 30 ppm chlorine. Kinetic studies of the effect of 0.5 ppm chlorine also showed that immunodetection and PCR efficiently detected both dead and viable bacteria, whereas culture and viability testing were both affected after, respectively, a few hours and 2 days of treatment. Viable bacteria detected by TVC labeling remained infective, since they were able to multiply in A. polyphaga and recovered their culturability.
Chlorine is the most widely used disinfectant for drinking water, cooling towers, and wastewater. In France and elsewhere in Europe, concentrations of 1 to 2 (sometimes 3) ppm chlorine are used for continuous treatment, in order to obtain around 1 ppm at the point of use (DGS circular 2002/243 [22 April 2002]; 9a). “Shock” treatment of drinking water networks uses 10 to 20 ppm chlorine, followed by washes. As much as 50 ppm chlorine can be used to decontaminate cooling towers. Although in natural environments, temperature, pH, and organic matter can affect the availability of the active form of chlorine and modify the resistance of the bacteria to chlorination (9, 29, 33), here we used experimental concentrations compatible with those used for continuous treatment of drinking-water networks and for shock decontamination.
Chlorine treatment markedly influenced the results of the methods used here to quantify L. pneumophila organisms. The concentration of 0.5 ppm chlorine abolished culturability (changing bacteria into VBNC forms) and reduced viability after 24 h. Culture was the least sensitive detection and quantification technique. The PCR method was less sensitive (detection limit, 170 GU/liter, according to the manufacturer's instructions) than immunodetection and TVC, which can detect 1 stained bacterium per filtered sample (17). PCR is therefore less precise than solid-phase cytometry when the number of bacteria with amplifiable DNA is too low or the bacteria are unsuitable for any efficient DNA extraction and/or amplification due to their poor quality. These results imply that a small number of bacteria, probably including some viable cells, were present in some PCR-negative samples treated with 3 ppm chlorine. However, the discrepancy between the TVC signal (viable bacteria) and the immunodetection signal (intact bacteria) at the same chlorine concentration was even larger, suggesting that at 3 ppm chlorine, immunodetection detects mainly dead bacteria compared to TVC and PCR techniques. It has been shown that the bacterial membrane can be permeabilized by chlorine in distilled water (28, 33). As with other permeabilizing agents, such as Triton, antibodies can still detect the bacterial membrane or even potentially functional intracellular enzymes (5, 13, 20, 22, 32). However, this permeabilization process, when used for prolonged periods of time, could allow the bacterial contents, including DNA, to escape or could allow chlorine to damage bacterial DNA. Indeed, HOCl, the more reactive form of chlorine in aqueous environments, causes lethal DNA damage even at the concentrations used in drinking water (9, 28). Bacterial “ghost” cells have been described as stable immunogenic bacterial cell envelopes that have been emptied of their cytoplasmic contents through a lysis pore created by expression of the cloned phage ϕX174 lysis gene E. Thus, an intact bacterial envelope does not necessarily imply bacterial viability or the presence of amplifiable DNA, as shown by the PCR, TVC, and immunodetection results in the presence of 3 ppm chlorine. Altogether, chlorine could act in the following sequence: (i) culturability is lost due to degradation of surface components (as in the presence of 0.5 ppm chlorine); (ii) chlorine gradually penetrates the cell and directly degrades intracellular esterases and/or allows the release of the fluorochrome after pore formation, thus explaining the gradual fall in the TVC signal with increasing concentrations of chlorine; (iii) chlorine degrades the bacterial DNA, leading to extinction of the PCR signal (as in the presence of 3 ppm chlorine); and (iv) chlorine abolishes immunodetection signals by destroying the physical integrity of the bacterial cell (as in the presence of 30 ppm chlorine). Taken together, these data show that PCR signals include viable bacteria, detected by TVC, and some dead bacteria. In contrast with PCR, whose signal is negative when the number of viable bacteria is null or very low, immunodetection detects mainly dead bacteria at 1 and 3 ppm chlorine.
L. pneumophila enters the VBNC state in order to survive in starvation environments (4, 30). VBNC legionellae have been detected in natural water samples (8). Chlorination results in the complete loss of culturability of both starved and nonstarved L. pneumophila organisms (12). Heat treatment also abolishes culturability without affecting bacterial integrity. Steinert et al. were the first to demonstrate that VBNC legionellae could be resuscitated by adding an amoeba, Acanthamoeba castellani (30), and this finding was subsequently confirmed by other authors with VBNC legionellae obtained after starvation, heat, or treatment with chlorine and other biocides (4, 12). TVC with ChemChrome V6 detects intracellular esterase activity in various bacterial species, including L. pneumophila (6, 8, 26). Nonspecific viability labeling methods such as TVC staining must be combined with a specific identification technique. Delgado-Viscogliosi et al. used anti-Legionella antibodies and ChemChrome V6 staining with epifluorescence microscopy to detect total and viable L. pneumophila bacteria in natural water samples. They also found that chlorine abolished viability staining without affecting antibody reactivity (8).
In conclusion, our findings suggest that PCR results include viable (TVC-positive) but noncultivable bacteria. These viable bacteria were able to multiply in A. polyphaga and to recover their culturability. This explains the large discrepancies between PCR and culture for L. pneumophila quantification in chlorine-treated water samples (2, 3, 7, 14, 23-25, 34-36). Different environmental conditions and disinfection processes could have different effects on VBNC generation. Our results also suggest that dead bacteria were detected by PCR at 0.5 and 1 ppm chlorine in water together with VBNC forms, and not at higher concentrations (≥3 ppm chlorine), as PCR signals were not observed. Since the application domain of the viability and immunodetection quantification tools is still limited to filterable water, research should also be done to develop similar methods that can be applied to nonfilterable environmental or clinical samples.
Acknowledgments
We thank EDF, in particular the Medical Studies Department, for lending us the Chemscan (AES Chemunex).
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Albert-Weissenberger, C., C. Cazalet, and C. Buchrieser. 2007. Legionella pneumophila—a human pathogen that co-evolved with fresh water protozoa. Cell. Mol. Life Sci. 64:432-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behets, J., P. Declerck, Y. Delaedt, B. Creemers, and F. Ollevier. 2007. Development and evaluation of a TaqMan duplex real-time PCR quantification method for reliable enumeration of Legionella pneumophila in water samples. J. Microbiol. Methods 68:137-144. [DOI] [PubMed] [Google Scholar]
- 3.Catalan, V., F. Garcia, C. Moreno, M. J. Vila, and D. Apraiz. 1997. Detection of Legionella pneumophila in wastewater by nested polymerase chain reaction. Res. Microbiol. 148:71-78. [DOI] [PubMed] [Google Scholar]
- 4.Chang, C. W., Y. H. Hwang, W. Y. Cheng, and C. P. Chang. 2007. Effects of chlorination and heat disinfection on long-term starved Legionella pneumophila in warm water. J. Appl. Microbiol. 102:1636-1644. [DOI] [PubMed] [Google Scholar]
- 5.Cimino, M., L. Alamo, and L. Salazar. 2006. Permeabilization of the mycobacterial envelope for protein cytolocalization studies by immunofluorescence microscopy. BMC Microbiol. 6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cools, I., E. D'Haese, M. Uyttendaele, E. Storms, H. J. Nelis, and J. Debevere. 2005. Solid phase cytometry as a tool to detect viable but non-culturable cells of Campylobacter jejuni. J. Microbiol. Methods 63:107-114. [DOI] [PubMed] [Google Scholar]
- 7.Declerck, P., J. Behets, E. Lammertyn, I. Lebeau, J. Anne, and F. Ollevier. 2006. Detection and quantification of Legionella pneumophila in water samples using competitive PCR. Can. J. Microbiol. 52:584-590. [DOI] [PubMed] [Google Scholar]
- 8.Delgado-Viscogliosi, P., T. Simonart, V. Parent, G. Marchand, M. Dobbelaere, E. Pierlot, V. Pierzo, F. Menard-Szczebara, E. Gaudard- Ferveur, K. Delabre, and J. M. Delattre. 2005. Rapid method for enumeration of viable Legionella pneumophila and other Legionella spp. in water. Appl. Environ. Microbiol. 71:4086-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dukan, S., and D. Touati. 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 178:6145-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.European Working Group for Legionella Infections. January 2005. European guidelines for control and prevention of travel associated Legionnaires' disease, supplement 1B. Treatment methods. http://www.ewgli.org/data/european_guidelines/eg_supplement1b.pdf.
- 10.Gao, L. Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect. Immun. 65:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, L. Y., O. S. Harb, and Y. Abu Kwaik. 1998. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect. Immun. 66:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia, M. T., S. Jones, C. Pelaz, R. D. Millar, and Y. Abu Kwaik. 2007. Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ. Microbiol. 9:1267-1277. [DOI] [PubMed] [Google Scholar]
- 13.Ghrebi, S. S., G. R. Owen, and D. M. Brunette. 2007. Triton X-100 pretreatment of LR-white thin sections improves immunofluorescence specificity and intensity. Microsc. Res. Tech. 70:555-562. [DOI] [PubMed] [Google Scholar]
- 14.Joly, P., P. A. Falconnet, J. Andre, N. Weill, M. Reyrolle, F. Vandenesch, M. Maurin, J. Etienne, and S. Jarraud. 2006. Quantitative real-time Legionella PCR for environmental water samples: data interpretation. Appl. Environ. Microbiol. 72:2801-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jules, M., and C. Buchrieser. 2007. Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics. FEBS Lett. 581:2829-2838. [DOI] [PubMed] [Google Scholar]
- 16.La Scola, B., G. Michel, and D. Raoult. 1999. Isolation of Legionella pneumophila by centrifugation of shell vial cell cultures from multiple liver and lung abscesses. J. Clin. Microbiol. 37:785-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemarchand, K., N. Parthuisot, P. Catala, and P. Lebaron. 2001. Comparative assessment of epifluorescence microscopy, flow cytometry and solid-phase cytometry used in the enumeration of specific bacteria in water. Aquat. Microb. Ecol. 25:301-309. [Google Scholar]
- 18.Leoni, E., and P. P. Legnani. 2001. Comparison of selective procedures for isolation and enumeration of Legionella species from hot water systems. J. Appl. Microbiol. 90:27-33. [DOI] [PubMed] [Google Scholar]
- 19.Leoni, E., P. P. Legnani, M. A. Bucci Sabattini, and F. Righi. 2001. Prevalence of Legionella spp. in swimming pool environment. Water Res. 35:3749-3753. [DOI] [PubMed] [Google Scholar]
- 20.Masson, C., C. Bouniol, N. Fomproix, M. S. Szollosi, P. Debey, and D. Hernandez-Verdun. 1996. Conditions favoring RNA polymerase I transcription in permeabilized cells. Exp. Cell Res. 226:114-125. [DOI] [PubMed] [Google Scholar]
- 21.Meenhorst, P. L., A. L. Reingold, D. G. Groothuis, G. W. Gorman, H. W. Wilkinson, R. M. McKinney, J. C. Feeley, D. J. Brenner, and R. van Furth. 1985. Water-related nosocomial pneumonia caused by Legionella pneumophila serogroups 1 and 10. J. Infect. Dis. 152:356-364. [DOI] [PubMed] [Google Scholar]
- 22.Molmeret, M., M. Santic, R. Asare, R. A. Carabeo, and Y. Abu Kwaik. 2007. Rapid escape of the dot/icm mutants of Legionella pneumophila into the cytosol of mammalian and protozoan cells. Infect. Immun. 75:3290-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morio, F., S. Corvec, N. Caroff, F. Le Gallou, H. Drugeon, and A. Reynaud. 2007. Real-time PCR assay for the detection and quantification of Legionella pneumophila in environmental water samples: utility for daily practice. Int. J. Hyg. Environ. Health. doi: 10.1016/j.ijheh.2007.06.002. [DOI] [PubMed]
- 24.Ng, D. L., B. B. Koh, L. Tay, and B. H. Heng. 1997. Comparison of polymerase chain reaction and conventional culture for the detection of legionellae in cooling tower waters in Singapore. Lett. Appl. Microbiol. 24:214-216. [DOI] [PubMed] [Google Scholar]
- 25.Nintasen, R., F. Utrarachkij, K. Siripanichgon, A. Bhumiratana, Y. Suzuki, and O. Suthienkul. 2007. Enhancement of Legionella pneumophila culture isolation from microenvironments by macrophage infectivity potentiator (mip) gene-specific nested polymerase chain reaction. Microbiol. Immunol. 51:777-785. [DOI] [PubMed] [Google Scholar]
- 26.Parthuisot, N., P. Catala, K. Lemarchand, J. Baudart, and P. Lebaron. 2000. Evaluation of ChemChrome V6 for bacterial viability assessment in waters. J. Appl. Microbiol. 89:370-380. [DOI] [PubMed] [Google Scholar]
- 27.Patterson, W. J., D. V. Seal, E. Curran, T. M. Sinclair, and J. C. McLuckie. 1994. Fatal nosocomial Legionnaires' disease: relevance of contamination of hospital water supply by temperature-dependent buoyancy-driven flow from spur pipes. Epidemiol. Infect. 112:513-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phe, M. H., M. Dossot, H. Guilloteau, and J. C. Block. 2005. Nucleic acid fluorochromes and flow cytometry prove useful in assessing the effect of chlorination on drinking water bacteria. Water Res. 39:3618-3628. [DOI] [PubMed] [Google Scholar]
- 29.Shang, C., and E. R. Blatchley III. 2001. Chlorination of pure bacterial cultures in aqueous solution. Water Res. 35:244-254. [DOI] [PubMed] [Google Scholar]
- 30.Steinert, M., L. Emody, R. Amann, and J. Hacker. 1997. Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl. Environ. Microbiol. 63:2047-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinert, M., K. Heuner, C. Buchrieser, C. Albert-Weissenberger, and G. Glockner. 2007. Legionella pathogenicity: genome structure, regulatory networks and the host cell response. Int. J. Med. Microbiol. 297:577-587. [DOI] [PubMed] [Google Scholar]
- 32.Trejo-Tapia, G., J. Cuevas-Celis, G. Salcedo-Morales, J. L. Trejo-Espino, M. L. Arenas-Ocampo, and A. Jimenez-Aparicio. 2007. Beta vulgaris L. suspension cultures permeabilized with Triton X-100 retain cell viability and betacyanines production ability: a digital image analysis study. Biotechnol. Prog. 23:359-363. [DOI] [PubMed] [Google Scholar]
- 33.Virto, R., P. Manas, I. Alvarez, S. Condon, and J. Raso. 2005. Membrane damage and microbial inactivation by chlorine in the absence and presence of a chlorine-demanding substrate. Appl. Environ. Microbiol. 71:5022-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanez, M. A., C. Carrasco-Serrano, V. M. Barbera, and V. Catalan. 2005. Quantitative detection of Legionella pneumophila in water samples by immunomagnetic purification and real-time PCR amplification of the dotA gene. Appl. Environ. Microbiol. 71:3433-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaradou, D. F., S. Hallier-Soulier, S. Moreau, F. Poty, Y. Hillion, M. Reyrolle, J. Andre, G. Festoc, K. Delabre, F. Vandenesch, J. Etienne, and S. Jarraud. 2007. Integrated real-time PCR for detection and monitoring of Legionella pneumophila in water systems. Appl. Environ. Microbiol. 73:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]