Abstract
Shiga toxin 2e (Stx2e)-producing strains from food (n = 36), slaughtered pigs (n = 25), the environment (n = 21), diseased pigs (n = 19), and humans (n = 9) were investigated for production of Stx2e by enzyme-linked immunosorbent assay, for virulence markers by PCR, and for their serotypes to evaluate their role as potential human pathogens. Stx2e production was low in 64% of all 110 strains. Stx2e production was inducible by mitomycin C but differed considerably between strains. Analysis by nucleotide sequencing and transcription of stx2e genes in high- and low-Stx2e-producing strains showed that toxin production correlated with transcription rates of stx2e genes. DNA sequences specific for the int, Q, dam, and S genes of the stx2e bacteriophage P27 were found in 109 strains, indicating cryptic P27-like prophages, although 102 of these were not complete for all genes tested. Genes encoding intimin (eae), enterohemorrhagic Escherichia coli hemolysin (ehx), or other stx1 or stx2 variants were not found, whereas genes for heat-stable enterotoxins STI, STII, or EAST1 were present in 54.5% of the strains. Seven major serotypes that were associated with diseased pigs (O138:H14, O139:H1, and O141:H4) or with slaughter pigs, food, and the environment (O8:H4, O8:H9, O100:H30, and O101:H9) accounted for 60% of all Stx2e strains. The human Stx2e isolates did not belong to these major serotypes of Stx2e strains, and high production of Stx2e in human strains was not related to diarrheal disease. The results from this study and other studies do not point to Stx2e as a pathogenicity factor for diarrhea and hemolytic uremic syndrome in humans.
Production of Shiga toxins (Stx), or Vero toxins, is associated with certain Escherichia coli strains designated Shiga toxin-producing E. coli (STEC) strains or Vero toxin-producing E. coli strains. STEC is commonly found in the intestines of ruminant animals such as cattle, sheep, and goats (11). In these domestic animals, STEC normally does not cause disease, and some STEC types were shown to be closely associated with certain animal host species and populations (9, 10, 27, 45). The situation is different in monogastric animals such as pigs. These do not harbor STEC as part of their normal gut flora but may occasionally excrete these bacteria with their feces (16, 54).
In contrast to the case in ruminant animals, STEC plays an important role as pathogens in humans and in pigs. Humans can be infected by the oral route through ingestion of food containing STEC, by contact with an STEC-contaminated environment, or by direct transmission of STEC from infected animals or humans (19). In the human host, STEC may cause diarrhea, and certain STEC strains, designated enterohemorrhagic E. coli (EHEC), cause life-threatening hemorrhagic colitis and hemolytic uremic syndrome (HUS) (47). In weaned pigs, STEC plays an important role as agents of edema disease, which is characterized by high contagiousness, neurological disorders, hemorrhagic lesions, and often fatal outcome (1, 19).
Analysis of Shiga toxins produced by different strains of E. coli revealed two toxin families, called Stx1 and Stx2, which are genetically and immunologically distinct from each other (35). Toxins of the Stx1 and Stx2 families can be divided further into subtypes differing in the composition of their A or B subunit (40). Studies on human virulent EHEC have revealed that certain toxin types, such as Stx2, Stx2c, and activatable Stx2d, are significantly associated with higher virulence of the bacteria for humans, which may cause severe diseases such as hemorrhagic colitis and HUS (4, 7, 17, 22, 36). In pigs, the toxin type Stx2e was identified as a key factor contributing to the pathogenicity of porcine STEC and therefore was designated the edema disease principle (1). In contrast, Stx2e-producing STEC bacteria are rarely isolated from humans and account for only 0.9 to 1.7% of human STEC isolates (4, 42). A significant association between Stx2e and human diarrhea was not found, and it was suggested that these STEC bacteria are not important as human pathogens (4, 17, 19, 42).
The Shiga toxin variant 2e is produced by STEC isolated from various sources, such as animals, food, the environment, and humans. Stx2e-producing STEC strains belong to different serotypes, and the presence of Stx2e and other virulence attributes was found in a variety of chromosomal backgrounds (12, 33). The occurrence of the stx2e gene in genetically distinct types of E. coli could indicate that these are spread by bacteriophages, similar to what was found for other genes of the Stx1 and Stx2 families (21). A lambdoid phage called P27 carrying an stx2e gene was recently isolated from STEC originating from a human patient, but inducible stx-carrying phages were not found in other types of Stx2e-producing strains (31, 39). Stx2e-producing strains from humans were shown to differ from porcine pathogenic Stx2e-producing strains in their serotypes and adhesion to epithelial cell lines from humans and pigs (42). Additionally, none of the virulence factors hitherto described for this group of STEC, including production of Stx2e, could be clearly associated with diarrheal disease in humans (15, 42).
However, humans are likely to have contact with Stx2e-producing STEC, as these strains were shown to constitute about 19% of the STEC isolated from meat and milk products in Germany (5). Stx2e-producing strains also frequently occur in pigs (16), in the environment of pig farms (46), and as contaminants of wastewater from abattoirs (26). The low incidence of Stx2e strains in human patients could indicate that most of these strains cannot colonize or may infect but do not cause disease in humans. This is supported by epidemiological findings indicating that Stx2e strains are very rarely isolated from humans, and those patients show mild diarrheal symptoms or are asymptomatic carriers (4, 17, 39, 42). In a previous study we have found that Stx2e-producing strains differ largely in the amount of Stx2e produced (2). In this study, we investigated whether the level of Stx expression is related to serotypes and sources of the Stx2e strains and serves as an indicator for human pathogenicity. We compared Stx2e-producing STEC strains differing in their origin, virulence attributes, and geno- and phenotypes to evaluate the public health impact of this particular group of STEC, which accounts for a major part of STEC isolated from food in Germany.
MATERIALS AND METHODS
Bacteria.
A total of 110 Stx2e-producing E. coli strains from the collection of the NRL-E. coli were investigated. The strains were isolated from human feces (n = 9; six patients with uncomplicated diarrhea and three asymptomatic individuals), the environment (n = 21; seven strains from pig farms and 14 from river water), food (n = 36; 33 strains from meat and meat products and 3 from milk), feces from pigs at slaughter (n = 25), and feces or organ samples from pigs with edema disease or diarrhea (n = 19) between 1997 and 2005. The origins of some strains from human patients (4, 15), food samples (5), pig farm environments (46), and diseased pigs (50) have been described elsewhere. Detection and characterization of Shiga toxin genes (stx) in the strains was performed by PCR following previously published protocols (5). Serotyping of O:H antigens and molecular typing of the flagellar (fliC) genes were performed as described previously (5). All strains were investigated for cytotoxicity with the Vero cell toxicity test and for adherence to HEp-2 cells as previously described (4, 42).
The E. coli reference strains used as positive controls for PCR detection of virulence markers investigated in this study were used as described previously (3, 20, 50). The human fecal E. coli strain 2771/97 (ONT:NM), which carries the Stx2e-encoding bacteriophage P27, was used as positive control for detection of P27-associated genes (31, 39). The properties of representative Stx2e-producing strains that were investigated for stx2e and associated gene sequences are summarized in Table 1.
TABLE 1.
Origins and properties of STEC strains investigated for stx2e and associated gene sequences
| Strain | Serotype | Sourcea | Stx2e productionb | Virulence marker(s)c | GenBank accession no. |
|---|---|---|---|---|---|
| CB10282 | O8:H9 | Milk | Low | — | AM937001 |
| CB10284 | O8:H9 | Water | Low | — | AM939641 |
| CB10394 | O101:H9 | HF, D | High | — | AM939642 |
| CB10402 | O100:[H30] | Pork | High | STI-p | AM939643 |
| CB8771 | O159:H21 | PF | High | DA, STI-p, STII | AM940005 |
| CB7671 | ONT:H19 | HF, As | High | DA | AM940007 |
| CB8770 | ONT:H21 | PF | High | — | AM940006 |
| CB8810 | O8:H9 | PF | High | DA | AM940004 |
| 2771/97d | ONT:NM | HF, D | High | ND | AJ298298 |
HF, human feces; D, diarrhea; PF, fecal samples from pigs at slaughter; As, asymptomatic.
P1-g-EIA results for strains grown in presence of mitomycin C.
—, negative for virulence markers investigated here; STI-p, heat-stable enterotoxin STI (animal); DA, diffuse adherence to HEp-2 cells; STII, heat-stable enterotoxin STII; ND, not done.
Reference strain carrying the stx2e bacteriophage P27 (39).
Production of Shiga toxins.
All strains were tested for cytotoxic activity in the Vero cell test, which was performed as described previously (2). The strains were investigated quantitatively for production of Stx1 and Stx2 by the P1-glycoprotein receptor enzyme immunoassay (P1-g-EIA) (2). This assay is based on binding of the Stx B subunit to the P1-glycoprotein receptor. Inducible production of Stx was measured by comparing P1-g-EIA results obtained with bacteria grown in tryptic soy broth (TSB) to those obtained with cultures growing in TSB supplemented with 50 ng/ml mitomycin C. EIA results were recorded photometrically as described previously (2). Test results were recorded as negative (0) if the extinction was ≤0.1 above the negative control, as weakly positive (1+) if the extinction was >0.1 to 0.5 above, as moderately positive (2+) if the extinction was >0.5 to 1.0 above, and as strongly positive (3+ or 4+) if the extinction was >1.0 to 2.0 above or extinction >2.0 above. Cultures of representative strains belonging to toxicity groups 1+ to 4+ were examined by end point titration in the P1-g-EIA. Twofold dilutions of broth cultures still showing a positive result in the P1-g-EIA were as follows: 1+, 1:0 to 1:8; 2+, 1:16 to 1:64; 3+, 1:128 to 1:512; and 4+, ≥1:1,024.
Preparation of total DNA, RNA, and cDNA from bacteria.
Total DNA of bacteria was prepared from 1-ml overnight cultures (approximately 1 × 109 bacteria) with the RT Spin Bacteria DNA minikit (Invitek, Berlin, Germany). DNA preparations were stored at 4°C for use. For preparation of RNA, bacteria were grown to late-exponential phase in TSB without and with mitomycin C as described above. Total RNA was isolated from 5-ml bacterial cultures with the RNeasy minikit (Qiagen, Hilden, Germany). RNA preparations were repeatedly digested with RNase-free DNase I (Roche Applied Systems, Mannheim, Germany) for 30 min at 37°C as described by the manufacturer. Next, DNase I was inactivated by heat treatment for 10 min at 75°C, and the samples were purified on RNeasy columns following the instructions of the provider (Qiagen). The absence of DNA from RNA samples was controlled by real-time PCR (35 cycles) amplification of the icdA and the stx2e genes. The RNA preparations were stored at −20°C for use. Preparation of cDNA from the RNA samples was performed with the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) following the instructions supplied by the provider.
Detection of virulence genes in E. coli by PCR.
PCRs for detection of genes of the Stx1 and Stx2 families and for detection of the eae gene, encoding intimin, were performed as described previously (5). In addition, a number of virulence genes known to be associated with porcine pathogenic E. coli strains were investigated. PCR for specific detection of genes encoding Stx2e (stx2e), alpha-hemolysin (α-hlyA), EHEC hemolysin (ehxA), heat-labile enterotoxin LT-I, heat-stable enterotoxin STI (STa), cytolethal distending toxins (cdt), cytotoxic necrotizing factors (cnf), cell cycle inhibition factor (cif), and intimin (eae) was performed as described previously (3, 6, 50). PCR protocols for detection of genes encoding fimbrial adhesins F4 (K88), F5 (K99), F6 (987p), F18 (F107), and F41 and heat-stable enterotoxins (STIh, STIp, STII and EAST1) are listed in Table 2. PCR primers were developed with the help of Accelrys Gene v2.5 software (Accelrys Inc., Cambridge, United Kingdom).
TABLE 2.
Detection of virulence genes by PCR
| Target gene (phenotype) | Accession no. | Primer | Nucleotide sequence (5′→3′) | PCR conditions | No. of PCR cycles | PCR product (bp) | Reference |
|---|---|---|---|---|---|---|---|
| stx2e (Stx2e, B subunit) | M21534 | SLT-IIBv1 | ATGAAGAAGATGTTTATAGCG | 94.0°C, 30 s; 62.0°C, 60 s; 72.0°C, 30 s | 30 | 261 | 2 |
| SLT-IIBv2 | GTTAAACTTCACCTGGGCAAAG | ||||||
| astA (EAST1) | L11241 | EAST11a | CCATCAACACAGTATATCCGA | 95°C, 30 s; 55°C, 120 s; 72°C, 120 | 30 | 111 | 52 |
| EAST11b | GGTCGCGAGTGACGGCTTTGT | ||||||
| STII (STb) | M35586 | STb-f | CGCATTTCTTCTTGCATCTATG | 94°C, 40 s; 55°C, 40 s; 72°C, 40 s | 30 | 189 | This work |
| STb-r | TGCTGCAACCATTATTTGGG | ||||||
| STI-h (human) | M34916 | STIh-f | TCCCTCAGGATGCTAAAC | 94°C, 40 s; 47°C, 60 s; 72°C, 40 s | 30 | 244 | 6 |
| ST1h-b | GCAACAGGTACATACGTT | ||||||
| STI-p (animal) | M25607 | STIp-f | TTCTGTATTATCTTTCCCC | 94°C, 40 s; 51.3°C, 40 s; 72°C, 40 s | 30 | 258 | 6 |
| ST1p-b | TTATGATTTTCTCAGCACC | ||||||
| faeG (F4 [K88]) | AJ616236 | K88-f | GGTGATTTCAATGGTTCGGTC | 94°C, 30 s; 61.8°C 60 s; 72°C 45 s | 30 | 766 | This work |
| K88-r | ATTGCTACGTTCAGCGGAGCG | ||||||
| fan (F5 [K99], fimbrial subunit) | M35282 | K99-f | GACTACCAATGCTTCTGCGA | 95.0°C, 30 s; 60.0°C 45 s; 72.0°C 45 s | 30 | 459 | This work |
| K99-b | GGTGGATATAAAGCTGGCGT | ||||||
| fasA (F6 [987p], fimbrial subunit) | M35257 | 987p-f | CTGCCAGTCTATGCCAAGTG | 94°C, 30 s; 61.8°C, 60 s; 72°C, 45 s | 30 | 459 | This work |
| 987p-b | ACGGTGTACCTGCTGAACGAATAG | ||||||
| fedA (F18 [F107], fimbrial subunit) | M61713 | F107-f | CTTTCACATTGCGTGTGGAG | 94°C, 30 s; 61.8°C, 60 s; 72°C, 45 s | 30 | 513 | This work |
| F107-r | ACCACCTTTCAGTTGAGCAG | ||||||
| Fim41a (F41, fimbrial subunit) | X14354 | F41-f | GCATCAGCGGCAGTATCT | 95.0°C, 30 s; 60.0°C 45 s; 72.0°C 45 s | 30 | 380 | 14 |
| F41-b | AGGTGATAATACTGAGCTAGGGAC |
Detection of gene sequences associated with stx2e-carrying bacteriophage P27 and insertion of P27 into the E. coli chromosome.
The presence of stx2e-carrying bacteriophage P27-associated gene sequences was investigated by PCR. PCR primers specific for the corresponding gene sequences of P27 which did not amplify related genes of other (stx-carrying) bacteriophages were selected by BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/). Strain C600 (E. coli K-12) (2) was used as a negative control and strain 2771/97 (31) as a positive control. The open reading frame labels (L01, L21, L24-25, and L26-27) are from the published P27 bacteriophage sequence (39). Primers P27-1 (5′ GTTATTTTTCTGTATCGGCTGC 3′) and P27-2 (5′ TGTTGAGTCGAAAAGTCTATCG 3′) were used for amplification of a 772-bp region flanked by the phage DNA methylase gene (L24) and the stxA2e subunit gene (L25). Primers P27-3 (5′ GAGAATACTGGACGAACAGATG 3′) and P27-4 (5′ CACTGTCAAACCCTGTGT 3′) were used to amplify the 799-bp downstream region between the stxB2e subunit gene (L26) and the S (holin) gene (L28) of P27. The amplification conditions for both PCRs were 30 cycles of 94°C for 40 s, 57.7°C for 40 s, and 72°C for 60 s.
The P27 antitermination Q gene (L21)-specific PCR (30 cycles of 94°C for 40 s, 50.5°C for 40 s, and 72°C for 60 s), yielding a 204-bp amplicon, was performed with primers P27-5 (5′ GTTTACTACTCACCGATAGCAG 3′) and P27-6 (5′ CCTTGTCTTTTGGCGATATTTC 3′). The insertion of P27 in the chromosomal E. coli yecE gene was investigated by PCR with primer P27-7 (5′ TGCGGGTCAAAAATTCAGTCAC 3′), located in the yecE gene, and primer P27-8 (5′ GGCATCGAAAGGCATACCAGTC 3′), located in the P27 integrase (L01) gene. The PCR was performed for 30 cycles (94°C for 40 s, 56.5°C for 40 s, and 72°C for 60 s), resulting in a 450-bp fragment with strains carrying the phage P27 inserted into the yecE gene. The presence of an intact yecE gene in P27 integrase-negative strains was investigated by PCR with primers P27-9 (5′ AATGGTCGCATCCTAAATGG 3′) and P27-10 (5′ GTACAGCATGTACCGGAAC 3′), resulting in a 549-bp fragment spanning the insertion site of P27 into the yecE gene (39). The PCR was performed for 30 cycles (94°C for 40 s. 56.6°C for 40 s, and 72°C for 60 sec).
Transcriptional analysis of stxA2e genes by qRT-PCR.
Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed with the Applied Biosystems 7500 real-time PCR system (Applied Biosystems) with cDNA samples from bacteria. Transcription rates of the stxA2e gene in bacteria were compared to those of the icdA housekeeping gene (51). Primers and TaqMan MGB probes were developed with ABI Prism Primer software (Applied Biosystems) and produced by Applied Biosystems (Applera Deutschland, Darmstadt, Germany). Primers icdA-F and icdA-R were used as described previously (51) together with the 6-carboxyfluorescein-labeled icdA MGB probe (5′ ACCCTGCAAAACGGCAA 3′). Primers Stx2-f (5′ CAGGCAGATACAGAGAGAATTTCG 3′) and Stx2-b (5′ CCGGCGTCATCGTATACACA 3′) were used together with the VIC-labeled Stx2-specific TaqMan MGB probe (5′ ACTGTCTGAAACTGCTC 3′) for amplification of the stxA2e gene. Real-time PCR amplifications were performed for 35 cycles in 50-μl reaction volumes using TaqMan universal PCR master mix (Applied Biosystems). Relative quantification assays were performed with dilutions of cDNA in an icdA and stx2e multiplex assay. qRT-PCR assays were analyzed with the 7500 system SDS software version 1.4 (Applied Biosystems).
Determination of the sequence of the region upstream of the stx2e gene in low- and high-toxin-producing strains.
The complete stx2e genes and adjacent DNA segments of two low (CB10282 and CB10284)- and two high (CB10394 and CB10402)-Stx2e-producing E. coli strains were analyzed for their nucleotide sequences (Table 1). A 2,385-bp DNA stretch encompassing the homologous region between the putative DNA methylase gene (L24) and the putative holin gene (L28) of stx2e bacteriophage P27 (accession no. AJ298298) was sequenced in Stx2e-producing strains CB10282 and CB10284 using primers and PCR products derived from the sequence under accession no. AJ298298. Strains CB10394 and CB10402 were found negative for sequences homologous to AJ298298 in the region upstream of the stxA2e gene as tested by PCR using primers P27-1 and P27-2 (see above). To obtain the nucleotide sequence of the DNA region upstream of the stxA2e gene in strains CB10394 and CB10402, total DNA was isolated and digested with restriction enzymes which cut only once each in different positions downstream of the stxA2e gene. Restriction-digested DNA fragments were ligated with T4 ligase and used as templates in a PCR using primer Stx2e out1 (5′ GTAACAGGCACAGTATCCAC 3′) (positions 571 to 552 in the sequence under accession no. AM939642) in combination with primer Stx2e out2 (5′ GACAACTATTTCCATGACAACG 3′) (positions 906 to 927 in the AM939642 sequence) or with stx2e out3 (5′ CAGCCATTGTTACAAAGTGC 3′) (positions 1666 to 1685 in the AM939642 sequence). A PCR product derived from amplification with these primers is generated only if the DNA sequences adjacent to the 3′ ends of the primers are joined to form a circular DNA molecule. PCR products were obtained with ligation mixtures of NciI- and PstI-digested total DNA. The PCR products were purified and analyzed for their nucleotide sequences. The nucleotide sequence data were used to generate primer Stx2e 4f (5′ AGGATAATAAGACTCTCTCGCC 3′), which was used in combination with primer P27-2 (see above) to generate a 643-bp amplicon of the region upstream of the stxA2e gene in strains CB10394 and CB10402. The same primers were also used for amplification and nucleotide sequencing of this region present in the Stx2e-producing strains CB8770, CB8771, CB8810, and CB7671. Oligonucleotides for sequencing of the stx2e gene and the region downstream of stxB2e in strains CB10394, CB10402, CB10394, and CB10402 were derived from the published sequence of the stx2e bacteriophage P27 from strain 2771/97 (accession no. AJ298298) (39).
PCR products were purified and used for sequencing by applying the dye terminator chemistry (PE Applied Biosystems, Darmstadt, Germany) and separated on an automated DNA sequencer (ABI PRISM 3100 Genetic Analyzer; Applied Biosystems, Foster City, CA). The sequences were analyzed using the Lasergene software (DNASTAR, Madison, WI) and Accelrys Gene v2.5 software.
Statistical analysis.
The H test (Kruskal-Wallis) followed by the U test (Mann-Whitney) was performed. The Holm correction method (α-correction) assesses significance assuming that each test is not independent from the other. We selected the Holm correction method to identify significant differences between the strains according to their source. We also applied the Bonferroni correction by adjusting the P value. Bonferroni corrections gave similar results.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the sequences determined in this study are listed in Table 1.
RESULTS
Presence and expression of stx genes in stx2e-positive STEC.
All 110 strains showed cytotoxic activity in the Vero cell assay. Investigation for genes encoding Shiga toxins (Stx) was performed as described previously (5). The 110 strains were all positive in a PCR specific for the stx2e gene (Table 2), and other stx genes of the Stx1 or Stx2 family were not detected. Production of Stx was measured quantitatively in the P1-g-EIA, and the bacteria were investigated for induction of Stx production by use of mitomycin C as described in Materials and Methods. The results are summarized in Table 3. When grown without induction, about one-third of the strains showed no detectable Stx and another third showed only weak (1+) production of toxin in the P1-g-EIA. The remaining strains showed moderate (2+) to high (3+ to 4+) production of Stx2e. Growing bacteria in TSB plus mitomycin C resulted in increased toxin production in all but one strain. Stx2e production was enhanced more than 1,000-fold in some strains, and 1,000-fold differences in toxin production were detected between the strains (Table 4 and unpublished data). Strains were divided in low producers (0 and 1+) and high producers (3+ and 4+) according to their Stx2e production upon mitomycin induction. High toxin producers were frequently found among strains belonging to serotypes O101:H9 (100%), O141:H4 (100%), and O100:[H30] (94.4%). Low toxin producers were more frequent among serotype O8:H4 (87.5%), O138:H14 (80%), and O8:H9 (40%) isolates (data not shown). Interestingly, Stx2e production was significantly higher in strains isolated from humans than in those isolated from diseased pigs (P < 0.001 with and without mitomycin C induction).
TABLE 3.
Relative amounts of Stx produced by stx2e strains as measured with the P1-g-EIA
| Stx productiona | No. (%) of strains grown in:
|
|
|---|---|---|
| TSB | TSB + mitomycin C | |
| Undetectable (0) | 34 (30.9) | 1 (0.9) |
| Weak (1+) | 37 (33.6) | 25 (22.7) |
| Moderate (2+) | 20 (18.2) | 9 (8.2) |
| High (3+) | 13 (11.8) | 14 (12.7) |
| High (4+) | 6 (5.5) | 61 (55.5) |
0 to 4+ correspond to the P1-g-EIA extinction rates as listed in Materials and Methods. Stx2e production was significantly higher (P < 0.01 to P < 0.001) in strains from all sources when grown in the presence of mitomycin C.
TABLE 4.
Production of Stx2e and relative quantification of stxA2e gene transcription in high- and low-Stx2e-producing strains
| Strain | TSB
|
TSB + mitomycin C
|
||
|---|---|---|---|---|
| P1-g-EIAa | Relative quantification by RT-PCR (ratio of stxA2e to icdA gene expression)b | P1-g-EIA | Relative quantification by RT-PCR (ratio of stxA2e to icdA gene expression) | |
| CB10282 | 0 (negative) | 1.0 ± 0.0 | 1+ (1:0) | 1.0 ± 0.2 |
| CB10284 | 0 (negative) | 0.16 ± 0.1 | 1+ (1:0) | 1.0 ± 0.2 |
| CB10394 | 1+ (1:4) | 2.01 ± 0.4 | 4+ (1:2,048) | 1,179.0 ± 286 |
| CB10402 | 1+ (1:2) | 1,19 ± 0.0 | 4+ (1:1,024) | 9.84 ± 0.8 |
| 2771/97c | 2+ (1:32) | 5.71 ± 2.9 | 4+ (1:4,096) | 151.86 ± 17.7 |
The highest dilution of culture medium showing a positive result in the P1-g-EIA is indicated in parentheses.
Means and standard deviations from two separate experiments with measurements performed in duplicate.
Reference strain carrying the stx2e bacteriophage P27.
Nucleotide sequence analysis of the Stx2e-encoding region in low- and high-Stx2e-producing STEC strains.
We were interested in whether the major differences found between Stx2e strains for Stx production could be related to alterations in the nucleic acid sequences of stx2e and adjacent genes. In order to explore this, we analyzed representative low- and high-Stx2e-producing strains (Table 1) for the nucleotide sequences of stx2e and adjacent genes. The 2,385-bp region encompassing the stx2e gene (see Materials and Methods) was identical in low-Stx2e-producing strains CB10282 (accession no. AM937001) and CB10284 (AM939641) and differed in only four nucleotides (99.8% similarity) from the corresponding region of stx2e from phage P27 (AJ298298) (Fig. 1A).
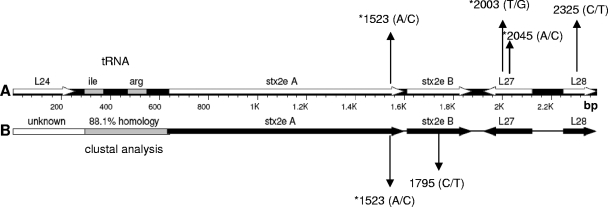
FIG. 1.
A DNA segment corresponding to positions 16866 to 19250 of the bacteriophage P27 genome (accession no. AJ298298) was investigated in strains CB10282 (AM937001) and CB10284 (AM939641) (A) as well as in strains CB10394 (AM939642) and CB10402 (AM939643) (B). (A) The low-Stx2e-producing strains CB10282 and CB10284 differed in only four nucleotides, at positions 1523, 2003, 2045, and 2325, from the AJ299298 sequence. Nucleotide changes leading to alterations in the amino acid sequence are indicated by asterisks. (B) The high-Stx2e-producing strains CB10304 and CB10402 differed in two nucleotides (positions 1523 and 1795) in the 1,749-bp stretch between stxA2e and L28. These strains, as well as the high Stx2e producers CB8771 (AM940005), CB7671 (AM940007), CB8770 (AM940006), and CB8810 (AM940004), shared only 88.1% genetic homology with AJ298298 in the 344-bp tRNA region located upstream of stxA2e (for details, see the Clustal analysis in Fig. 2). The “unknown” 241-bp region upstream left of the tRNA region (identical in strains CB10304, CB10402, CB8771, CB7671, CB8770, and CB8810) does not show similarity to any sequence deposited in GenBank.
The 2,274 bp region encompassing the stx2e gene was identical in both high-Stx2e-producing strains CB10394 (accession no. AM939642) and CB10402 (AM939643). High (CB10394 and CB10402)- and low (CB10282 and CB10284)-Stx2e-producing strains were identical in their stxA2e gene sequences and differed from the P27 sequence (strain 2771/97) by one nucleotide at position 1523, leading to change in the amino acid composition (lysine to threonine) (Fig. 1A and B). The high- and low-Stx2e-producing strains were not different from phage P27 in the amino acid sequence of their Stx2e B subunit.
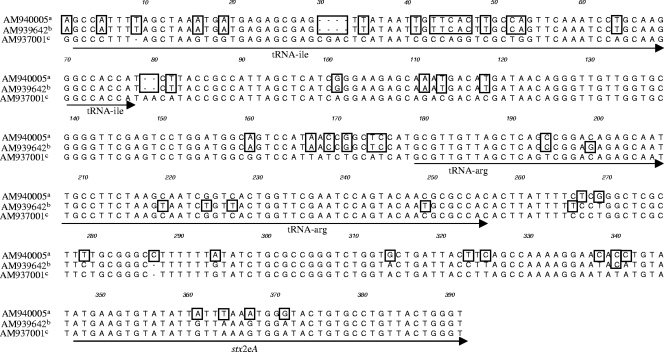
On the other hand, the high-Stx2e-producing strains CB10394 and CB10402 differed in the region upstream of the stxA2e gene from the corresponding sequences of 2771/97 (phage P27), with CB10282 and CB10284 showing only 88.1% homology in the region starting with the tRNA-Ile to the beginning of the stxA2e gene. Nineteen alterations were observed in the region encoding tRNA-Ile (76 nucleotides) and six changes in the region encoding tRNA-Arg (77 nucleotides) (Fig. 2). No homology to any DNA sequences deposited in GenBank was found for the 192-bp DNA stretch upstream of the tRNA-Ile region present in strains CB10394 and CB10402 (Fig. 1).
FIG. 2.
(a and b) Clustal analysis of the tRNA region present in high-Stx2e-producing strains CB7671 (accession no. AM940007) (a) and CB10394 (AM939642) (b) and identical sequences present in CB10402 (AM939643), CB8771 (AM940005), CB8770 (AM940006), and CB8810 (AM940004). (c) Sequence present in low-Stx2e-producing strains CB10282 (AM937001) and CB10284 (AM939641) and in the high Stx2e producer 2771/97, which harbors the stx2e-carrying phage P27 (AJ298298). The positions of tRNA genes and the start of the stxA2e gene are indicated by bold arrows. Nucleotide changes and gaps are indicated by frames at the corresponding positions.
Besides CB10394 and CB10402, 13 other strains from our study belonging to different serotypes were found to be altered in the region upstream of the stxA2e gene compared to phage P27. All these strains were high (4+) Stx2e producers when grown in TSB with mitomycin C. In order to investigate the possible genetic changes in these strains more specifically, we determined the nucleotide sequences in this region of four of these strains, CB8770, CB8810, CB7671, and CB8771 (Table 1), that were unrelated in their origins and serotypes. The sequences of the regions upstream of the stxA2e genes of strains CB10394, CB10402, CB8770, CB8810, and CB7671 were found to be identical. Strain CB8771 showed the same alterations in the tRNA-Ile region but differed from all other strains in the region downstream of tRNA-Ile into the stxA2e gene (Fig. 2).
Transcription of stxA2e genes in low- and high-Stx2e-producing strains.
The similarity of the stx2e coding sequences present in high and low Stx2e producers (see above) indicates that the differences in Stx production are not due to mutations in the Stx2e-coding sequences. In order to investigate whether the differences resulted from transcriptional regulation, we compared the mRNA transcription level of the stxA2e gene with that of the icdA housekeeping gene as a standard by RT-PCR in a relative quantification assay as described in Materials and Methods. Two low (CB10282 and CB10284)- and two high (CB10394 and CB10402)-Stx2e-producing strains were compared. Strain 2771/97, which has previously been described to be inducible for Stx2e production by mitomycin C (31), was used as positive control. The results are presented in Table 4. When grown in TSB without mitomycin C, only strains 2771/97, CB10394, and CB10402 showed detectable Stx2e production with the P1-g-EIA. Growth in TSB supplemented with mitomycin C enhanced Stx2e production clearly in these strains; in contrast, only small amounts of Stx2e (1+ in the P1-g-EIA) became detectable with low toxin producers CB10282 and CB10284. The differences in Stx2e production in these strains were found to relate to the transcription rates of the stxA2e gene in relative-quantification RT-PCR assays. Upon mitomycin C induction, the transcription rates of the stxA2e gene were increased between 10- and 1,000-fold over that of the icdA gene in the high-Stx2e-producing strains (CB10394, CB10402, and 2771/97), whereas the low Stx2e producers CB10282 and CB10284 did not show an increase in stxA2e transcription.
Relationship of Stx2e production to presence of bacteriophage P27-specific gene sequences.
The transducible bacteriophage P27 was previously isolated from STEC 2771/97 and found to carry an Stx2e-encoding gene (31). The complete sequence of bacteriophage P27 was published, and corresponding sequences located upstream and downstream of the stx2e gene were found to be present in other Stx2e-producing strains (39). We were interested in whether the presence and expression of the stxA2e gene are correlated with the presence of bacteriophage P27 sequences in the 110 E. coli strains investigated in this work. The occurrence of phage P27-related sequences was tested in PCRs amplifying four different gene loci of the phage genome (Table 5). Only one (CB8810 [Table 1]) of the 110 strains was negative for all P27 phage-specific genes (except the stx2e gene). The 772-bp region between the phage DNA-methylase gene (L24) and the stxA2e gene (L25) was found in 95 (86.4%) of the strains. All 15 strains that were negative for this specific region of the P27 genome gave a PCR product of 643 bp with primers Stx2e 4f and P27-2. The same primers were used to characterize the altered stxA2e upstream regions of strains CB10394 and CB10402 by nucleotide sequence analysis. The specificity of the Stx2e 4f and P27-2 PCR products in another four Stx2e-producing strains (CB8770, CB8771, CB8810, and CB7671) was tested by nucleotide sequencing (Fig. 2).
TABLE 5.
Presence of stx2e bacteriophage-specific genes in stx2e-positive STEC strains
| No. of P27 gene loci | PCR detection of bacteriophage P27 genes outside stxAB2ea
|
No. of positive strains | |||
|---|---|---|---|---|---|
| Integrase (yecE-int [L01]) (0-283)b | Putative P27 Q gene (L21) (13854-14056) | DNA methylase- stxA2e (L24) (16828-17599) | Holin (stxB2e-S [L28]) (18607-19405) | ||
| 0 | 0 | 0 | 0 | 0 | 1 |
| 1 | + | 0 | 0 | 0 | 6 |
| 1 | 0 | 0 | + | 0 | 46 |
| 2 | 0 | 0 | + | + | 23 |
| 2 | + | 0 | 0 | + | 8 |
| 3 | + | 0 | + | + | 6 |
| 3 | + | + | + | 0 | 13 |
| 4 | + | + | + | + | 7 |
| No. of positive strains | 40 | 20 | 95 | 44 | |
0, PCR negative; +, PCR positive.
PCR product position in bacteriophage P27 sequence (accession no. AJ298298).
A region homologous to phage P27 downstream from stxB2e (L26) to the S (holin) gene (L27) was present in 44 (40.0%) of the strains. Forty strains (36.6%) gave a PCR product spanning from the P27 integrase gene (L01) to the chromosomal yecE gene. Twenty strains (18.2%) carried the P27 Q gene homologue (L21). On the other hand, only seven of the strains (6.4%) were positive for all P27-specific sequences investigated here. Most of the strains carried one to three of the gene loci tested in different combinations (Table 5). The results indicate that a P27-like phage was probably present in all of the strains at an earlier stage but that changes resulting in loss or modification of P27-specific genes have occurred in at least 93.6% of the 110 strains investigated.
We were interested in whether the amount of Stx2e production is correlated with the integrity of the P27 genome in the strains. Of the 61 strains producing large amounts (4+) of Stx2e in TSB plus mitomycin C, one (CB8810) was negative for all, 25 were positive for one, 20 for two, 10 for three, and 5 for all four P27 gene loci, as listed in Table 5. On the other hand, all 15 strains that were altered in the upstream region of the stx2e gene (see above) were significantly higher (4+) Stx2e producers (P < 0.001) than the 95 strains that carried the region which was homologous to the P27 sequence.
Virulence profiles and serotypes associated with Stx2e-producing strains.
Besides Shiga toxin genes, we investigated the presence of the intimin gene (eae) and the plasmid-carried EHEC hemolysin (ehxA) gene, as both of these genes are closely associated with human EHEC strains. None of the 110 Stx2e-producing strains was positive for eae or for ehxA. Similarly, none of the strains belonged to E. coli serotypes that are frequently associated with typical or atypical EHEC strains that are pathogenic for humans. By serotyping, the 110 STEC strains could be divided into nine major serotypes representing 72 (65.5%) of the strains (Table 6). Thirty-one (28.2%) of the strains were not typeable (ONT) with O antisera covering O1 to O181. The remaining seven strains belonged to rare E. coli serotypes, and one was O-rough.
TABLE 6.
Serotypes, origins, and virulence markers of Stx2e-producing strains from this study
| Serotype | Total no. of strains | No. of strains from:
|
No. of strains with the following virulence markera
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Slaughter pigs | Foodb | Environmentc | Diseased pigsd | Humanse | α-hlyA | F18 | STI-p | STII | EAST1 | ||
| O2:[H32] | 4 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| O141:H4 | 10 | 1 | 1 | 5 | 3 | 0 | 10 | 8 | 1 | 1 | 0 |
| O139:H1 | 10 | 0 | 0 | 0 | 10 | 0 | 10 | 9 | 0 | 0 | 1 |
| O138:H14 | 5 | 0 | 0 | 0 | 5 | 0 | 4 | 5 | 3 | 2 | 0 |
| O101:H9 | 5 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 3 |
| O100:H30 | 18 | 6 | 5 | 7 | 0 | 0 | 0 | 0 | 16 | 0 | 2 |
| O8:H9 | 10 | 3 | 6 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 2 |
| O8:H4 | 8 | 1 | 6 | 1 | 0 | 0 | 0 | 0 | 7 | 0 | 1 |
| O8:H19 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ONTf | 31 | 10 | 12 | 4 | 0 | 5 | 0 | 1 | 4 | 1 | 12 |
| Singleg | 7 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 2 |
| Total | 110 | 25 | 36 | 21 | 19 | 9 | 26 | 24 | 37 | 5 | 24 |
Virulence markers were investigated by PCR as described in Materials and Methods. Virulence markers which tested negative for all strains are listed in Results.
Strains from meat (n = 33) and milk (n = 3).
Strains from pig farms (n = 7) and river water (n = 14).
Strains isolated from feces or organs of pigs with edema disease.
Strains from feces of human asymptomatic carriers (n = 3) and patients with diarrhea (n = 6).
O-untypeable strains.
Single isolates of the following serotypes, with the origin and virulence marker(s) in parentheses: O36:19 (meat, α-hlyA), O59:H21 (meat, STI-p, STII), O60:H4 (human diarrhea, EAST1), O137:H1 (diseased pig), O121:H19 (pig farm environment, F18, EAST1), O159:H21 (slaughter pig, STI-p, STII), and Or:H19 (asymptomatic human).
We investigated virulence markers that are frequently found in human and animal pathogenic STEC and enterotoxigenic E. coli strains, such as adhesins, cytotoxins, and enterotoxins. Except for F18 fimbriae (21.8% positive), all strains were negative for genes encoding adhesins K88, K99, 987p, and F41, which play a role as colonization factors for cattle and pigs. Diffuse adherence (DA) was observed with 31 strains (28.2%) in assays performed on cultured HEp-2 cells. The DA type was not associated with other phenotypical traits and adhesins investigated here or with the origin of the strains. By performing the HEp-2 cell assay, complete detachment of HEp-2 cells from coverslips within a 3-h incubation period with bacteria was found with 11 (10.0%) of the strains. We have looked for the genes encoding cytotoxins, such as α-hlyA, cnf, cdt, and cif, which could be responsible for this effect, but only alpha-hemolysin was found to be present in 7 (63.6%) of these 11 strains.
Among the heat-stable enterotoxins STI-p, STI-h, STII, and EAST1, only STI-p (33.6%), EAST1 (21.8%), and STII (5.4%) were detected in some Stx2e strains. No strain was positive for heat-labile enterotoxin LT-I. The results are summarized in Table 6.
Twenty-six (23.6%) of the STEC strains belonged to serotypes that are commonly associated with porcine pathogenic E. coli types (O137:H1, O138:H14, O139:H1, and O141:H4). Most of these strains (73.1%) were isolated from diseased pigs, and virulence attributes such as alpha-hemolysin (92.3%) and F18 fimbriae (84.6%) were frequently associated with these strains. Seven (26.9%) of strains belonging to porcine pathogenic types (two O138, two O139, and three O141) provoked rapid detachment of HEp-2 cells in adherence assays (see above). In contrast, alpha-hemolysin and F18 fimbriae were present each in only two (2.3%) and rapid detachment of HEp-2 cells was observed with only four (4.8%) of the 84 Stx2e strains from other sources.
Strains belonging to serotypes O8:H4, O8:H9, and O100:[H30] (n = 36) were frequently isolated from pigs at slaughter, from food, and from surface water and soil. All these were negative for the virulence markers tested except for STI-p (75.0%) and EAST1 (13.9%). Serotypes O8:H9 and O100:H30 were also described as frequent in swine feces in a nationwide study performed in the United States (16).
The nine Stx2e-producing strains originating from humans were heterogeneous for their serotypes, and only three of these (two O101:H9 and one O60:H4) were typeable for their O antigen. Five of the human Stx2e strains showed DA and one showed rapid killing of HEp-2, cells as described above. Other virulence markers were not present, except for EAST1 in two (22.2%) of the strains.
DISCUSSION
Shiga toxin 2e-encoding genes are present in E. coli strains from various sources and with different genetic backgrounds, indicating that the stx2e genes are spread horizontally in E. coli. A lysogenic stx2e bacteriophage designated P27 was recently identified and shown to transduce the stx2e gene to stx-negative E. coli strains (31). Although P27 shows a typical lambdoid structure, it is only remotely related to other lambdoid stx-carrying phages (39). The presence of bacteriophage P27-specific genes in almost all of the strains investigated in this study indicates that the stx2e determinant is spread in E. coli by P27-like phages. It was previously suggested that P27-like stx2e phages were acquired early in the evolution of Stx2e strains and have undergone multiple recombination events since then (39). This is in agreement with the finding that most (93.6%) of the 110 Stx2e strains from this study lacked one or more of the P27-specific genes, perhaps by successive deletions of phage sequences. It is therefore likely that the P27-like phages are present as cryptic prophages or phage remnants in most of the strains. This suggestion is also supported by the finding that stx2e genes are generally not transducible from their host strains, which is in contrast to the case for other types of stx genes (29, 31).
Besides the genomic differences, the host range within the E. coli strains of the stx2e phage type differs from that of strains of the other stx phage types. The stx2e gene is associated with E. coli serotypes that have not been described as bacterial hosts for other types of Stx so far (4, 16, 17, 39, 42). Additionally, the stx2e gene was not reported to be present in typical and atypical EHEC strains causing bloody diarrhea and HUS, except for one case reported in 1994 (11, 36, 43).
In our study the stx2e gene was not found in combination with other stx genes in the bacteria. Similar findings were reported from a study on STEC prevalence in pigs in the United States; the stx2e determinant was rarely found in association with other stx genes (16). In contrast to that, other variants of the stx2 family occur frequently in combination with stx1 and stx2 genes in STEC from patients and ruminant animals (11). The reason for these differences between Stx2e-producing and other STEC strains is not known.
The stx2e-positive strains investigated here differed largely in the amount of Stx produced by the bacteria. Without induction, two-thirds of the strains were identified as poor producers of Stx (0 to 1+), but 99% of the strains were inducible for Stx2e production by mitomycin C. Mitomycin C was chosen as a potent inductor for phage-encoded gene expression in vitro. The inducibility of Stx production is likely to play a role in vivo for pathogenesis, since Stx production was shown to be triggered by neutrophils and norepinephrine in the gastrointestinal tract (13, 49). The inducible phenotype could not be related to phage production (data not shown) or to the integrity of the phage P27 genome in these strains. The presence of the phage-carried Q-gene was reported to be required for effective transcription and expression of stx2 genes on stx-carrying phages of O157:H7 strains (25). In our study, the absence of the P27 Q gene homologue was not associated with low Stx2e production (P = 0.292), since it was absent in 82.6% of the high-producing (3+ and 4+) Stx2e strains. Our data confirm previous findings indicating that the P27 Q gene is frequently disconnected genetically from the stx2e gene (39). It cannot be excluded that the P27 Q gene is replaced by another phage Q homologue by recombination. It is also possible that induction of Stx2e production in the strains is generally triggered by phage-independent functions such as RecA, as previously described for EHEC O157 strains (18).
The amount of Stx2e produced by the strains was measured with the P1-g-EIA, a binding assay that measures the quantity and function of the various allelic Stx B proteins. However, it is not likely that the differences found in the amount of Stx2e produced by the strains are attributable to mutations in the stx2e genes, since translation of the nucleotide sequence of the stxAB2e genes in high- and low-Stx2e-producing strains resulted in identical amino acid sequences. On the other hand, production of Stx2e was associated with the transcription efficiency of the stx2e gene in these strains, which was found to be drastically enhanced by mitomycin C. Similar findings were reported previously for stx2d and stx2e variants (53). Only five (4.5%) of the 110 Stx2e strains showed high production of Stx2e with noninduced and induced cultures, indicating that constitutive high expression of stx2e genes is not common in natural isolates.
The upstream regions flanking the 5′ ends of the stx genes were reported to be conserved and continuous with phage sequences in a number of Shiga toxin-encoding bacteriophages (32, 39, 44). Genes for rare tRNAs are located in the neighborhood of the stx2 genes, and it was suggested that these could play a role in effective expression of Stx2 (23, 38). Correspondingly, the region located upstream from the stxA2e gene was found to be conserved in 95 (86.4%) of the Stx2e strains investigated here, pointing to a common origin of the stx2e genes in most of the strains. However, large differences in Stx2e production were observed despite identical tRNA regions (Table 4 and Fig. 1 and 2), indicating that other functions interfere with the expression of stx2e genes. The 15 stx2e strains that were genetically different from P27 in this region were all high Stx2e producers, indicating that the conserved tRNA region of the P27 genome is not essential for production of Stx2e in these strains. Although most of these 15 strains were unrelated in their serotypes, virulence attributes, and origins (data not shown), they were highly similar in the DNA region upstream of stxA2e. This could indicate that these were spread by a recombinant type of P27 phage to genetically different E. coli host strains and supports the suggestion of an early uptake of stx2e phages in the evolution of this group of STEC strains (39).
Evaluation of Stx2e strains as potential human diarrheal pathogens.
There are only few data available on the role of Shiga toxins in human diarrhea. In vitro production of Stx has been shown to be positively correlated with the severity of diarrheal symptoms in patients infected with EHEC O157 (30, 34), and Stx2 was found to cause damage and fluid secretion in intestinal epithelial cells (41). Therefore, it cannot be excluded that Stx2e might have an effect on fluid secretion in the human intestine.
There are no data available on the toxicity of Stx2e for the human organism. Three of the nine human Stx2e strains produced large amounts (3+ to 4+) of Stx2e when grown without mitomycin C and eight strains (all 4+) did so when grown with mitomycin C, indicating that most of the human strains were good Stx2e producers. Stx2e differs from other Shiga toxins by having an altered receptor specificity (Gb4) (24), and effective binding of Stx2e to porcine red blood cells was found to be essential for development of edema disease in pigs (28). Stx2e was also found to bind as efficiently as Sx1 and Sx2 to human erythrocytes (8), but in contrast to Stx2, Stx2e is not associated with bloody diarrhea and HUS in humans (4, 17, 36). In the pig intestine, Stx2e does not cause fluid accumulation, which indicates that the toxin itself does not cause diarrhea in the animals (48). It was suggested that the lack of enterotoxicity of Stx2e in pigs is due to the absence of toxin receptors in the villus absorptive enterocytes (48). Accordingly, diarrhea is not a typical sign in pigs with edema disease, and it was reported to precede edema disease only in infections with strains producing enterotoxins besides Stx2e (1). The finding that high-Stx2e-producing strains were isolated from humans with no or only mild symptoms of diarrhea indicates that Sx2e has no or little potential to elicit diarrhea in humans, possibly because of a lack of receptors Gb4 and Gb3, which could serve as targets for Stx2e (24) on human enterocytes.
Are Stx2e strains pathogenic for humans? Six of the human excretors suffered from uncomplicated diarrhea, and three individuals showed no symptoms of enteric disease (data not shown). The amounts of Stx2e produced in strains from humans with diarrhea and from asymptomatic cases were similar (data not shown). These findings correspond to other studies reporting that Stx2e-producing strains are rarely isolated from humans, are not significantly associated with diarrhea, and do not cause severe disease (4, 15, 17, 37, 39, 42). The low prevalence of these strains in humans is in clear contrast to their high prevalence in meat and its products in Germany (5). This indicates that most of the Stx2e strains present in food are not good colonizers of the human intestine. Stx2e strains from humans were found to differ from the corresponding porcine pathogenic strains by their adherence to human and swine epithelial cells, but the adhesins of human Stx2e strains were not identified (42). DA to HEp-2 cells, which was used as an indicator for colonization of human epithelial cells, was observed with five human Stx2e strains in this study. The DA type was found in 28.2% of all Stx2e strains investigated here but could not be associated with a particular serotype or with other virulence markers (42). Toxicity resulting in rapid detachment of epithelial cells was reported to be associated with porcine pathogenic types of Stx2e strains (42) and was found to be associated with O138, O139, O141, and O101 strains in our study. On the other hand, only one of the nine human Stx2e strains (O60:H4) provoked rapid detachment of HEp-2 cell monolayers.
Certain more rarely occurring serogroups of Stx2e strains, such as O60 and O101, were reported more frequently in association with human infections (15, 17, 37, 39). It is possible that some of the Stx2e-producing strains have the ability to cause diarrhea in humans, but the underlying mechanism remains unknown. Enterotoxins, which could be a cause of diarrhea in humans, were not investigated in previous studies dealing with human STEC infections (15, 37, 39). It is conceivable that enterotoxins may play a role as a cause for human diarrhea. In contrast to all other types of STEC, more than 50% of Stx2e-producing strains from this study were positive for heat-stable enterotoxins such as STI-p, STII, and EAST1. The majority of the Stx2e strains from food carried STI-p and/or EAST1 genes, and it is thus likely that human consumers frequently come in contact with ST- or EAST1-positive Stx2e strains. In our study, EAST1 was present in two of six Stx2e strains (ONT:H4 and O60:H4) from human patients with diarrhea (4, 17, 42). We also cannot exclude that genetic variants of enterotoxins and adhesion factors that were not detected with the PCR primers used in the study are present in some of the strains.
In conclusion, the results from this study complement others indicating that Stx2e is not a pathogenicity factor for diarrhea and HUS in humans. This finding is important for assessment of the public health impact of STEC from food, because Stx2e-producing E. coli accounted for 19% of all STEC isolated from food in a previous study (5).
Acknowledgments
We thank John Fairbrother (University of Montreal, Montréal, Canada), Bela Nagy (Hungarian Academy of Sciences, Budapest, Hungary), and Herbert Schmidt (University of Hohenheim, Stuttgart, Germany) for supplying some of the E. coli reference strains used in this work.
We are grateful to Katja Steege, Karin Pries, Sabine Haby, and Nadine Albrecht for technical assistance.
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Bertschinger, H. U., and C. L. Gyles. 1994. Oedema disease of pigs, p. 193-219. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 2.Beutin, L. H. Steinruck, G. Krause, K. Steege, S. Haby, G. Hultsch, and B. Appel. 2007. Comparative evaluation of the Ridascreen((R)) verotoxin enzyme immunoassay for detection of Shiga-toxin producing strains of Escherichia coli (STEC) from food and other sources. J. Appl. Microbiol. 102:630-639. [DOI] [PubMed] [Google Scholar]
- 3.Beutin, L., S. Kaulfuss, S. Herold, E. Oswald, and H. Schmidt. 2005. Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L., G. Krause, S. Zimmermann, S. Kaulfuss, and K. Gleier. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin, L., A. Miko, G. Krause, K. Pries, S. Haby, K. Steege, and N. Albrecht. 2007. Identification of human pathogenic strains of Shiga toxin-producing Escherichia coli from food by combination of serotyping and molecular typing of Shiga toxin genes. Appl. Environ. Microbiol. 73:4769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin, L., J. Tao, L. Feng, G. Krause, S. Zimmermann, K. Gleier, Q. Xia, and L. Wang. 2005. Sequence analysis of the Escherichia coli O15 antigen gene cluster and development of a PCR assay for rapid detection of intestinal and extraintestinal pathogenic E. coli O15 strains. J. Clin. Microbiol. 43:703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bielaszewska, M., A. W. Friedrich, T. Aldick, R. Schurk-Bulgrin, and H. Karch. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160-1167. [DOI] [PubMed] [Google Scholar]
- 8.Bitzan, M., S. Richardson, C. Huang, B. Boyd, M. Petric, and M. A. Karmali. 1994. Evidence that verotoxins (Shiga-like toxins) from Escherichia coli bind to P blood group antigens of human erythrocytes in vitro. Infect. Immun. 62:3337-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brett, K. N., V. Ramachandran, M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2003. stx1c is the most common Shiga toxin 1 subtype among Shiga toxin-producing Escherichia coli isolates from sheep but not among isolates from cattle. J. Clin. Microbiol. 41:926-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brett, K. N., M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2003. Bovine non-O157 Shiga toxin 2-containing Escherichia coli isolates commonly possess stx2-EDL933 and/or stxvhb subtypes. J. Clin. Microbiol. 41:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caprioli, A., S. Morabito, H. Brugereb, and E. Oswald. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289-311. [DOI] [PubMed] [Google Scholar]
- 12.Chapman, T. A., X. Y. Wu, I. Barchia, K. A. Bettelheim, S. Driesen, D. Trott, M. Wilson, and J. J. Chin. 2006. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 72:4782-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowd, S. E. 2007. Escherichia coli O157:H7 gene expression in the presence of catecholamine norepinephrine. FEMS Microbiol. Lett. 273:214-223. [DOI] [PubMed] [Google Scholar]
- 14.Franck, S. M., B. T. Bosworth, and H. W. Moon. 1998. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J. Clin. Microbiol. 36:1795-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke, S., D. Harmsen, A. Caprioli, D. Pierard, L. H. Wieler, and H. Karch. 1995. Clonal relatedness of Shiga-like toxin-producing Escherichia coli O101 strains of human and porcine origin. J. Clin. Microbiol. 33:3174-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fratamico, P. M., L. K. Bagi, E. J. Bush, and B. T. Solow. 2004. Prevalence and characterization of Shiga toxin-producing Escherichia coli in swine feces recovered in the National Animal Health Monitoring System's Swine 2000 study. Appl. Environ. Microbiol. 70:7173-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs, S., I. Muhldorfer, A. Donohue-Rolfe, M. Kerenyi, L. Emody, R. Alexiev, P. Nenkov, and J. Hacker. 1999. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb. Pathog. 27:13-23. [DOI] [PubMed] [Google Scholar]
- 19.Gyles, C. L. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85:E45-E62. [DOI] [PubMed] [Google Scholar]
- 20.Han, W., B. Liu, B. Cao, L. Beutin, U. Kruger, H. Liu, Y. Li, Y. Liu, L. Feng, and L. Wang. 2007. DNA microarray-based identification of serogroups and virulence gene patterns of Escherichia coli isolates associated with porcine postweaning diarrhea and edema disease. Appl. Environ. Microbiol. 73:4082-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herold, S., H. Karch, and H. Schmidt. 2004. Shiga toxin-encoding bacteriophages—genomes in motion. Int. J. Med. Microbiol. 294:115-121. [DOI] [PubMed] [Google Scholar]
- 22.Jelacic, J. K., T. Damrow, G. S. Chen, S. Jelacic, M. Bielaszewska, M. Ciol, H. M. Carvalho, A. R. Melton-Celsa, A. D. O'Brien, and P. I. Tarr. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719-729. [DOI] [PubMed] [Google Scholar]
- 23.Kanjo, N., and H. Inokuchi. 1999. Genes for tRNA(Arg) located in the upstream region of the Shiga toxin II operon in enterohemorrhagic Escherichia coli O157:H7. DNA Res. 6:71-73. [DOI] [PubMed] [Google Scholar]
- 24.Keusch, G. T., M. Jacewicz, D. W. Acheson, A. Donohue-Rolfe, A. V. Kane, and R. H. McCluer. 1995. Globotriaosylceramide, Gb3, is an alternative functional receptor for Shiga-like toxin 2e. Infect. Immun. 63:1138-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koitabashi, T., V. Vuddhakul, S. Radu, T. Morigaki, N. Asai, Y. Nakaguchi, and M. Nishibuchi. 2006. Genetic characterization of Escherichia coli O157: H7/− strains carrying the stx2 gene but not producing Shiga toxin 2. Microbiol. Immunol. 50:135-148. [DOI] [PubMed] [Google Scholar]
- 26.Loukiadis, E., M. Kerouredan, L. Beutin, E. Oswald, and H. Brugere. 2006. Characterization of Shiga toxin gene (stx)-positive and intimin gene (eae)-positive Escherichia coli isolates from wastewater of slaughterhouses in France. Appl. Environ. Microbiol. 72:3245-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mainil, J. G., and G. Daube. 2005. Verotoxigenic Escherichia coli from animals, humans and foods: who's who? J. Appl. Microbiol. 98:1332-1344. [DOI] [PubMed] [Google Scholar]
- 28.Matise, I., N. A. Cornick, J. E. Samuel, and H. W. Moon. 2003. Binding of Shiga toxin 2e to porcine erythrocytes in vivo and in vitro. Infect. Immun. 71:5194-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muniesa, M., J. E. Blanco, M. De Simon, R. Serra-Moreno, A. R. Blanch, and J. Jofre. 2004. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 150:2959-2971. [DOI] [PubMed] [Google Scholar]
- 30.Muniesa, M., M. De Simon, G. Prats, D. Ferrer, H. Panella, and J. Jofre. 2003. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71:4554-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muniesa, M., J. Recktenwald, M. Bielaszewska, H. Karch, and H. Schmidt. 2000. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect. Immun. 68:4850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muniesa, M., R. Serra-Moreno, and J. Jofre. 2004. Free Shiga toxin bacteriophages isolated from sewage showed diversity although the stx-genes appeared conserved. Environ. Microbiol. 6:716-725. [DOI] [PubMed] [Google Scholar]
- 33.Nagy, B., R. A. Wilson, and T. S. Whittam. 1999. Genetic diversity among Escherichia coli isolates carrying f18 genes from pigs with porcine postweaning diarrhea and edema disease. J. Clin. Microbiol. 37:1642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishikawa, Y., Z. Zhou, A. Hase, J. Ogasawara, T. Cheasty, and K. Haruki. 2000. Relationship of genetic type of Shiga toxin to manifestation of bloody diarrhea due to enterohemorrhagic Escherichia coli serogroup O157 isolates in Osaka City, Japan. J. Clin. Microbiol. 38:2440-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Persson, S., K. E. Olsen, S. Ethelberg, and F. Scheutz. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierard, D., L. Huyghens, S. Lauwers, and H. Lior. 1991. Diarrhoea associated with Escherichia coli producing porcine oedema disease verotoxin. Lancet 338:762. [DOI] [PubMed] [Google Scholar]
- 38.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Recktenwald, J., and H. Schmidt. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage φP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70:1896-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheutz, F., and N. A. Strockbine. 2005. Genus I. Escherichia, p. 607-624. In G. M. Garrity, D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology. Springer, New York, NY.
- 41.Schuller, S., G. Frankel, and A. D. Phillips. 2004. Interaction of Shiga toxin from Escherichia coli with human intestinal epithelial cell lines and explants: Stx2 induces epithelial damage in organ culture. Cell. Microbiol. 6:289-301. [DOI] [PubMed] [Google Scholar]
- 42.Sonntag, A. K., M. Bielaszewska, A. Mellmann, N. Dierksen, P. Schierack, L. H. Wieler, M. A. Schmidt, and H. Karch. 2005. Shiga toxin 2e-producing Escherichia coli isolates from humans and pigs differ in their virulence profiles and interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 71:8855-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, A., T. Cheasty, H. Chart, and B. Rowe. 1994. Isolation of Vero cytotoxin-producing Escherichia coli serotypes O9ab:H− and O101:H− carrying VT2 variant gene sequences from a patient with haemolytic uraemic syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 13:1074-1076. [DOI] [PubMed] [Google Scholar]
- 44.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urdahl, A. M., L. Beutin, E. Skjerve, S. Zimmermann, and Y. Wasteson. 2003. Animal host associated differences in Shiga toxin-producing Escherichia coli isolated from sheep and cattle on the same farm. J. Appl. Microbiol. 95:92-101. [DOI] [PubMed] [Google Scholar]
- 46.Vernozy-Rozand, C., M. P. Montet, Y. Bertin, F. Trably, J. P. Girardeau, C. Martin, V. Livrelli, and L. Beutin. 2004. Serotyping, stx2 subtyping, and characterization of the locus of enterocyte effacement island of Shiga toxin-producing Escherichia coli and E. coli O157:H7 strains isolated from the environment in France. Appl. Environ. Microbiol. 70:2556-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verweyen, H. M., H. Karch, M. Brandis, and L. B. Zimmerhackl. 2000. Enterohemorrhagic Escherichia coli infections: following transmission routes. Pediatr. Nephrol. 14:73-83. [DOI] [PubMed] [Google Scholar]
- 48.Waddell, T. E., C. A. Lingwood, and C. L. Gyles. 1996. Interaction of verotoxin 2e with pig intestine. Infect. Immun. 64:1714-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner, P. L., D. W. Acheson, and M. K. Waldor. 2001. Human neutrophils and their products induce Shiga toxin production by enterohemorrhagic Escherichia coli. Infect. Immun. 69:1934-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, L., B. Liu, Q. Kong, H. Steinruck, G. Krause, L. Beutin, and L. Feng. 2005. Molecular markers for detection of pathogenic Escherichia coli strains belonging to serogroups O 138 and O 139. Vet. Microbiol. 111:181-190. [DOI] [PubMed] [Google Scholar]
- 51.Wei, Y., J. M. Lee, D. R. Smulski, and R. A. LaRossa. 2001. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J. Bacteriol. 183:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto, T., and P. Echeverria. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, W., M. Bielaszewska, A. W. Friedrich, T. Kuczius, and H. Karch. 2005. Transcriptional analysis of genes encoding Shiga toxin 2 and its variants in Escherichia coli. Appl. Environ. Microbiol. 71:558-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zweifel, C., S. Schumacher, L. Beutin, J. Blanco, and R. Stephan. 2006. Virulence profiles of Shiga toxin 2e-producing Escherichia coli isolated from healthy pig at slaughter. Vet. Microbiol. 117:328-332. [DOI] [PubMed] [Google Scholar]