Abstract
Babesiosis is a tick-transmitted disease of veterinary and medical importance. The first Austrian case of human babesiosis was recently recorded. In the current study, ticks at all life cycle stages (instars), including 853 Ixodes ricinus and 11 Haemaphysalis concinna ticks, from sampling sites throughout Austria were examined for the presence of Babesia spp. by using 18S rRNA gene PCR and sequencing, and the overall mean infection rate was 51.04%. The infection rates for sampling sites were highly variable, ranging from 0% to almost 100%. Different instars and different sexes were infected almost equally. Babesia isolates occurring in Austrian ticks were identified as Babesia divergens, Babesia divergens-like, and Babesia sp. strain DD by sequencing a fragment of the heat shock protein 70 gene and internal transcribed spacer regions 1 and 2. To our knowledge, this is the first investigation of Babesia spp. in Austrian ticks.
Babesiosis, primarily known as a worldwide disease of veterinary importance mainly in cattle but also in horses, sheep, pigs, and dogs, has attracted increased notice in recent years as a zoonotic infection in humans. Babesia spp. are protozoan intraerythrocytic parasites belonging to the Apicomplexa. More than 100 species have been described so far, primarily based on morphological characteristics (i.e., a pear shape in erythrocytes) and the assumption that Babesia spp. are strictly host specific (31). However, the current species designations have not been supported by recent molecular studies. On the one hand, species have been shown to be molecularly identical, and therefore it has been suggested that they should be considered the same taxon parasitizing different hosts (13). On the other hand, the different strains currently classified as Babesia divergens have been shown to probably belong to more than one species (16).
The first case of human babesiosis was documented in 1957 in a splenectomized farmer in Croatia (35). Since then, several hundred cases have been reported, mainly from the United States (36), where the first case of human babesiosis occurred in 1966 (33). In Europe about 30 cases have been documented so far, mostly in France and Great Britain, probably due to greater medical and scientific interest. The main clinical manifestations of human babesiosis are intravascular hemolysis with hemoglobinuria and jaundice. Persistent high fever, shaking chills, sweating, headache, myalgia, and lumbar and abdominal pain have been described as concomitant symptoms. Splenectomy has been identified as the major predisposing factor. Babesiosis in patients with intact spleens usually is less severe. Interestingly, more than one-half of the European cases were described after 1985 (14). The possible reasons for the “emerging status” of the disease have been suggested to be ecological changes, increased awareness, and increased populations of susceptible individuals (e.g., AIDS patients) (23, 26). Human babesiosis in Europe is predominantly caused by the bovine species B. divergens (25); however, this species is not the only cause (16). Moreover, B. divergens-like Babesia spp. almost identical to the bovine species B. divergens have recently been detected in reindeer and cervids (10, 27). Ixodes ricinus is the European vector for B. divergens, as it is for the B. divergens-like species, for B. microti, and for the recently described organism Babesia sp. strain EU1 (2, 3, 8, 24). The latter taxon was identified as the causative agent in three European cases of human babesiosis in the recent past (19, 21); furthermore, two human infections in Europe were caused by the rodent species B. microti (22, 29). As in Europe, in the United States not all Babesia infections in humans have been caused by the same species; although B. microti has been responsible for the majority of cases in the United States, several cases have been caused by B. duncani and other Babesia strains of the WA and CA types, as reviewed by Conrad et al. (5), and even one infection caused by Babesia sp. strain MO1 has been described (20).
In the past few years, more cases of human babesiosis have been reported. Moreover, thorough investigations have shown that more Babesia species than previously assumed can be involved in human disease. However, information concerning the prevalence and species composition of Babesia spp. in Central European ticks is scarce (23). Recently, the first Austrian case of human babesiosis was described (21). The aim of the present study was to investigate Austrian ticks for the presence of Babesia spp.
MATERIALS AND METHODS
Tick collection.
Questing ticks were collected by blanket dragging from May to August 2005 throughout Austria. Species and life cycle stages (instars) were determined, and then ticks were stored in vials at −20°C. In order to cover Austria in a representative way, approximately equal numbers of all tick instars were obtained from two ecologically different sampling sites in each federal state (a total of 96 ticks per federal state). The aim was to focus on Ixodes ricinus, the vector for Babesia spp. However, at one sampling site, where the number of I. ricinus ticks found was too low to fulfill the sample size, Haemaphysalis concinna was included in the investigation. Altogether, 864 ticks (for I. ricinus, 257 adult ticks, 355 nymphs, and 241 larvae; for H. concinna, 11 larvae) were used in the investigation. The vegetation ranged from deciduous to mixed woodland, and the altitudes (151 to 827 m above sea level) and tick densities at the sampling sites differed (Table 1).
TABLE 1.
Sampling sites in Austria, including altitude, tick density, and numbers of adult ticks, nymphs, and larvae infected and investigated
| Federal state | Sampling site | Altitude (m above sea level) | Tick densitya | No. of ticks infected/no. of ticks investigated | No. of adult ticks infected/no. of adult ticks investigated | No. of nymphs infected/no. of nymphs investigated | No. of larvae infected/no. of larvae investigated |
|---|---|---|---|---|---|---|---|
| Vorarlberg | Thüringen | 573 | Medium | 32/48 | 16/16 | 0/16 | 16/16 |
| Klaus | 507 | High | 0/48 | 0/16 | 0/28 | 0/4 | |
| Tyrol | Imst | 827 | High | 47/48 | 15/16 | 16/16 | 16/16 |
| Fügen | 545 | Low | 0/48 | 0/16 | 0/16 | 0/16 | |
| Salzburg | Adnet | 484 | Medium to high | 35/48 | 12/16 | 11/16 | 12/16 |
| Goldegg | 822 | Medium | 47/48 | 16/16 | 23/24 | 8/8 | |
| Carinthia | St. Paul | 412 | Low | 14/48 | 2/16 | 10/16 | 2/16 |
| Drobollach | 501 | Low | 35/48 | 15/16 | 4/16 | 16/16 | |
| Upper Austria | Stallhofen | 444 | Low | 45/48 | 14/16 | 16/16 | 15/16 |
| Niederottensheim | 270 | High | 0/48 | 0/16 | 0/16 | 0/16 | |
| Styria | Mürzzuschlag | 670 | Medium | 36/48 | 4/16 | 16/16 | 16/16 |
| Graz-Umgebung | 526 | Low | 43/48 | 16/16 | 12/16 | 15/16 | |
| Lower Austria | Wofenreith | 647 | Low | 42/48 | 14/16 | 28/32 | 0/0 |
| Hüttendorf | 190 | Low | 0/48 | 0/6 | 0/26 | 0/16 | |
| Vienna | Wien-Prater | 160 | Medium | 17/48 | 4/16 | 13/16 | 0/16 |
| Wien-Lobau | 151 | Low | 0/48b | 0/16 | 0/16 | 0/16c | |
| Burgenland | Oberwart | 315 | Medium to high | 48/48 | 7/7 | 25/25 | 16/16 |
| Stoob | 265 | Low | 0/48 | 0/4 | 0/28 | 0/16 |
Low, 0 to 250 ticks; medium, 250 to 500 ticks; high, ≥500 ticks (per sampling site [∼100 m2]).
Including 11 H. concinna ticks.
Five I. ricinus larvae and 11 H. concinna larvae were investigated.
DNA isolation.
Prior to DNA isolation, ticks were thawed, immersed in 70% ethanol for 10 min, and air dried on microscope slides. Adult ticks and nymphs were cut lengthwise with sterile scalpel blades, and larvae were processed whole. Total DNA was isolated with a DNeasy blood and tissue kit (Qiagen GmbH, Germany) using a modified protocol of Beati and Keirans (1) and a final concentration of proteinase K (Roche, Austria) of 1.44 μg/μl. Isolated DNA was eluted in 40 μl AE buffer (Qiagen GmbH, Germany).
Primer design and PCR.
Altogether, four pairs of primers were designed (Table 2). The first primer pair (Babfor/Babrev) was designed to evaluate the overall rate of infection of Austrian I. ricinus ticks with Babesia spp. The 18S rRNA gene, which is a highly conserved gene and is the only gene whose sequence is available for various Babesia species, was chosen as the target region in order to cover all Babesia spp. known to date. Since the 18S rRNA gene is conserved not only in a genus but also across different phyla, primer sites were chosen so that sequences were identical for all Babesia spp. but markedly different from the sequences of other organisms, including I. ricinus.
TABLE 2.
PCR programs used for the detection and identification of Babesia spp. in ticks
| PCR | Program | Primer
|
||
|---|---|---|---|---|
| Designation | Sequence (5′-3′) | Approximate nucleotide position | ||
| 18S rRNA gene | 95°C for 1 min, 53°C for 1.50 min, | Babfor | GACTAGGGATTGGAGGTC | 970 |
| and 72°C for 1.50 min for 35 cycles | Babrev | GAATAATTCACCGGATCACTC | 1590 | |
| ITS1 | 95°C for 1 min, 52°C for 2 min, and | ITS1for | CGAGTGATCCGGTGAATTATTC | 1590 (18S rRNA gene) |
| 72°C for 3 min for 40 cycles | ITS1rev | CCTTCATCGTTGTGTGAGCC | Within 5.8S rRNA gene | |
| ITS2 and HSP70 | 95°C for 1 min, 54°C for 2 min, and | ITS2for | GGCTCACACAACGATGAAGG | Within 5.8S rRNA gene |
| 72°C for 3 min for 40 cycles | ITS2rev | CTCGCCGTTACTAAGGGAATC | 76 (28S rRNA gene) | |
| HSP70for | GCTATTGGTATTGACTTGGG | 13 | ||
| HSP70rev | CCTTCATCTTGATAAGGACC | 366 | ||
| Sequencing | 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min for 30 cycles | |||
For precise species differentiation within the genus Babesia, a second primer pair (HSP70for/HSP70rev) was designed to amplify the more variable heat shock protein 70 (HSP70) gene. Finally, in order to confirm the HSP70 results, two more primer pairs that amplified highly variable ribosomal internal transcribed spacer region 1 (ITS1) and ITS2 of Babesia (ITS1for/ITS1rev and ITS2for/ITS2 rev) were designed for the 18S rRNA gene, the 5.8S rRNA gene, and the 28S rRNA gene. ITS1for has the same nucleotide sequence as Babrev except that it has one additional nucleotide at the 5′ end; ITS1rev and ITS2for are identical.
Three controls, B. divergens DNA, B. microti DNA, and blood of cattle with a B. divergens infection, were used for primer testing and establishing the PCR. Total DNA was extracted with the DNeasy blood and tissue kit (Qiagen GmbH) as described above. Primer testing and establishing the PCR were performed both in a separate laminar flow and before tick DNA was isolated. Every tick PCR was performed with a negative control sample but without a positive control sample in order to avoid cross-contamination. Instead, all positive PCR products were confirmed by sequencing the 18S rRNA gene fragment.
PCR and sequencing.
All PCRs were performed using 3 μl of total DNA in a 50-μl PCR mixture and a Mastercycler ep gradient S (Eppendorf, Germany) with standard amplification programs (Table 2). PCR products were separated using an ethidium bromide-stained 3% agarose gel (3:1 sieve agarose; Biozym, Austria) in Tris-acetate-EDTA buffer and were visualized under UV light. Positive PCR products were purified using a PCR DNA and gel band purification kit (GE Healthcare Bio-Sciences AB, Austria) and were subjected to cycle sequencing using an ABI PRISM Big Dye kit (AB, Langen, Germany) and a standard sequencing program (Table 2). Some sequencing was carried out using a 310 ABI PRISM automated sequencer (AB, Langen, Germany), and some sequencing was performed by Ingenetix GmbH (Vienna, Austria). Sequences were aligned using the GeneDoc sequence editor computer program (30).
Sampling site analysis.
Data were analyzed using SPSS 14.0 software. Correlations were calculated using Pearson and Spearman coefficients.
Nucleotide sequence accession numbers.
HSP70, ITS1, and ITS2 sequences have been deposited in the GenBank database under accession numbers EU185801 to EU185804 (ITS1 and ITS2) and EU18505 to EU185815 (HSP70).
RESULTS
Overall infection rate.
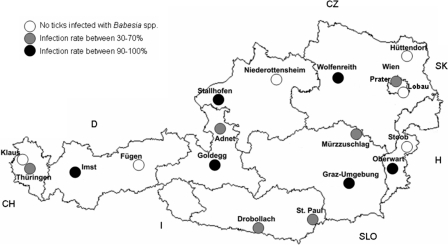
Altogether, 864 ticks were investigated, and positive PCR results were obtained for 441 of these ticks (51.04%). While all investigated H. concinna larvae (n = 11) were negative for Babesia spp., 51.70% of the 853 I. ricinus ticks investigated were PCR positive. Interestingly, all I. ricinus life stages, as well as both sexes, were infected almost equally. The results showed that 53.52% (135/257) of the adults were infected; about one-half of adults (122/257) were investigated to examine sex-specific differences, and it was found that 39.06% (25/64) of the females and 37.93% (22/58) of the males were infected with Babesia spp. When the Babesia-specific 18S rRNA PCR was used, 49.01% (174/355) of the nymphs and 54.77% of the larvae (132/241) were positive. Interestingly, however, the infection rates for sampling sites ranged from 0 to 100% (Table 1 and Fig. 1). One-third of the sampling sites were completely negative, with no investigated tick showing an infection with Babesia sp., while in another one-third of the sampling sites the infection rate ranged from ∼90 to 100%, meaning that 42 to 48 of the 48 ticks investigated for each sampling site were infected with Babesia sp. At the remaining one-third of the sampling sites the infection rates ranged from 30 to 70%. There was no geographical clustering of high or low infection rates (Fig. 1). However, a significant correlation between the altitude of the sampling site and the infection rate of ticks was observed; i.e., more ticks were infected with Babesia spp. at higher altitudes (Pearson coefficient = 0.628 [P = 0.005]; Spearman coefficient = 0.539 [P = 0.021]).
FIG. 1.
Sampling sites in Austria. CZ, Czech Republic; SK, Slovakia; H, Hungary; SLO, Slovenia; I, Italy; CH, Switzerland; D, Germany; Wien, Vienna.
Sequencing of the 18S rRNA gene fragment.
In order to verify the positive PCR results, all 441 PCR products were sequenced. In three cases sequencing failed, probably due to a DNA concentration in the amplicon that was too low; however, for the remaining 438 cases with positive PCR results, the 612-bp 18S rRNA amplicons could be sequenced successfully, and all of the DNA sequences obtained were proven to be Babesia sequences by a BLAST search. Almost all sequences obtained (428/441) showed the highest level of similarity to the B. divergens-B. capreoli-Babesia sp. strain MO1 cluster in a multiple-sequence alignment. Of the eight B. divergens sequences, one B. capreoli sequence, and one Babesia sp. strain MO1 sequence available in the GenBank database, four B. divergens sequences and the B. capreoli and Babesia sp. strain MO1 sequences are identical to one another for the 612-bp gene fragment (accession numbers U16370, AY046576, AY789076, AY144688, AY048113, and AY726009); the remaining sequences (accession numbers AJ439713, Z48751, U07885, and AY572456) differ by 6, 3, 8, and 2 bp from each other and from the six identical sequences. However, the Babesia sp. strain EU1 and Babesia sp. strain DD sequences (accession numbers AY046575, EF185818, and AY553915), which are identical to one another in the 612-bp region, differ from the B. divergens-B. capreoli-Babesia sp. strain MO1 sequence by no more than 5 bp in the gene fragment. One sequence was identical to the Babesia sp. strain EU1-Babesia sp. strain DD sequence. Even after both strands were sequenced several times, five sequences had ambiguous bases at two sequence positions. GeneDoc alignment revealed that ambiguous bases occurred at two positions where the B. divergens-B. capreoli-Babesia sp. strain MO1 and Babesia sp. strain EU1-Babesia sp. strain DD sequences differed in the 18S rRNA gene fragment. Sequence analysis revealed superimposed peaks for the two versions. Four sequences were proven to be Babesia spp. sequences but could not be further classified.
Precise species differentiation was not possible using the 612-bp 18S rRNA gene fragment; consequently, other, more variable molecular markers were chosen in order to obtain more detailed information.
Identification at the species level.
Seven strains, whose 18S rRNA gene fragments were identical to those of the B. divergens positive control (cattle) and B. divergens-B. capreoli-Babesia sp. strain MO1 sequences, were chosen for HSP70 gene PCR and sequencing in order to detect whether there was sequence identity in this more variable gene. Moreover, the HSP70 gene fragments of two of the strains, which had ambiguous bases in the 18S rRNA gene fragment (strains MzN6 and AdN3), as well as the strain whose 18S rRNA gene fragment was identical to the Babesia sp. strain EU1-Babesia sp. strain DD 18S rRNA gene fragment (strain GoA3), were amplified and sequenced.
Six of the seven 333-bp HSP70 gene fragments investigated showed 100% homology with each other and also with the B. divergens positive control and with the B. divergens HSP70 reference sequence (accession number AB248739), which had been obtained from a naturally infected cow from the United Kingdom (38). One strain differed by two bases (G instead of A at nucleotide positions 65 and 163) from the sequence mentioned above; thus, the sequence identity was 99.40%.
The sequences of strains MzN6, AdN3, and GoA3 were identical to one another and showed the highest level of similarity (96.10%) to the B. odocoilei HSP70 sequence (accession number AB248740) but only 92.49% similarity to the B. divergens HSP70 sequence (accession number AB248739). Unfortunately, the B. capreoli, Babesia sp. strain MO1, Babesia sp. strain EU1, and Babesia sp. strain DD HSP70 gene sequences are not available in the GenBank database.
ITS1 and ITS2 sequencing.
Since strains AdN3 and MzN6 had ambiguous bases at two sequence positions in the 18S rRNA gene and their HSP70 gene fragments were identical to the strain GoA3 HSP70 gene fragment, an even more variable molecular marker that included the end of the 18S rRNA gene (118 bp), the complete ITS1 region, the 5.8S rRNA gene, the complete ITS2 region and ∼80 bp of the 28S rRNA gene was chosen for further testing of these strains. Therefore, the ITS1 and ITS2 sequences of strains AdN3 and MzN6 and, for comparison, strain GoA3 and the B. divergens positive control were amplified and determined.
While the entire 978-bp fragments of strains GoA3 and MzN6 were identical, strain AdN3 differed by two bases (G instead of A at nucleotide positions 205 and 407); thus, for strain AdN3 the sequence identity with strains GoA3 and MzN6 was 99.80%. Sequence data for the ITS regions of Babesia sp. strain EU1 and Babesia sp. strain DD are not available in the GenBank database; thus, the 18S rRNA gene, ITS1, 5.8S rRNA gene, ITS2, and 28S rRNA gene of all three strains showed the highest level of homology (89.38%) with B. divergens (accession number EF458177) in a BLAST search. However, the 118-bp 18S rRNA part of the 978-bp fragment (which was subjected to a separate BLAST search) of all three strains showed 100% identity with the Babesia sp. strain DD sequence (accession number AY553519).
Sequencing of the B. divergens positive control gave a 1,044-bp sequence, which showed the highest level of homology (99.80%) to B. divergens (accession number EF458184) for 986 bp of the 18S rRNA gene, ITS1, the 5.8S rRNA gene, ITS2, and the 28S rRNA gene. (The B. divergens positive control sequence differed by 2 bp at nucleotide positions 237 and 823 wobble base M [A and C] instead of A; moreover, the 18S rRNA gene part of the reference sequence is 58 bp shorter and the 28S rRNA gene is 9 bp longer compared with the B. divergens sequence found in the present study.)
DISCUSSION
Infection rates.
The random sample of Austrian ticks (n = 864) investigated for the presence of Babesia spp. had a surprisingly high overall mean infection rate, 51.04%. To our knowledge, this is the first study on the prevalence of Babesia spp. in Austrian ticks. Edelhofer and Baumgartner (11) showed that B. divergens babesiosis is widespread among Austrian cattle, reaching a prevalence of 50%. This is in good agreement with the prevalence in ticks detected in the present study. Studies in other European countries have revealed lower rates of infection of ticks with Babesia spp., with values ranging from 0.9 to 20.6% (4, 8, 17, 18, 32, 34). Tick density is probably the factor determining the infection rate. In our study, we aimed to collect approximately equal numbers of ticks from every sampling site throughout Austria. While at some sites the tick densities were high, at other sites hundreds of square meters had to be covered to obtain the assigned sample size. Thus, ticks (especially the larvae) from a collection site with a high tick density, collected by several blanket drags, were likely to be members of the same population, while at sites with lower tick densities, where many blanket drags and large collection areas were required, ticks were presumably members of several different populations. This would explain why the infection rate was highly variable from collection site to collection site throughout Austria, ranging from 0 to 100%. Typically, at sampling sites with high tick densities either all individuals or no individuals were infected with Babesia spp., while at sampling sites with medium or low tick densities the infection rates were more variable (Table 1 and Fig. 1). Foppa et al. (12) reported a similar observation for B. microti in Switzerland, and they suggested that B. microti is maintained in small focal areas and that the risk of human infection depends on the tick density. As B. divergens and Babesia sp. strain EU1, the two major disease-causing Babesia taxa in Europe, have been shown to be transmitted not only transstadially but also transovarially (2, 3, 15), these taxa are passed on for generations and all offspring of an infected female tick are positive. Thus, one Babesia-infected blood host can be a source of infection that causes a focal distribution of Babesia sp. This conclusion is corroborated by our observation that ticks collected at higher altitudes were more likely to be infected with Babesia spp. than ticks from sampling sites at lower altitudes. At higher altitudes the tick habitat differs from habitats at lower altitudes in terms of both the vegetation and the availability of blood hosts. Ticks are probably dependent on a small number of hosts, and if one of the few host individuals (a single deer can serve as a blood source for a great number of adult ticks [15]) is infected with Babesia spp., it will introduce the pathogen into the entire local tick population and, due to transovarial transmission, the infection will remain in this tick population and will be passed on to any other host individual entering the local area. Interestingly, two recent investigations, one in Slovenia on cervids and one in France on roe deer, showed that in Slovenia 76.50% of the roe deer investigated were infected with either B. divergens (54.90%) or Babesia sp. strain EU1 (21.60%) and 16.70% of the red deer investigated were infected with B. divergens exclusively; the two taxa were nearly identical to bovine B. divergens and a human isolate of Babesia sp. strain EU1 (10). In France, 31 of 79 roe deer were infected with Babesia spp. (3); thus, the risk that female adult ticks would acquire a Babesia infection from cervids is high.
All positive ticks belonged to the species I. ricinus. Of the 853 I. ricinus ticks investigated, 441 were PCR positive for Babesia spp. (51.70%). The 11 H. concinna larvae investigated were negative for Babesia spp. H. concinna has not been demonstrated to be a vector for Babesia spp. yet, although other Haemaphysalis species are known to be vectors for Babesia spp. (6, 7, 28, 37, 39).
Species composition.
It was shown that almost all Babesia spp. found in Austrian ticks belong to the B. divergens-B. capreoli-Babesia sp. strain MO1 cluster. For the seven strains of this cluster for which HSP70 sequences were obtained (all of which had the same 18S rRNA gene fragment), six sequences were identical to one another and also to the B. divergens positive control and the HSP70 B. divergens reference sequence. The remaining strain had a sequence that differed by 2 bp, and thus this strain is classified as B. divergens-like. It cannot be classified as B. divergens as all other B. divergens strains for which sequence data are available are 100% identical for this gene fragment. Altogether, Austrian ticks seem to be predominantly infected with B. divergens or B. divergens-like species rather than with B. capreoli or Babesia sp. strain MO1.
Three strains, strains GoA3, MzN6, and AdN3, were identified as Babesia sp. strain DD since the sequences of the ends of their 18S rRNA genes were identical to the Babesia sp. DD sequence in the GenBank database; the other Babesia species, including the closely related strain EU1, differ by several base pairs in this region. Interestingly, Babesia sp. strain DD has been isolated from I. ricinus ticks in Slovenia, a neighbor of Austria (9). The HSP70 genes of these three strains are identical; however, no sequence data for any other Babesia sp. strain DD isolate are available. The next closest Babesia sp. for which sequence data for HSP70 are available is B. odocoilei. In the ITS regions strain AdN3 differs by 2 bp.
In five cases, including strains MzN6 and AdN3, ticks were infected with two different Babesia species (mixed infections), one belonging to the B. divergens-B. capreoli-Babesia sp. strain MO1 cluster and one belonging to the Babesia sp. strain EU1-Babesia sp. strain DD cluster, as determined by 18S rRNA gene sequencing.
To date, there has been only one documented case of human babesiosis in Austria, and this case was caused by Babesia sp. strain EU1 (21).
In conclusion, about 50% of the investigated Austrian I. ricinus ticks (n = 853) were infected with Babesia spp. Altogether, three different Babesia taxa were detected in the ticks: B. divergens, B. divergens-like, and Babesia sp. strain DD. Moreover, some ticks probably had mixed infections. To our knowledge, this is the first investigation of Babesia spp. in Austrian ticks. Babesia sp. strain DD was detected for the first time in Austria. Furthermore, the sequences described here are the first sequences for Babesia sp. strain DD for ITS1, the 5.8S rRNA gene, ITS2 the beginning of the 28S rRNA gene, and the HSP70 gene.
This study presents important epidemiological data on the prevalence of Babesia spp. in Austrian ticks and should contribute to medical awareness.
Acknowledgments
This study was funded by the European Union (512598-BOVAC; grant FA794A0101).
We thank S. Ölzant, S. Eberhard, N. Ruckenbauer, N. Knopp, M. Köhsler, U. Fürnkranz, G. Duscher, B. Blaschitz, and S. Payrböck for collecting ticks, S. Eberhard for creating the map of sampling sites, M. Zahler of the Institute for Comparative Tropical Medicine and Parasitology, University of Munich, for providing the B. divergens and B. microti DNA, and R. Edelhofer of the Institute for Parasitology and Zoology, University of Veterinary Medicine Vienna, for providing blood of cattle with a B. divergens infection.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Beati, L., and J. E. Keirans. 2001. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J. Parasitol. 87:32-48. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet, S., M. Jouglin, L. Malandrin, C. Becker, A. Agoulon, M. L'Hostis, and A. Chauvin. 2007. Transstadial and transovarial persistence of Babesia divergens DNA in Ixodes ricinus ticks fed on infected blood in a new skin-feeding technique. Parasitology 134:197-207. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet, S., M. Jouglin, M. L'Hostis, and A. Chauvin. 2007. Babesia sp. EU1 from roe deer and transmission within Ixodes ricinus. Emerg. Infect. Dis. 13:1208-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casati, S., H. Sager, L. Gern, and J. C. Piffaretti. 2006. Presence of potentially pathogenic Babesia sp. for humans in Ixodes ricinus in Switzerland. Ann. Agric. Environ. Med. 13:65-70. [PubMed] [Google Scholar]
- 5.Conrad, P. A., A. M. Kjemtrup, R. A. Carreno, J. Thomford, K. Wainwright, M. Eberhard, R. Quick, S. R. Telford III, and B. L. Herwaldt. 2006. Description of Babesia duncani n. sp. (Apicomplexa: Babesiidae) from humans and its differentiation from other piroplasms. Int. J. Parasitol. 36:779-789. [DOI] [PubMed] [Google Scholar]
- 6.Curioni, V., S. Cerquetella, P. Scuppa, L. Pasqualini, T. Beninati, and G. Favia. 2004. Lyme disease and babesiosis: preliminary findings on the transmission risk in highly frequented areas of the Monti Sibillini National Park (central Italy). Vector Borne Zoonotic Dis. 4:214-220. [DOI] [PubMed] [Google Scholar]
- 7.Darghouth, M. A. 2004. Piroplasmids of livestock in Tunisia. Arch. Inst. Pasteur Tunis 81:21-25. [PubMed] [Google Scholar]
- 8.Duh, D., M. Petrovec, and T. Avsic-Zupanc. 2001. Diversity of Babesia infecting European sheep ticks (Ixodes ricinus). J. Clin. Microbiol. 39:3395-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duh, D., M. Petrovec, and T. Avsic-Zupanc. 2005. Molecular characterization of human pathogen Babesia EU1 in Ixodes ricinus ticks from Slovenia. J. Parasitol. 91:463-465. [DOI] [PubMed] [Google Scholar]
- 10.Duh, D., M. Petrovec, A. Bidovec, and T. Avsic-Zupanc. 2005. Cervids as Babesiae hosts, Slovenia. Emerg. Infect. Dis. 11:1121-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edelhofer, R., and W. Baumgartner. 1996. Seroepidemiological studies of bovine Anaplasma marginale and Babesia divergens in Austria, involving autochthonous infections, p. 472-476. In Proceedings of the World Association for Buiatrics Congress, Edinburgh, Scotland, vol. 2. [Google Scholar]
- 12.Foppa, I. M., P. J. Krause, A. Spielman, H. Goethert, L. Gern, B. Brand, and S. R. Telford III. 2002. Entomologic and serologic evidence of zoonotic transmission of Babesia microti, eastern Switzerland. Emerg. Infect. Dis. 8:722-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Sanmartin, J., O. Aurtenetxe, M. Barral, I. Marco, S. Lavin, A. L. Garcia-Perez, and A. Hurtado. 2007. Molecular detection and characterization of piroplasms infecting cervids and chamois in northern Spain. Parasitology 134:391-398. [DOI] [PubMed] [Google Scholar]
- 14.Gorenflot, A., K. Moubri, E. Precigout, B. Carcy, and T. P. Schetters. 1998. Human babesiosis. Ann. Trop. Med. Parasitol. 92:489-501. [DOI] [PubMed] [Google Scholar]
- 15.Gray, J. S. 2002. Biology of Ixodes species ticks in relation to tick-borne zoonoses. Wien Klin. Wochenschr. 114:473-478. [PubMed] [Google Scholar]
- 16.Gray, J. S. 2006. Identity of the causal agents of human babesiosis in Europe. Int. J. Med. Microbiol. 296(Suppl. 40):131-136. [DOI] [PubMed] [Google Scholar]
- 17.Halos, L., T. Jamal, R. Maillard, F. Beugnet, A. Le Menach, H. J. Boulouis, and M. Vayssier-Taussat. 2005. Evidence of Bartonella sp. in questing adult and nymphal Ixodes ricinus ticks from France and co-infection with Borrelia burgdorferi sensu lato and Babesia sp. Vet. Res. 36:79-87. [DOI] [PubMed] [Google Scholar]
- 18.Hartelt, K., R. Oehme, H. Frank, S. O. Brockmann, D. Hassler, and P. Kimmig. 2004. Pathogens and symbionts in ticks: prevalence of Anaplasma phagocytophilum (Ehrlichia sp.), Wolbachia sp., Rickettsia sp., and Babesia sp. in southern Germany. Int. J. Med. Microbiol. 293(Suppl. 37):86-92. [DOI] [PubMed] [Google Scholar]
- 19.Häselbarth, K., A. M. Tenter, V. Brade, G. Krieger, and K. P. Hunfeld. 2007. First case of human babesiosis in Germany—clinical presentation and molecular characterisation of the pathogen. Int. J. Med. Microbiol. 297:197-204. [DOI] [PubMed] [Google Scholar]
- 20.Herwaldt, B., D. H. Persing, E. A. Precigout, W. L. Goff, D. A. Mathiesen, P. W. Taylor, M. L. Eberhard, and A. F. Gorenflot. 1996. A fatal case of babesiosis in Missouri: identification of another piroplasm that infects humans. Ann. Intern. Med. 124:643-650. [DOI] [PubMed] [Google Scholar]
- 21.Herwaldt, B. L., S. Caccio, F. Gherlinzoni, H. Aspock, S. B. Slemenda, P. Piccaluga, G. Martinelli, R. Edelhofer, U. Hollenstein, G. Poletti, S. Pampiglione, K. Loschenberger, S. Tura, and N. J. Pieniazek. 2003. Molecular characterization of a non-Babesia divergens organism causing zoonotic babesiosis in Europe. Emerg. Infect. Dis. 9:942-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hildebrandt, A., K. P. Hunfeld, M. Baier, A. Krumbholz, S. Sachse, T. Lorenzen, M. Kiehntopf, H. J. Fricke, and E. Straube. 2007. First confirmed autochthonous case of human Babesia microti infection in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 26:595-601. [DOI] [PubMed] [Google Scholar]
- 23.Hildebrandt, A., A. M. Tenter, E. Straube, and K. P. Hunfeld. 15 December 2007. Human babesiosis in Germany: just overlooked or truly new? Int. J. Med. Microbiol. doi: 10.1016/j.ijmm.2007.11.001. [DOI] [PubMed]
- 24.Hilpertshauser, H., P. Deplazes, M. Schnyder, L. Gern, and A. Mathis. 2006. Babesia spp. identified by PCR in ticks collected from domestic and wild ruminants in southern Switzerland. Appl. Environ. Microbiol. 72:6503-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunfeld, K. P., and V. Brade. 2004. Zoonotic Babesia: possibly emerging pathogens to be considered for tick-infested humans in Central Europe. Int. J. Med. Microbiol. 293(Suppl. 37):93-103. [DOI] [PubMed] [Google Scholar]
- 26.Kjemtrup, A. M., and P. A. Conrad. 2000. Human babesiosis: an emerging tick-borne disease. Int. J. Parasitol. 30:1323-1337. [DOI] [PubMed] [Google Scholar]
- 27.Langton, C., J. S. Gray, P. F. Waters, and P. J. Holman. 2003. Naturally acquired babesiosis in a reindeer (Rangifer tarandus tarandus) herd in Great Britain. Parasitol. Res. 89:194-198. [DOI] [PubMed] [Google Scholar]
- 28.Mazyad, S. A., and S. A. Khalaf. 2002. Studies on Theileria and Babesia infecting live and slaughtered animals in Al Arish and El Hasanah, North Sinai Governorate, Egypt. J. Egypt. Soc. Parasitol. 32:601-610. [PubMed] [Google Scholar]
- 29.Meer-Scherrer, L., M. Adelson, E. Mordechai, B. Lottaz, and R. Tilton. 2004. Babesia microti infection in Europe. Curr. Microbiol. 48:435-437. [DOI] [PubMed] [Google Scholar]
- 30.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. Embnet News 4:1-4. [Google Scholar]
- 31.Palmer, G. H. 2002. Babesiosis, p. 1-8. In Encyclopedia of life sciences. Macmillan Publishers Ltd., Nature Publishing Group, London, United Kingdom. www.els.net.
- 32.Piccolin, G., G. Benedetti, C. Doglioni, C. Lorenzato, S. Mancuso, N. Papa, L. Pitton, M. C. Ramon, C. Zasio, and G. Bertiato. 2006. A study of the presence of B. burgdorferi, Anaplasma (previously Ehrlichia) phagocytophilum, Rickettsia, and Babesia in Ixodes ricinus collected within the territory of Belluno, Italy. Vector Borne Zoonotic Dis. 6:24-31. [DOI] [PubMed] [Google Scholar]
- 33.Scholtens, R. G., E. H. Braff, G. A. Healey, and N. Gleason. 1968. A case of babesiosis in man in the United States. Am. J. Trop. Med. Hyg. 17:810-813. [DOI] [PubMed] [Google Scholar]
- 34.Skotarczak, B., and A. Cichocka. 2001. Isolation and amplification by polymerase chain reaction DNA of Babesia microti and Babesia divergens in ticks in Poland. Ann. Agric. Environ Med. 8:187-189. [PubMed] [Google Scholar]
- 35.Skrabalo, Z., and Z. Deanovic. 1957. Piroplasmosis in man; report of a case. Doc. Med. Geogr. Trop. 9:11-16. [PubMed] [Google Scholar]
- 36.Telford, S. R., III, and H. K. Goethert. 2004. Emerging tick-borne infections: rediscovered and better characterized, or truly ‘new’? Parasitology 129(Suppl.):S301-S327. [DOI] [PubMed] [Google Scholar]
- 37.Tsuji, N., B. Battsetseg, D. Boldbaatar, T. Miyoshi, X. Xuan, J. H. Oliver, Jr., and K. Fujisaki. 2007. Babesial vector tick defensin against Babesia sp. parasites. Infect. Immun. 75:3633-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamasaki, M., H. Inokuma, C. Sugimoto, S. E. Shaw, M. Aktas, M. J. Yabsley, O. Yamato, and Y. Maede. 2007. Comparison and phylogenetic analysis of the heat shock protein 70 gene of Babesia parasites from dogs. Vet. Parasitol. 145:217-227. [DOI] [PubMed] [Google Scholar]
- 39.Yin, H., W. Lu, J. Luo, Q. Zhang, W. Lu, and H. Dou. 1996. Experiments on the transmission of Babesia major and Babesia bigemina by Haemaphysalis punctata. Vet. Parasitol. 67:89-98. [DOI] [PubMed] [Google Scholar]