Abstract
Cationic biocides (CBs) are widely used in domestic and public hygiene and to control biofouling and microbial contamination in industry. The increased use of biocides has led to concern regarding possible reductions in biocide effectiveness. Domestic drain microcosms were stabilized for 5 months and then exposed to polyhexamethylene biguanide (PHMB) at 0.1, 0.2, and 0.4g liter−1 over 6 months and characterized throughout by differential culture, together with eubacterial-specific PCR-denaturing gradient gel electrophoresis. Additionally, MICs and minimal bactericidal concentrations (MBCs) for bacteria previously isolated from a domestic drain (n = 18) and the human skin (n = 13) were determined before, during, and after escalating, sublethal exposure (14 passages) to two quaternary ammonium compounds (QAC1 and QAC2), the bisbiguanide chlorhexidine (CHX), and PHMB. Exposure of the drain microcosm to PHMB did not decrease the total viable count although significant (P < 0.01) decreases in recovery were observed for the gram-positive cocci with associated clonal expansion of pseudomonads (from ca. 0.1% of the population to ca. 10%). This clonal expansion was also manifested as elevations in bacteria that could grow in the presence of PHMB, CHX, and QAC1. Decreases in susceptibility (greater than twofold) occurred for 10/31 of the test bacteria for QAC1, 14/31 for QAC2, 10/31 for CHX, and 7/31 for PHMB. Exposure of microcosms to PHMB targeted gram-positive species and caused the clonal expansion of pseudomonads. In terms of prolonged-sublethal passage on CBs, exposure to all the biocides tested resulted in susceptibility decreases for a proportion of test bacteria, but refractory clones were not generated.
Cationic biocides (CBs) have been in use since the 1930s for surface disinfection and topical antisepsis (for a review, see reference 15). Broad-spectrum activity and relatively low toxicity have led to increased deployment of these compounds in medicated dressings (37), in contact lens cleaning solutions (5), in swimming pools to control microbial growth (16), and in domestic cleaning products (29). While CBs are a chemically diverse group of compounds, their mode of action normally involves interaction with the cell envelope, displacing divalent cations. Subsequent interactions with membrane proteins and the lipid bilayer depend upon the specific nature of the biocide, but generally CB exposure results in membrane disruption and lethal leakage of cytoplasmic materials (22).
CBs have a range of structures that can be categorized according to the number of cationic groupings per molecule. The quaternary ammonium compounds ([QACs] e.g., cetrimide) are often monocationic surfactants generally containing one quaternary nitrogen associated with at least one major hydrophobic component. The bisbiguanides (e.g., chlorhexidine [CHX]) have two cationic groups separated by a hydrophobic bridging structure (hexamethylene) while the polymeric biguanides (e.g., polyhexamethylene biguanide [PHMB]) are polycationic linear polymers with a hydrophobic backbone and multiple cationic groupings separated by hexamethylene chains (15).
The safety of certain biocide applications has been questioned over the last decade because of the theoretical possibility that chronic exposure of environmental bacteria might select for strains less susceptible either to the agent itself or to other compounds including antibiotics (31, 41). The phenomenon of selectable biocide resistance has been convincingly demonstrated by McMurry et al. (32) using the hydroxydiphenylether biocide triclosan (TCS) to select for mutants of Escherichia coli that exhibit TCS resistance. Other investigators have subsequently repeated this observation (23). These reports have raised the possibility that biocides other than TCS might contribute to the emergence of resistance (2, 12, 43). While the interaction of TCS with a specific target (the enoyl acyl carrier protein reductase) of E. coli may be unique, it is possible to generate clones of some species with decreased QAC susceptibility in pure culture training experiments whereby bacteria are passaged on escalating concentrations of the microbial (8, 17, 19, 20, 26, 39).
Such clones are normally altered in membrane phospholipid content (15) or upregulate the expression of multidrug efflux pumps (36). Since multidrug efflux pump expression has been associated with changes in the MICs of therapeutically important antibiotics, there is the potential for decreases in CB susceptibility, accompanied by reductions in the effectiveness of antibiotics (13, 14) or other biocides (2, 18). A number of studies, however, have questioned such associations (25, 43). Since the 1970s there have been sporadic reports of bacteria with reduced susceptibility within clinical environments that would normally receive long-term exposure to these biocides (7, 10, 42). Importantly, certain bacterial species are innately less susceptible than others; examples include providencia and the pseudomonads (40). Surveying environments for susceptibility profiles is complicated by the fact that it is difficult to differentiate between clonal expansion of these preexisting, innately refractory clones from the arguably more serious mutation and selection for resistance in hitherto susceptible bacteria. Since most biocidal treatments are not limited by considerations of toxicity, moderate changes in susceptibility are unlikely to result in treatment failure.
The increased use of CBs in the domestic, clinical, and industrial environment will expose a wide range of environmental and potentially pathogenic bacteria to these biocides, often at sublethal concentrations. Due diligence clearly calls for the microecological and susceptibility effects of such exposures to be investigated. The aim of the present study, therefore, was to investigate the effect of simulated exposure of environmental and commensal bacteria to QACs, bisbiguanides (CHX), and the polymeric biguanide PHMB. A domestic drain microcosm (27, 28) was chosen to be representative of microbial communities associated with aquatic habitats such as recreational waters and those found in the home and in industry, e.g., food processing. These environments, together with human skin, were identified as being high risk since they all undergo repeated exposure to CBs. In order to investigate the effects of CB exposure on environmental biofilm communities, the domestic drain microcosm was exposed to PHMB for 6 months, and biocide susceptibility profiles and population dynamics were monitored using differential plate counts and PCR-denaturing gradient gel electrophoresis (DGGE). Isolates from the domestic drain microcosm (n = 18) and human skin (n = 13) were repeatedly sublethally exposed to CBs in pure culture using a 14-passage training regime and tested for changes in susceptibility.
MATERIALS AND METHODS
Chemicals.
CHX digluconate was purchased from Sigma (Poole, Dorset, United Kingdom). Bardac 2250 (QAC1), a twin-chain dimethyl ammonium chloride (average molecular weight, 361), and Barquat MB80 (QAC2), a blend of alkyl dimethyl benzyl ammonium chlorides (average molecular weight, 357), were obtained from the Lonza group (Fair Lawn, NJ). Vantocil IB, a 20% aqueous solution of polyhexamethylene biguanide (molecular weight, 2,667; polydispersity, 1.82) was obtained from Arch Chemicals Inc. (Blackley, Manchester, United Kingdom). Formulated bacteriological media were purchased from Oxoid (Basingstoke, United Kingdom). All other chemicals were obtained from Sigma (Poole, Dorset, United Kingdom).
Bacteria.
Achromobacter xylosoxidans MBRG 4.31, Aeromonas hydrophila MBRG 4.3, Aeromonas jandaei MBRG 4.2, Aranicola proteolyticus MBRG 20.1, Bacillus cereus MBRG 4.21, Chryseobacterium sp. strain MBRG 4.28, Chryseobacterium indologenes MBRG 4.29, Citrobacter sp. strain MBRG 20.9, Enterococcus saccharolyticus MBRG 20.4, Eubacterium sp. strain MBRG 4.14, Halonella gallinarum MBRG 4.27, Microbacterium phyllosphaerae MBRG 4.30, Pseudomonas nitroreductans MBRG 4.6, Pseudomonas sp. strain MBRG 4.7, Pseudoxanthomonas sp. strain MBRG 40.1, Ralstonia sp. strain MBRG 4.13, Sphingobacterium multivorum MBRG 30.1, and Stenotrophomonas maltophilia MBRG 4.17 were isolated from a domestic drain microcosm (28). Corynebacterium pseudogenitalum MBRG 9.24, Corynebacterium renale MBRG 9.26, Micrococcus luteus MBRG 9.25, Staphylococcus capitis MBRG 9.34, Staphylococcus capral MBRG 9.3, Staphylococcus cohnii MBRG 9.31, Staphylococcus epidermidis MBRG 9.33, Staphylococcus haemolyticus MBRG 9.35, Staphylococcus hominis MBRG 9.37, Staphylococcus kloosii MBRG 9.28, Staphylococcus lugdenesis MBRG 9.36, Staphylococcus saccharolyticus MBRG 9.32, Staphylococcus saprophyticus MBRG 9.29, and Staphylococcus warneii MBRG 9.27 were obtained from the axillae of three male volunteers, ranging from 25 to 30 years old (23).
Domestic drain microcosms.
Domestic drain microcosms were established as described previously (28). Developed communities were characterized periodically over the course of the investigation by differential culture at the time of sampling; samples were also archived for subsequent PCR-DGGE analysis.
Addition of PHMB to microcosms.
Microcosms were stabilized for 6 months, after which dilutions of PHMB (0.1 g liter−1, 0.2 g liter−1, and 0.4 g liter−1) were sequentially added for 2 months each using a peristaltic pump for 10 min at 6-h intervals at a flow rate of 55.2 ml h−1.
Bacterial characterization by culture.
Constant-depth film fermentor plugs (two) were macerated with a sterile mortar and pestle, homogenized, and diluted 1:10. For enumeration, dilutions of macerated drain or model biofilm (1:10) were serially diluted with prereduced half-strength peptone-water (7.5 g liter−1). During long-term experiments in order to minimize variation due to sampling of immature biofilms, only those constant-depth film fermentor pans that had been in situ for at least 1 month were removed for analysis. Aliquots (0.1 ml) of appropriate dilutions were plated in triplicate onto a variety of selective and nonselective media (Oxoid, Basingstoke, United Kingdom) as follows: MacConkey agar number 3 (enteric organisms), mannitol salts agar (gram-positive cocci), pseudomonas isolation agar (total pseudomonads), R2A (aerobic and facultative heterotrophs), and R2A supplemented with Bardac (100 μg ml−1), CHX (100 μg ml−1), or PHMB (100 μg ml−1). Plates were incubated aerobically for up to 5 days, except for the Wilkins Chalgren plates, which were incubated in an anaerobic cabinet (atmosphere, 10:10:80, H2-CO2-N2).
Direct bacterial cell counts.
The proportion of the viable bacterial communities that could be cultured by the methods used above was estimated by comparison with vital staining and direct microscopy. A subsample (100 μl) of an appropriate dilution of macerated domestic drain microcosm was stained with a live-dead bacterial viability kit (BacLight; Molecular Probes, Leiden, The Netherlands) and counted with an improved Neubauer counting chamber in conjunction with fluorescence microscopy with a 100-W mercury vapor lamp. Live (green fluorescent) and dead (red fluorescent) cells were visualized separately with fluorescein and Texas red band-pass filters, respectively, in accordance with the manufacturer's instructions.
DNA extraction from microcosm biofilms.
Archived biofilm material (0.2 to 0.5 g) was mixed with 1 ml of Tris buffer (0.12 M; pH 8.0), vortex mixed, and subjected to two cycles of freezing and heating (−60°C for 10 min and 60°C for 2 min). Samples were then transferred to a bead-beater vial containing 0.3 g of sterile zirconia beads (0.1-mm diameter). Tris-equilibrated phenol (pH 8.0; 150 μl) was added, and the suspension was shaken three times for 80 s at maximum speed (Mini-Bead-Beater; Biospec Products, Bartlesville, OK). After 10 min of centrifugation at 13,000 × g, the supernatant was extracted three times with an equal volume of phenol-chloroform and once with chloroform-isoamyl alcohol (24:1 [vol/vol]). The DNA was precipitated from the aqueous phase with 3 volumes of ethanol, air dried, and resuspended in 100 μl of deionized water. The amount and quality of DNA extracted were estimated by electrophoresis of 5-μl aliquots on a 0.8% agarose gel and by comparison to a molecular weight standard (stained with ethidium bromide). DNA extracts were stored at −60°C prior to analysis.
PCR amplification for DGGE analysis.
The V2-V3 variable region of the 16S rRNA gene (corresponding to positions 339 to 539 of E. coli) was amplified with the eubacterium-specific primers HDA1-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′) as previously described (44). The reactions were performed in 0.2-ml tubes with a DNA thermal cycler (model 480; Perkin-Elmer, Cambridge, United Kingdom). In all cases, reactions were carried out with Red Taq DNA polymerase ready mix (25 μl; Sigma, Poole, Dorset, United Kingdom), HDA primers (2 μl of each; 5 μM), nanopure water (16 μl), and extracted community DNA (5 μl). Quantification and standardization of extracted DNA were achieved with a fluorescence assay (DNA Quantitation Kit; Sigma, Poole, Dorset, United Kingdom), in accordance with the manufacturer's instructions. The thermal program was as follows: 94°C for 4 min, followed by 30 thermal cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 60 s. The final cycle incorporated a 7-min chain elongation step (68°C).
DGGE analysis.
Biofilm samples were analyzed by DGGE with a D-Code Universal Mutation Detection System (Bio-Rad, Hemel Hempstead, United Kingdom). Polyacrylamide (8%) gels (16 by 16 cm; 1 mm deep) were run with 1× Tris-acetate-EDTA (TAE) buffer diluted from 50× TAE buffer (40 mM Tris base, 20 mM glacial acetic acid, 1 mM EDTA). Initially, separation parameters were optimized by running PCR products from selected pure cultures of drain bacteria and PCR amplicons from extracted drain DNA on gels with a 0 to 100% denaturation gradient, perpendicular to the direction of electrophoresis (a 100% denaturing solution contained 40% [vol/vol] formamide and 7.0 M urea). Denaturing gradients were formed with two 10% acrylamide (acrylamide-bisacrylamide ratio, 37.5:1) stock solutions (Sigma, Poole, Dorset, United Kingdom). On this basis, a denaturation gradient for parallel DGGE analysis ranging from 30 to 60% was selected for community analyses. DNA for loading onto gels was quantified and, when necessary, standardized between samples with a fluorescence assay (see above). Electrophoresis was carried out at 150 V at 60°C for approximately 4.5 h. All gels were stained with Sybr Gold stain (diluted to 10−4 in 1×TAE buffer; Molecular Probes, Leiden, The Netherlands) for 30 min. Gels were viewed, and images were documented with a BioDocit system (UVP, CA).
Sequencing of bacterial isolates and excised gel bands.
For analysis of the major resolved DGGE amplicons, selected resolved bands were cut out of the polyacrylamide gels with a sterile scalpel under UV illumination and incubated at 4°C for 20 h together with 20 μl of nanopure water in nuclease-free universal bottles. Portions (5 μl) were removed and used as a template for a PCR identical to that outlined for DGGE analysis. PCR products were purified with QIAquick PCR purification kits (Qiagen Ltd., West Sussex, United Kingdom) and sequenced with the reverse (non-GC clamp) primer (HDA2). The sequencing was at 94°C for 4 min followed by 25 cycles of 96°C for 30 s, 50°C for 15 s, and 60°C for 4 min. Once chain termination was complete, sequencing was done in a Perkin-Elmer ABI 377 sequencer. DNA sequences were compiled with Chromas-Lite (Technelysium Pty. Ltd., Helensville, Queensland, Australia) to obtain consensus sequences or to check and edit unidirectional sequences. For excised DGGE band PCRs, the fidelity of derived sequences was used as an indictor of the purity of the target sequence, and the presence of a GC clamp upon sequence analyses confirmed that the correct target, rather than a contaminant, had been reamplified.
Phylogenetic identification of DGGE amplicons.
BLAST searches on each sequence were done against sequences in the EMBL nucleotide sequence database.
Repeated exposure of bacterial monocultures to biocides.
A model CU spiral plater (Spiral Systems, Cincinnati, OH) was used to create reproducible radial biocide concentration gradients across agar plates. This spiral plater acts as a dispenser, which distributes liquid onto the surface of a rotating agar plate from the center to the edge in an Archimedes spiral, establishing an approximately 100-fold concentration gradient, with antibacterial concentration decreasing from the center to the edge. To enable repeated, sublethal exposure of the test bacteria to the biocides, aqueous stock solutions of test agents QAC1, QAC2, CHX, and PHMB were prepared, filter sterilized (0.2-μm pore size; Millipore, United Kingdom), and stored at −60°C. Petri dishes (10-cm diameter) were filled with 27.5 ± 1.0 ml of nutrient agar (skin isolates) or R2A agar (environmental species) to produce a mean agar depth of ca. 3.5 mm. The plates were kept for 2 days at room temperature prior to use to ensure dryness of the agar surface. Volumes of biocide stock solutions (50 μl) were then deposited onto the agar surface using the variable cam of the spiral plater. Plates were then dried for up to 1 h at room temperature prior to radial deposition of inocula (20 μl of a ca. 108 CFU ml−1 culture) using a sterile inoculation loop. After further drying (1 h), plates were inverted and incubated appropriately for up to 4 days at 37°C. After incubation, bacterial growth observed near to the endpoint in the transition between the growth and growth-inhibition area was aseptically removed and homogenized in Muller Hinton broth (1 ml). A 20-μl portion was then transferred to the next gradient plate (prepared as above) until 14 passages had been completed. Initially, biocides were deposited at 1 mg ml−1. Where no endpoint was achieved either on first exposure or at any point during the process, biocide concentration was increased by 10 mg ml−1, and exposure was repeated. Unexposed bacteria (before) and those harvested after 7 and 14 biocide passages (P7 and P14, respectively) were frozen at −60°C for subsequent MIC and minimum bactericidal concentration (MBC) testing.
MIC determination.
Inocula for microtiter plate determination of bacterial antimicrobial susceptibility were prepared as follows: single colonies of test bacteria were inoculated into sterile Muller Hinton broth (10 ml) within sterile plastic universal bottles (25-ml total volume) and incubated in a standard aerobic incubator. Based on previous validation studies, batch cultures were incubated at 37°C for 24 h (±2 h), until they were entering early stationary phase. The cultures were then diluted to ca. 105 CFU ml−1 in sterile broth for use as inocula in the MIC and MBC tests (see below). Stock solutions (4 mg ml−1) of antimicrobials were prepared in distilled water. Test agents were as follows: QAC1, QAC2, CHX, and PHMB. In order to reduce experimental variation, the total volume of each antimicrobial solution for these studies was prepared in advance, sterilized by filtration through single cellulose acetate filters (0.2-μm pore size; Millipore, United Kingdom), separated into aliquots (1.5 ml), and stored at −60°C. Testing was performed in 96-well microtiter plates (Becton Dickinson, Oxford, United Kingdom). Initial concentrations of the antimicrobial agent were 1 mg ml−1. Diluted overnight culture (100 μl) was delivered to each test well. Antimicrobial solution (100 μl) was added to the first column of the test organism and mixed. Doubling dilutions were then carried out across the plate using a multichannel pipette, changing the tips at each dilution step. The plates were then incubated for 48 h in a standard incubator at 37°C. MICs were expressed as the lowest concentrations of antimicrobial at which growth did not occur. Growth was detected as turbidity (495 nm) relative to an uninoculated well using a microtiter plate reader (Anthos HTII; Anthos-Labtec Instruments, Salzburg, Austria). Each MIC determination was carried out in triplicate (in the same 96-well plate). Negative controls were performed with only sterile broth in each well, and positive controls were performed with only overnight culture in the wells.
MBC determinations.
MBC testing was carried out using the microtiter plates set up for the MIC determinations. Aliquots (10 μl) taken from each well up to and including the MIC endpoint were transferred and spot plated onto the appropriate agar and incubated overnight. MBCs were expressed as the lowest concentration of biocide at which growth was not observed after 5 days of incubation.
RESULTS AND DISCUSSION
In this study, a dual approach was used to study the effect of CB exposure upon biofilms and pure cultures of commonly exposed bacteria. First, a domestic drain biofilm microcosm was dosed with increasing concentrations of PHMB over 6 months, allowing effects on community dynamics and biocide-resistant populations to be determined. Second, bacteria that had been isolated from two high-risk environments, the skin and a domestic drain biofilm, were repeatedly exposed to either QACs, bisbiguanides, or polymeric biguanides in vitro. The microcosm system has been previously used to investigate the effects of proprietary TCS (27) and QAC-containing detergent on domestic drain biofilm communities (29). The monoculture passage-based training regime has previously been validated for TCS, where the method selected for TCS-refractory clones of E. coli in two passages (23, 30). By applying both methods independently, it is possible to gain insights into the potential effects of prolonged sublethal exposure of biofilms to PHMB and to determine whether any changes in the population dynamics observed can be attributed to either clonal expansion of innately insusceptible bacteria or a change in the susceptibility of the original population. Since the majority of bacteria exposed to biocides in the environment will be associated with communities, the first approach has the advantage of closely replicating the real-life situation, enabling ecological effects of antimicrobial treatments to be tested. For reasons such as biocide penetration gradients, competitive phenomena, and technical issues relating to microbial analysis of complex communities, however, they give less information about the possible occurrence of resistance at the level of individual clones.
Effects of PHMB exposure on biofilm microcosms.
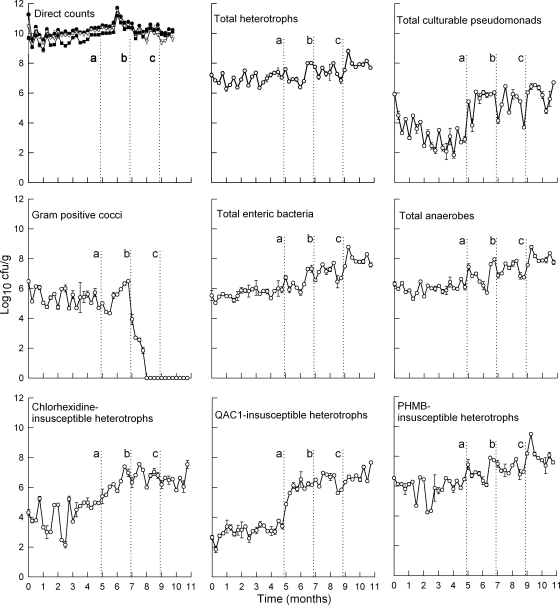
Direct live and dead counts, together with viable counts of functional groups of bacteria of the PHMB-exposed microcosms, are presented in Fig. 1. The total count reached a steady state after 2 to 3 weeks and remained in a dynamic steady state throughout the various exposures to PHMB. Differential viable counting over 6 months, together with PCR-DGGE analysis (Fig. 2), confirmed that the microcosms achieved dynamic steady states prior to dosing. These data also show that overall bacterial viability remained largely unaffected by the addition of PHMB to the system. Considerable fluxes occurred, however, within the pseudomonads, which increased on first exposure to PHMB from ca. 0.1% of the population to ca. 10%, while numbers of the highly susceptible gram-positive cocci were markedly reduced. The recovery of enteric and anaerobic bacteria remained stable after PHMB addition, suggesting that these groups of bacteria are largely unaffected by prolonged PHMB exposure. In order to monitor the number of less-susceptible species present, viable counts were carried out on agars containing biocides (Fig. 1); interestingly the number of viable heterotrophs capable of tolerating 100 μg ml−1 of CHX and PHMB markedly increased over the PHMB exposure period, as did those that can grow in the presence of 100 μg ml−1 of QAC1 (1,000-fold; P < 0.05). This increase was rapidly (within 3 weeks) manifested after PHMB dosing commenced and remained relatively constant when the concentration of PHMB was increased to 0.4 mg ml−1 (Fig. 1). Congruence between these data and total, viable pseudomonad counts further demonstrates the relatively low susceptibility of certain gammaproteobacteria toward CBs (3, 6, 40). The fact that such bacteria were present before dosing, albeit at low levels, suggests that these fluxes did not result from clonal changes in susceptibility. Importantly, the numbers of bacteria able to grow in the presence of 100 μg ml−1 PHMB were relatively high within the microcosms before dosing commenced, increasing from approximately 10-fold after the addition of PHMB to the bioreactor. These ecological effects can be largely predicted based on the relative susceptibility of these groups of bacteria. The MIC and MBC data for the largely gram-negative environmental isolates (Tables 1 to 4) and the gram-positive skin isolates (Tables 5 to 8) indicate that many of the gram-negative environmental isolates are more than 10 times less susceptible than the staphylococci. They also highlight the fact that while simple total counts may appear stable, they may mask major dynamic changes within microbial communities.
FIG. 1.
Total and viable counts of domestic drain microcosm biofilms before and throughout exposure to increasing amounts of PHMB, as indicated by the dotted lines (mg ml−1): a, 0.1; b, 0.2; and c, 0.4. Data are means ± standard deviations from two separate sample pans analyzed in triplicate. Filled circles, total cell count; open triangles, vital cell count; filled squares, dead cell count (BacLight); open circles, viable cell count (plate counts).
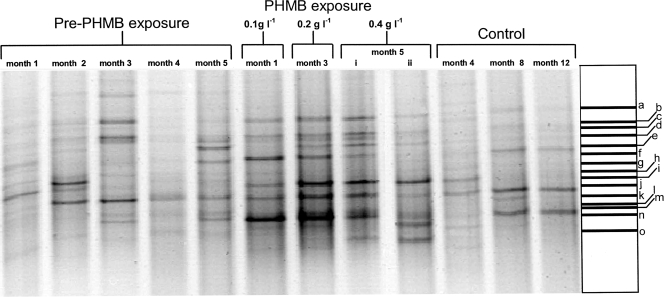
FIG. 2.
Negative image of a parallel DGGE gel showing eubacterial community fingerprints for microcosm samples before PHMB addition and after a 2-month exposure to PHMB at 0.1, 0.2, and 0.4 g liter−1 and control untreated fermentor. Excised band identities (a to o) are given in Table 9.
TABLE 1.
MICs and MBCs of bacteria isolated from a domestic drain biofilm before and after chronic QAC1 exposurea
| Strain | MIC (μg ml−1)
|
MBC (μg ml−1)
|
||||
|---|---|---|---|---|---|---|
| Before treatment | P7 | P14 | Before treatment | P7 | P14 | |
| Achromobacter xylosoxidans MBRG 4.31 | 3.9 | 3.9 | 3.9 | 3.9 | 3.9 | 7.8 |
| Aeromonas hydrophila MBRG 4.3 | 15.6 | 15.6 | 15.6 | 31.2 | 41.6 (18) | 31.2 |
| Aeromonas jandaei MBRG 9.11 | 15.6 | 15.6 | 15.6 | 52 (18) | 31.2 | 83 (36) |
| Aranicola proteolyticus MBRG 9.12 | 3.9 | 31.2 | 125 | 48.4 (23.8) | 83.3 (36) | 125 |
| Bacillus cereus MBRG 4.21 | 3.9 | 3.9 | 3.9 | 31.2 | 31.2 | 62.5 |
| Chryseobacterium indologenes MBRG 9.15 | 15.6 | 15.6 | 15.6 | 31.2 | 26 (9) | 31.2 (27) |
| Chryseobacterium sp. strain MBRG 9.17 | 3.9 | 3.9 | 3.9 | 3.9 | 3.9 | 41.6 (18) |
| Citrobacter sp. strain MBRG 9.18 | 7.8 | 7.8 | 7.8 | 10.4 (4.5) | 44.2 (31.5) | 7.8 |
| Enterococcus saccharolyticus MBRG 9.16 | 31.2 | 7.8 | 7.8 | 61.2 | 61.2 | 61.2 |
| Eubacterium strain MBRG 4.14 | 15.6 | 15.6 | 31.2 | 20.8 (9) | 31.2 | 31.2 |
| Halonella gallinarum MBRG 4.27 | 15.6 | 15.6 | 15.6 | 20.8 (9) | 15.6 | 20.8 (9) |
| Microbacterium phyllosphaerae MBRG 4.30 | 3.9 | 3.9 | 3.9 | 14.3 (14.7) | 15.6 (13.5) | 7.8 |
| Pseudomonas nitroreductans MBRG 4.6 | 15.6 | 7.8 | 7.8 | 57.2 (59.1) | 31.2 (27) | 57.2 (59.1) |
| Pseudomonas sp. strain MBRG 9.14 | 15.6 | 15.6 | 31.2 | 52 (63.1) | 88.5 (63.1) | 72.9 (47.7) |
| Pseudoxanthomonas sp. strain MBRG 9.20 | 7.8 | 3.9 | 3.9 | 15.6 | 3.9 | 3.9 |
| Ralstonia sp. strain MBRG 4.13 | 7.8 | 31.2 | 125 | 125 | 104.1 (36) | 187 (108.2) |
| Sphingobacterium multivorum MBRG 9.19 | 3.9 | 1.9 | 7.8 | 3.9 | 1.9 | 62.5 |
| Stenotrophomonas maltophilia MBRG 9.13 | 7.8 | 7.8 | 7.8 | 31.2 | 31.2 | 31.2 |
Data were determined by broth dilution endpoint (doubling dilutions). Data show means from duplicate experiments, each performed in replicate (n = 3). Where data varied between replicates, standard deviations are given in parenthesis. MICs and MBCs that increased more than twofold are indicated in boldface.
TABLE 4.
MICs and MBCs of bacteria isolated from a domestic drain biofilm before and after chronic PHMB exposurea
| Strain | MIC (μg ml−1)
|
MBC (μg ml−1)
|
||||
|---|---|---|---|---|---|---|
| Before treatment | P7 | P14 | Before treatment | P7 | P14 | |
| Achromobacter xylosoxidans MBRG 4.31 | 15.6 | 3.9 | 3.9 | 15.6 | 7.8 | 3.9 |
| Aeromonas hydrophila MBRG 4.3 | 31.2 | 31.2 | 31.2 | 52 (36) | 52 (36) | 31.2 |
| Aeromonas jandaei MBRG 9.11 | 31.2 | 31.2 | 31.2 | 41.6 (36) | 41.6 (36) | 208 (226) |
| Aranicola proteolyticus MBRG 9.12 | 7.8 | 125 | 125 | 20.8 (9) | 125 | 166 (144) |
| Bacillus cereus MBRG 4.21 | 20.8 (9) | 7.8 | 20.8 (9) | 31.25 | 62.5 | 62.5 |
| Chryseobacterium indologenes MBRG 9.15 | 3.9 | 3.9 | 3.9 | 7.8 | 7.8 | 7.8 |
| Chryseobacterium sp. strain MBRG 9.17 | 15.6 | 15.6 | 15.6 | 15.6 | 15.6 | 15.6 |
| Citrobacter sp. strain MBRG 9.18 | 31.2 | 15.6 | 15.6 | 31.2 | 7.8 | 7.8 |
| Enterococcus saccharolyticus MBRG 9.16 | 31.2 | 15.6 | 20.8 (9) | 166 (144) | 166 (144) | 208 (72) |
| Eubacterium strain MBRG 4.14 | 7.8 | 7.8 | 7.8 | 20.8 (9) | 20.8 (9) | 7.8 |
| Halonella gallinarum MBRG 4.27 | 7.8 | 3.9 | 3.9 | 44.25 (63) | 104 (36) | 13 (9) |
| Microbacterium phyllosphaerae MBRG 4.30 | 7.8 | 15.6 | 15.6 | 36.4 (23) | 15.6 | 15.6 |
| Pseudomonas nitroreductans MBRG 4.6 | 15.6 | 15.6 | 15.6 | 15.6 | 15.6 | 52 (36) |
| Pseudomonas sp. strain MBRG 9.14 | 7.8 | 7.8 | 7.8 | 20.5 (18) | 20.5 (18) | 31 (54) |
| Pseudoxanthomonas sp. strain MBRG 9.20 | 15.6 | 7.8 | 7.8 | 20.5 (18) | 7.8 | 7.8 |
| Ralstonia sp. strain MBRG 4.13 | 7.8 | 7.8 | 3.9 | 41.6 (36) | 15.6 | 26 (63.1) |
| Sphingobacterium multivorum MBRG 9.19 | 20.8 (9) | 7.8 | 7.8 | 7.8 | 7.8 | 7.8 |
| Stenotrophomonas maltophilia MBRG 9.13 | 3.9 | 7.8 | 7.8 | 7.8 | 7.8 | 15.6 |
For an explanation of the data, see the footnote to Table 1.
TABLE 5.
MICs and MBCs of bacteria isolated from human skin organisms before and after chronic QAC1 exposurea
| Strain | MIC (μg ml−1)
|
MBC (μg ml−1)
|
||||
|---|---|---|---|---|---|---|
| Before treatment | P7 | P14 | Before treatment | P7 | P14 | |
| Corynebacterium pseudogenitalum MBRG 9.24 | 7.8 | 31.2 | 62.5 | 13 (9) | 83 (36) | 62.5 |
| Corynebacterium renale group MBRG 9.13 | 3.9 | 3.9 | 15.6 | 31.2 | 62.5 | 125 |
| Micrococcus luteus MBRG 9.25 | 0.45 | 0.45 | 1.6 (0.56) | 6.4 (2.4) | 3.9 | 3.9 |
| Staphylococcus capitis MBRG 9.34 | 1.3 (0.56) | 0.97 | 2.6 (1.1) | 4.5 (2.9) | 11 (3.9) | 10 (4.5) |
| Staphylococcus capral MBRG 9.30 | 0.81 (0.2) | 0.65 (0.2) | 1.3 (0.56) | 3.2 (1.1) | 1.9 | 1.9 |
| Staphylococcus cohnii MBRG 9.31 | 0.45 | 0.81 (0.2) | 0.45 | 6.4 (2.4) | 5.2 (2.2) | 2.6 (1.1) |
| Staphylococcus epidermidis M 9.33 | 0.48 | 0.65 (0.2) | 0.45 | 2.6 (1.1) | 0.97 | 3.9 |
| Staphylococcus haemolyticus MBRG 9.35 | 1.6 (0.56) | 0.97 | 1.9 | 1.9 | 11.7 (6.7) | 9.1 (5.9) |
| Staphylococcus hominis MBRG v9.37 | 0.48 | 0.48 | 0.81 | 1.9 | 1.9 | 1.9 |
| Staphylococcus kloosii MBRG 9.28 | 0.65 (0.2) | 0.45 | 0.45 | 5.2 (2.2) | 6.4 (2.4) | 5.2 (2.2) |
| Staphylococcus lugdenensis MBRG 9.36 | 0.81 (0.2) | 0.48 | 1.9 | 3.9 | 1.9 | 18 (11) |
| Staphylococcus saprophyticus MBRG 9.29 | 0.45 | 0.45 | 0.45 | 6.4 (2.4) | 1.9 | 6.4 (2.4) |
| Staphylococcus warneii MBRG 9.27 | 1.3 (0.5) | 0.48 | 0.45 | 1.9 (3.3) | 0.97 | 1.9 |
For an explanation of the data, see the footnote to Table 1.
TABLE 8.
MICs and MBCs of bacteria isolated from human skin before and after chronic PHMB exposurea
| Strain | MIC (μg ml−1)
|
MBC (μg ml−1)
|
||||
|---|---|---|---|---|---|---|
| Before treatment | P7 | P14 | Before treatment | P7 | P14 | |
| Corynebacterium pseudogenitalum MBRG 9.24 | 1.9 | 1.9 | 1.9 | 7.8 | 7.8 | 7.8 |
| Corynebacterium renale group MBRG 9.13 | 3.9 | 3.9 | 10 (4) | 31.2 | 31.2 | 62.5 |
| Micrococcus luteus MBRG 9.25 | 0.97 | 0.97 | 0.97 | 3.9 (6.7) | 5.2 (2.2) | 3.9 (6.7) |
| Staphylococcus capitis MBRG 9.34 | 3.9 | 3.9 | 1.9 | 36 (23) | 41 (18) | 13 (9) |
| Staphylococcus capral MBRG 9.30 | 7.8 | 1.9 | 3.9 | 72 (47) | 41 (18) | 41 (18) |
| Staphylococcus cohnii MBRG 9.31 | 1.9 | 0.97 | 6.4 (2.4) | 23 (13) | 0.97 | 33 (27) |
| Staphylococcus epidermidis MBRG 9.33 | 1.9 | 3.9 | 3.9 | 13 (9) | 15.6 | 26 (18) |
| Staphylococcus haemolyticus MBRG 9.35 | 7.8 | 7.8 | 7.8 | 13 (9) | 104 (36) | 83 (36) |
| Staphylococcus hominis MBRG 9.37 | 7.8 | 3.9 | 3.9 | 26 (18) | 41 (18) | 15.6 |
| Staphylococcus kloosii MBRG 9.28 | 3.9 | 3.9 | 3.9 | 15.6 | 31.2 | 23 (13) |
| Staphylococcus lugdenensis MBRG 9.36 | 3.9 | 7.8 | 3.9 | 36 (23) | 62 (54) | 31.2 |
| Staphylococcus saprophyticus MBRG 9.29 | 3.9 | 1.9 | 3.9 | 20.8 (9) | 18 (10) | 13 (9) |
| Staphylococcus warneii MBRG 9.27 | 7.8 | 7.8 | 7.8 | 72 (47) | 62.5 | 31.2 |
For an explanation of the data, see the footnote to Table 1.
DGGE analysis.
Fig. 2 shows DGGE gel images corresponding to selected time points and exposure conditions. Control DGGE bacterial fingerprints, along with those from microcosms exposed to PHMB, are presented together with the assignments for the dominant bands made on the basis of sequence homology. Data suggest that the microcosm maintained dynamic stability prior to the addition of PHMB and that biocide exposure increasing from 0.1 g liter−1 to 0.2 g liter−1 and then 0.4 g liter−1 over 6 months caused some alterations in community profile, including increases in bacteria related to Herbaspirillum sp., Arcicella sp., and Cupriavadus necator. Detection of these bacteria during PHMB exposure may be indicative of general increases in gram-negative bacteria which are known to exhibit lower PHMB susceptibility. Data in Table 9 show closest relatives on the basis of BLAST searches with DNA sequences obtained from major DGGE gel bands. The major amplicons detected once PHMB was added to the system correspond to gram-negative environmental isolates such as Chryseobacterium sp., Cytophaga sp., Deinococcus sp., Flectobacillus sp., and Herbaspirillum sp.
TABLE 9.
Characterization of dynamic changes in a domestic drain microcosm on the basis of sequences of dominant PCR amplicons derived from DGGE gels
| Band | Closest relative (% sequence similarity)a | Sequence length (bp)b |
|---|---|---|
| a | Flectobacillus sp. strain AY308840 (97) | 173 (13) |
| b | Flectobacillus sp. strain AY423898 (100) | 176 (3) |
| c | Herminiimonas arsenicoxydans CU207211 (61) | 185 (0) |
| d | Deinococcus cellulolytica DQ883809 (80) | 175 (5) |
| e | Flectobacillus sp. strain AY30884085 (98) | 177 (0) |
| f | Bacteroidetes strain AY509355 (100) | 180 (3) |
| g | Burkholderia xenovorans AB186240 (71) | 179 (2) |
| h | Chryseobacterium sp. strain AY779536 (89) | 181 (2) |
| i | Herbaspirillum sp. strain AY766356 (96) | 176 (2) |
| j | Microscilla sp. strain AY849869 (96) | 180 (1) |
| k | Cytophaga sp. strain AF125326 (100) | 180 (1) |
| l | Deinococcus sp. strain AY826637 (72) | 190 (2) |
| m | Flavobacterium sp. strain AJ508710 (72) | 180 (2) |
| n | Arcicella sp. strain AJ746140 (80) | 187 (2) |
| o | Cupriavidus necator AB015605 (100) | 180 (3) |
Based on EMBL database searches.
The number of ambiguous bases are given in parenthesis.
Previous studies using domestic drain microcosms to investigate the effects of TCS (27) and QAC-containing domestic detergents (29) demonstrated that these formulations also caused clonal expansion of intrinsically nonsusceptible gram-negative organisms rather than changes in the susceptibility of established clones. In the current investigation, dosing of the drain microcosm indicated that recalcitrant organisms such as pseudomonads increased in number during exposure to PHMB (Fig. 1), but effects at the level of individual clones are better elucidated in pure culture. A series of experiments was therefore performed using bacteria isolated from a domestic drain (28) and human skin.
Effect of CB exposure of bacteria isolated from a domestic drain microcosm.
The average MICs prior to passage were as follows: 10.8 μg ml−1(QAC1), 24.6 μg ml−1 (QAC2), 16.2 μg ml−1 (CHX), and 15.7 μg ml−1 (PHMB). After exposing the test bacteria to a range of CBs over 14 passages, 12 species exhibited a more than twofold decrease in biocide susceptibility (Tables 1 to 4). These were Aeromonas hydrophila (QAC2), Aranicola proteolyticus (QAC1, QAC2, and CHX), Chryseobacterium (QAC1 and QAC2), Citrobacter sp. (QAC1), Eubacterium (QAC2), Halonella gallinarum (CHX), Pseudomonas sp. (QAC2 and CHX), Pseudoxanthomonas sp. (QAC2) Ralstonia sp. (QAC1 and CHX), Sphingobacterium multivorum (QAC1 and QAC2), and Stenotrophomonas maltophilia (CHX). In terms of exposing the test bacteria to PHMB, four species showed a greater than twofold decrease in their susceptibility after passage (Table 4). With respect to the identities of those organisms which changed susceptibility, members of the pseudomonads and related genera are known to be intrinsically less susceptible to biocides (40), and this is reflected in the distribution of CB susceptibilities among exposed bacteria (Tables 1 to 4). The mechanisms through which these species with lower susceptibility before CB exposure mediate further susceptibility reductions may relate to efflux pumps or cell surface characteristics (9, 24, 34, 38). Reductions in susceptibility could therefore arise from membrane alterations or from the up-regulation of efflux pumps (35). CB exposure also resulted in significant increases in susceptibility in some cases, for example, Pseudoxanthomonas sp. with CHX (Table 3), which is probably due to sublethal cell damage during the primary biocide exposure.
TABLE 3.
MICs and MBCs of bacteria isolated from a domestic drain biofilm before and after chronic CHX exposurea
| Strain | MIC (μg ml−1)
|
MBC (μg ml−1)
|
||||
|---|---|---|---|---|---|---|
| Before treatment | P7 | P14 | Before treatment | P7 | P14 | |
| Achromobacter xylosoxidans MBRG 4.31 | 15.6 | 31.2 | 31.2 | 166 (18) | 41.6 (36) | 31.2 |
| Aeromonas hydrophila MBRG 4.3 | 31.2 | 15.6 | 15.6 | 31.2 | 15.6 | 31.2 |
| Aeromonas jandaei MBRG 9.11 | 7.8 | 15.6 | 15.6 | 36.4(23) | 31.2 | 15.6 |
| Aranicola proteolyticus MBRG 9.12 | 7.8 | 62.5 | 125 | 52 (36) | 125 | 250 |
| Bacillus cereus MBRG 4.21 | 1.9 | 1.9 | 1.9 | 7.8 | 7.8 | 7.8 |
| Chryseobacterium indologenes SB1 MBRG 9.15 | 26 (18) | 31.2 | 31.2 | 20.5 (18) | 31.2 | 31.2 |
| Chryseobacterium sp. strain FR2 MBRG 9.17 | 3.9 | 7.8 | 7.8 | 20.8 (9) | 41.6 (36) | 31.2 |
| Citrobacter sp. strain MBRG 9.18 | 3.9 | 1.9 | 1.9 | 20.8 (9) | 20.8 (9) | 20 (18) |
| Enterococcus saccharolyticus MBRG 9.16 | 7.8 | 1.9 | 1.9 | 62.5 | 15.6 | 15.6 |
| Eubacterium strain MBRG 4.14 | 31.2 | 31.2 | 31.2 | 41.6 (36) | 31.2 | 31.2 |
| Halonella gallinarum MBRG 4.27 | 15.6 | 31.2 | 31.2 | 15.6 | 31.2 | 125 |
| Microbacterium phyllosphaerae MBRG 4.30 | 15.6 | 15.6 | 15.6 | 15.6 | 31.2 | 31.2 |
| Pseudomonas nitroreductans MBRG 4.6 | 15.6 | 3.9 | 3.9 | 15.6 | 5.2 (4.5) | 20 (18) |
| Pseudomonas sp. strain MBRG 9.14 | 1.9 | 7.8 | 15.6 | 20.8 (9) | 26 (18) | 31.2 |
| Pseudoxanthomonas sp. strain MBRG 9.20 | 62.5 | 0.97 | 0.97 | 62.5 | 0.97 | 7.8 |
| Ralstonia sp. strain U2 MBRG 4.13 | 7.8 | 31.2 | 167 (72) | 7.8 | 62.5 | 125 |
| Sphingobacterium multivorum MBRG 9.19 | 20.8 (9) | 15.6 | 15.6 | 15.6 | 15.6 | 62.5 |
| Stenotrophomonas maltophilia MBRG 9.13 | 15.6 | 62.5 | 62.5 | 62.5 | 166 (72) | 166 (144) |
For an explanation of the data, see the footnote to Table 1.
There are a number of reports in the literature showing comparable alterations in bacterial susceptibility after repeated passage in the presence of sublethal levels of TCS (23, 30, 32). Interestingly, however, in agreement with the present study reports of isolates refractory to QACs and bisbiguanides in exposed environments are less common. It has been reported that the exposure of E. coli to PHMB results in a change in transcriptional activity, leading to reduced susceptibility toward PHMB (1), although reports of resistance development in bacteria isolated from PHMB-exposed environments are scarce. PHMB possesses additional molecular activity compared to bisbiguanides such as CHX. For example, there is evidence that PHMB susceptibility is affected less by the induction or hyperexpression of multidrug efflux pumps. As with all membrane-active antimicrobials, small changes in MICs have been reported that correlate with alterations in envelope lipid composition and cation binding (4, 11).
Effect of CB exposure of bacteria isolated from human skin.
All of the bacteria isolated from human skin showed an overall greater susceptibility than the gram-negative organisms to the biocides tested in this study (Tables 5 to 8). This was unsurprising since gram-positive bacteria possess a permeable cell wall that does not restrict the penetration of antimicrobial agents (21). The average MICs prior to passage were as follows: 1.5 μg ml−1(QAC 1), 2.3 μg ml−1 (QAC 2), 8.7 μg ml−1 (CHX), and 4.4 μg ml−1 (PHMB).
The frequency of marked (greater than twofold) susceptibility decreases of the skin bacteria were as follows: QAC1 (6/13), QAC2 (6/13), CHX (4/13), and PHMB (3/13). Interestingly, the corynebacteria exhibited a decrease in susceptibility for every biocide tested in this study, possibly due to differential up-regulation of their efflux pumps (33). Pure culture selection though repeated passage results in very highly selective conditions since in the environment high-level exposure to CBs will generally be lethal to exposed cells while biocide penetration into complex, multispecies biofilms may be limited, and natural competition may select against any adaptation that affects fitness.
Conclusion.
Chronic exposure of domestic drain biofilms to PHMB resulted in clonal expansion of preexisting, nonsusceptible species. Repeated sublethal exposure to CBs resulted in selection for reduced susceptibility rather than nonsusceptibility. The propensity of the CBs to select for reduced susceptibility varied, with the highest frequency occurring for the QACs and the lowest for PHMB.
TABLE 2.
MICs and MBCs of bacteria isolated from a domestic drain biofilm before and after chronic QAC2 exposurea
| Strain | MIC (μg ml−1)
|
MBC (μg ml−1)
|
||||
|---|---|---|---|---|---|---|
| Before treatment | P7 | P14 | Before treatment | P7 | P14 | |
| Achromobacter xylosoxidans MBRG 4.31 | 31.2 | 3.9 | 3.9 | 15.6 | 15.6 | 20.5 (18) |
| Aeromonas hydrophila MBRG 4.3 | 31.2 | 62.5 | 125 | 31.2 | 62.5 | 125 |
| Aeromonas jandaei MBRG 9.11 | 31.2 | 31.2 | 62.5 | 52 (36) | 31.2 | 62.5 |
| Aranicola proteolyticus MBRG 9.12 | 3.9 | 62.5 | 125 | 62.5 (108) | 62.5 | 125 |
| Bacillus cereus MBRG 4.21 | 6.5 (2.2) | 5.2 (4.5) | 7.8 | 31.2 | 31.2 | 62.5 |
| Chryseobacterium indologenes MBRG 9.15 | 31.2 | 31.2 | 31.2 | 41.6 (36) | 31.2 | 31.2 |
| Chryseobacterium sp. strain MBRG 9.17 | 31.2 | 31.2 | 31.2 | 31.2 | 114 (236) | 62.5 (108) |
| Citrobacter sp. strain MBRG 9.18 | 26 (18) | 7.825 | 62.5 | 31.2 | 36.4 (23) | 292 (190) |
| Enterococcus saccharolyticus MBRG 9.16 | 31.2 | 1.9 | 31.2 | 62.5 | 7.8 | 62.5 |
| Eubacterium strain MBRG 4.14 | 31.2 | 31.2 | 31.2 | 31.2 | 31.2 | 146 (95) |
| Halonella gallinarum MBRG 4.27 | 31.2 | 31.2 | 31.2 | 31.2 | 31.2 | 31.2 |
| Microbacterium phyllosphaerae MBRG 4.30 | 15.6 | 3.9 | 31.2 | 31.2 | 7.8 | 62.5 |
| Pseudomonas nitroreductans MBRG 4.6 | 31.2 | 31.2 | 31.2 | 62.5 | 62.5 | 41.6 (36) |
| Pseudomonas sp. strain MBRG 9.14 | 31.2 | 31.2 | 62.5 | 31.2 | 41.6 (36) | 83.3 (72) |
| Pseudoxanthomonas sp. strain MBRG 9.20 | 7.8 | 15.6 | 31.2 | 15.6 | 52 (36) | 115 (236) |
| Ralstonia sp. strain MBRG 4.13 | 7.8 | 125 | 167 (72) | 31.2 | 125 | 205 |
| Sphingobacterium multivorum MBRG 9.19 | 31.2 | 15.6 | 31.2 | 65.2 | 26 (18) | 52 (36) |
| Stenotrophomonas maltophilia MBRG 9.13 | 31.2 | 7.8 | 31.2 | 41.6 (36) | 15.6 | 31.2 |
For an explanation of the data, see the footnote to Table 1.
TABLE 6.
MICs and MBCs of bacteria isolated from human skin before and after chronic QAC2 exposurea
| Strain | MIC (μg ml−1)
|
MBC (μg ml−1)
|
||||
|---|---|---|---|---|---|---|
| Before treatment | P7 | P14 | Before treatment | P7 | P14 | |
| Corynebacterium pseudogenitalum MBRG 9.24 | 15.6 | 31.2 | 15.6 | 15.6 | 104 (36) | 125 |
| Corynebacterium renale group MBRG 9.13 | 7.8 | 7.8 | 62.5 | 62.5 | 62.5 | 62.5 |
| Micrococcus luteus MBRG 9.25 | 0.45 | 0.97 | 0.97 | 4.5 (2.9) | 4.5 (2.9) | 5.2 (2.2) |
| Staphylococcus capitis MBRG 9.34 | 0.45 | 0.45 | 0.97 | 0.97 | 3.2 (1.1) | 1.9 (3.3) |
| Staphylococcus capral MBRG 9.30 | 0.45 | 0.97 | 0.97 | 3.2 (1.1) | 8.4 (6.8) | 5.2 (2.2) |
| Staphylococcus cohnii MBRG 9.31 | 0.45 | 0.45 | 0.45 | 3.2 (1.1) | 0.97 | 0.97 |
| Staphylococcus epidermidis M 9.33 | 0.45 | 0.45 | 0.45 | 1.9 | 6.4 (2.4) | 3.9 |
| Staphylococcus haemolyticus MBRG 9.35 | 0.45 | 15.6 | 0.97 | 3.9 | 3.9 | 3.9 |
| Staphylococcus hominis MBRG 9.37 | 0.45 | 0.45 | 0.97 | 5.2 (2.2) | 1.9 | 3.2 (1.1) |
| Staphylococcus kloosii MBRG 9.28 | 1.3 (0.5) | 0.45 | 0.97 | 5.2 (2.2) | 5.2 (2.2) | 1.9 |
| Staphylococcus lugdenensis MBRG 9.36 | 1.3 (0.5) | 0.45 | 0.97 | 3.9 (6.7) | 6.4(2.4) | 11.7 (6.7) |
| Staphylococcus saprophyticus MBRG 9.29 | 0.45 | 0.45 | 0.81 | 1.9 | 1.9 | 2.6 (1.1) |
| Staphylococcus warneii MBRG 9.27 | 0.45 | 0.45 | 0.97 | 6.4 (2.4) | 1.9 | 5.2 (2.2) |
For an explanation of the data, see the footnote to Table 1.
TABLE 7.
MICs and MBCs of bacteria isolated from human skin before and after chronic CHX exposurea
| Strain | MIC (μg ml−1)
|
MBC (μg ml−1)
|
||||
|---|---|---|---|---|---|---|
| Before treatment | P7 | P14 | Before treatment | P7 | P14 | |
| Corynebacterium pseudogenitalum MBRG 9.24 | 0.9 | 3.9 | 3.9 | 0.97 | 15.6 | 10 (4) |
| Corynebacterium renale group MBRG 9.13 | 7.8 | 31.2 | 31.2 | 62.5 | 62.5 | 104 (36) |
| Micrococcus luteus MBRG 9.25 | 3.9 | 7.8 | 7.8 | 31.2 | 36 (23) | 62.5 |
| Staphylococcus capitis MBRG 9.34 | 7.8 | 7.8 | 7.8 | 83 (36) | 62.5 | 114 (118) |
| Staphylococcus capral MBRG 9.30 | 7.8 | 7.8 | 7.8 | 93 (54) | 62.5 | 62.5 |
| Staphylococcus cohnii MBRG 9.31 | 10 (4) | 7.8 | 3.9 | 26 (18) | 83 (36) | 104 (36) |
| Staphylococcus epidermidis MBRG 9.33 | 7.8 | 7.8 | 7.8 | 93 (54) | 104 (36) | 31.2 |
| Staphylococcus haemolyticus MBRG 9.35 | 13 (9) | 15.6 | 15.6 | 145 (95) | 166 (72) | 104 (36) |
| Staphylococcus hominis MBRG 9.37 | 13 (4) | 7.8 | 7.8 | 83 (36) | 62.5 | 62.5 |
| Staphylococcus kloosii MBRG 9.28 | 7.8 | 13 (9) | 7.8 | 62.5 | 125 | 62.5 |
| Staphylococcus lugdenensis MBRG 9.36 | 13 (4) | 15.6 | 15.6 | 83 (36) | 166 (72) | 104 (36) |
| Staphylococcus saprophyticus MBRG 9.29 | 13 (4) | 3.9 | 3.9 | 31.2 | 31.2 | 31.2 |
| Staphylococcus warneii MBRG 9.27 | 7.8 | 13 (9) | 15.6 | 62.5 | 166 (72) | 208 (72) |
For an explanation of the data, see the footnote to Table 1.
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Allen, M. J., G. F. White, and A. P. Morby. 2006. The response of Escherichia coli to exposure to the biocide polyhexamethylene biguanide. Microbiology 152:989-1000. [DOI] [PubMed] [Google Scholar]
- 2.Braoudaki, M., and A. C. Hilton. 2004. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J. Clin. Microbiol. 42:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, M. R., W. M. Watkins, and J. H. Foster. 1969. Step-wise resistance to polymyxin and other agents by Pseudomonas aeruginosa. J. Gen. Microbiol. 55:17-18. [PubMed] [Google Scholar]
- 4.Broxton, P., P. M. Woodcock, F. Heatley, and P. Gilbert. 1984. Interaction of some polyhexamethylene biguanides and membrane phospholipids in Escherichia coli. J. Appl. Bacteriol. 57:115-124. [DOI] [PubMed] [Google Scholar]
- 5.Bruinsma, G. M., M. Rustema-Abbing, H. C. van der Mei, C. Lakkis, and H. J. Busscher. 30 January 2006. Resistance to a polyquaternium-1 lens care solution and isoelectric points of Pseudomonas aeruginosa strains. J. Antimicrob. Chemother. 57:764-766. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 6.Burdon, D. W., and J. L. Whitby. 1967. Contamination of hospital disinfectants with Pseudomonas species. Br. Med. J. 2:153-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carson, L. A., M. S. Favero, W. W. Bond, and N. J. Petersen. 1972. Factors affecting comparative resistance of naturally occurring and subcultured Pseudomonas aeruginosa to disinfectants. Appl. Microbiol. 23:863-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman, J. S. 1998. Characterizing bacterial resistance to preservatives and disinfectants. Int. Biodeterior. Biodegrad. 41:241-245. [Google Scholar]
- 9.Chuanchuen, R., K. Beinlich, T. T. Hoang, A. Becher, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 45:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dance, D. A., A. D. Pearson, D. V. Seal, and J. A. Lowes. 1987. A hospital outbreak caused by a chlorhexidine and antibiotic-resistant Proteus mirabilis. J. Hosp. Infect. 10:10-16. [DOI] [PubMed] [Google Scholar]
- 11.Das, J. R., M. Bhakoo, M. V. Jones, and P. Gilbert. 1998. Changes in the biocide susceptibility of Staphylococcus epidermidis and Escherichia coli cells associated with rapid attachment to plastic surfaces. J. Appl. Microbiol. 84:852-858. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, P., and A. J. McBain. 2001. Biocide usage in the domestic setting and concern about antibacterial and antibiotic resistance. J. Infect. 43:85-91. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, P., and A. J. McBain. 2001. Biofilms: their impact on health and their recalcitrance toward biocides. Am. J. Infect. Control 29:252-255. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, P., and A. J. McBain. 2002. Literature-based evaluation of the potential risks associated with impregnation of medical devices and implants with triclosan. Surg. Infect. 3:S55-63. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, P., and L. E. Moore. 2005. Cationic antiseptics: diversity of action under a common epithet. J. Appl. Microbiol. 99:703-715. [DOI] [PubMed] [Google Scholar]
- 16.Goeres, D. M., T. Palys, B. B. Sandel, and J. Geiger. 2004. Evaluation of disinfectant efficacy against biofilm and suspended bacteria in a laboratory swimming pool model. Water Res. 38:3103-3109. [DOI] [PubMed] [Google Scholar]
- 17.Guerin-Mechin, L., F. Dubois-Brissonnet, B. Heyd, and J. Y. Leveau. 2000. Quaternary ammonium compound stresses induce specific variations in fatty acid composition of Pseudomonas aeruginosa. Int. J. Food Microbiol. 55:157-159. [DOI] [PubMed] [Google Scholar]
- 18.Joynson, J. A., B. Forbes, and R. J. Lambert. 2002. Adaptive resistance to benzalkonium chloride, amikacin and tobramycin: the effect on susceptibility to other antimicrobials. J. Appl. Microbiol. 93:96-107. [DOI] [PubMed] [Google Scholar]
- 19.Karatzas, K. A., L. P. Randall, M. Webber, L. J. Piddock, T. J. Humphrey, M. J. Woodward, and N. G. Coldham. 2007. Phenotypic and proteomic characterization of MAR variants of Salmonella enterica serovar Typhimurium selected following exposure to disinfectants. Appl. Environ. Microbiol. 14:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karatzas, K. A., M. A. Webber, F. Jorgensen, M. J. Woodward, L. J. Piddock, and T. J. Humphrey. 2007. Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J. Antimicrob. Chemother. 60:947-955. [DOI] [PubMed] [Google Scholar]
- 21.Lambert, P. A. 2002. Cellular impermeability and uptake of biocides and antibiotics in gram-positive bacteria and mycobacteria. J. Appl. Microbiol. 92:46S-54S. [PubMed] [Google Scholar]
- 22.Lambert, P. A., and S. M. Hammond. 1973. Potassium fluxes, first indications of membrane damage in micro-organisms. Biochem. Biophys. Res. Commun. 54:796-799. [DOI] [PubMed] [Google Scholar]
- 23.Ledder, R. G., P. Gilbert, C. Willis, and A. J. McBain. 2006. Effects of chronic triclosan exposure upon the antimicrobial susceptibility of 40 ex-situ environmental and human isolates. J. Appl. Microbiol. 100:1132-1140. [DOI] [PubMed] [Google Scholar]
- 24.Levy, S. B. 2002. Active efflux, a common mechanism for biocide and antibiotic resistance. J. Appl. Microbiol. 92:65S-71S. [PubMed] [Google Scholar]
- 25.Loughlin, M. F., M. V. Jones, and P. A. Lambert. 2002. Pseudomonas aeruginosa cells adapted to benzalkonium chloride show resistance to other membrane-active agents but not to clinically relevant antibiotics. J. Antimicrob. Chemother. 49:631-639. [DOI] [PubMed] [Google Scholar]
- 26.Maxcy, R. B., N. P. Tiwari, and P. R. Soprey. 1971. Changes in Escherichia coli associated with acquired tolerance for quaternary ammonium compounds. Appl. Microbiol. 22:229-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, B. B. Price, and P. Gilbert. 2003. Exposure of sink drain microcosms to triclosan: population dynamics and antimicrobial susceptibility. Appl. Environ. Microbiol. 69:5433-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, A. H. Rickard, S. A. Symmons, and P. Gilbert. 2003. Microbial characterization of biofilms in domestic drains and the establishment of stable biofilm microcosms. Appl. Environ. Microbiol. 69:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBain, A. J., R. G. Ledder, L. E. Moore, C. E. Catrenich, and P. Gilbert. 2004. Effects of quaternary-ammonium-based formulations on bacterial community dynamics and antimicrobial susceptibility. Appl. Environ. Microbiol. 70:3449-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBain, A. J., R. G. Ledder, P. Sreenivasan, and P. Gilbert. 2004. Selection for high-level resistance by chronic triclosan exposure is not universal. J. Antimicrob. Chemother. 53:772-777. [DOI] [PubMed] [Google Scholar]
- 31.McBain, A. J., A. H. Rickard, and P. Gilbert. 2002. Possible implications of biocide accumulation in the environment on the prevalence of bacterial antibiotic resistance. J. Ind. Microbiol. Biotechnol. 29:326-330. [DOI] [PubMed] [Google Scholar]
- 32.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol. Lett. 166:305-309. [DOI] [PubMed] [Google Scholar]
- 33.Poole, K. 2005. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56:20-51. [DOI] [PubMed] [Google Scholar]
- 34.Poole, K. 2002. Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 92:55S-64S. [PubMed] [Google Scholar]
- 35.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 1:59-71. [DOI] [PubMed] [Google Scholar]
- 36.Randall, L. P., S. W. Cooles, N. G. Coldham, E. G. Penuela, A. C. Mott, M. J. Woodward, L. J. Piddock, and M. A. Webber. 2007. Commonly used farm disinfectants can select for mutant Salmonella enterica serovar Typhimurium with decreased susceptibility to biocides and antibiotics without compromising virulence. J. Antimicrob. Chemother. 60:1273-1280. [DOI] [PubMed] [Google Scholar]
- 37.Russell, A. D. 2002. Introduction of biocides into clinical practice and the impact on antibiotic-resistant bacteria. Symp. Ser. Soc. Appl. Microbiol. 31:121S-135S. [PubMed] [Google Scholar]
- 38.Russell, A. D. 2001. Mechanisms of bacterial insusceptibility to biocides. Am. J. Infect. Control 29:259-261. [DOI] [PubMed] [Google Scholar]
- 39.Rutala, W. A., M. M. Stiegel, F. A. Sarubbi, and D. J. Weber. 1997. Susceptibility of antibiotic-susceptible and antibiotic-resistant hospital bacteria to disinfectants. Infect. Control Hosp. Epidemiol. 18:417-421. [DOI] [PubMed] [Google Scholar]
- 40.Schweizer, H. P. 2003. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet. Mol. Res. 2:48-62. [PubMed] [Google Scholar]
- 41.Schweizer, H. P. 2001. Triclosan: a widely used biocide and its link to antibiotics. FEMS Microbiol. Lett. 202:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Shigeta, S., Y. Yasunaga, K. Honzumi, H. Okamura, R. Kumata, and S. Endo. 1978. Cerebral ventriculitis associated with Achromobacter xylosoxidans. J. Clin. Pathol. 31:156-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stickler, D. J., and G. L. Jones. 2008. Reduced susceptibility of Proteus mirabilis to triclosan. Antimicrob. Agents Chemother. 52:991-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]