Abstract
Classified as a distinct species in 1980, Lactobacillus reuteri strains have been used in probiotic formulations for intestinal and urogenital applications. In the former, the primary mechanism of action of L. reuteri SD2112 (ATCC 55730) has been purported to be its ability to produce the antibiotic 3-hydroxypropionaldehyde (3-HPA), also known as reuterin. In the vagina, it has been postulated that probiotic Lactobacillus reuteri RC-14 does not require reuterin production but mediates a restoration of the normal microbiota via hydrogen peroxide, biosurfactant, lactic acid production, and immune modulation. The aim of the present study was to determine whether strain RC-14 produced reuterin. Using PCR and DNA dot blot analyses, numerous Lactobacillus species, including RC-14, were screened for the presence of the gene encoding the large subunit of glycerol dehydratase (gldC), the enzyme responsible for reuterin production. In addition, lactobacilli were grown in glycerol-based media and both high-performance liquid chromatography and a colorimetric assay were used to test for the presence of reuterin. L. reuteri RC-14 was determined to be negative for gldC sequences, as well as for the production of reuterin when cultured in the presence of glycerol. These findings support that the probiotic effects of L. reuteri RC-14, repeatedly demonstrated during numerous studies of the intestine and vagina, are independent of reuterin production.
In 1980, Kandler et al. (16) reclassified Lactobacillus fermentum biotype II into Lactobacillus reuteri species, named after Gerhard Reuter, a German microbiologist who had worked on these organisms. Multiple studies have since shown L. reuteri to be a fairly universal species to the intestinal tracts of not only humans (30) but numerous animal species (23, 24, 32), and it is frequently isolated from natural environments as well as many meat and dairy products (31, 37). Based largely upon this ubiquitous and dominant nature, especially among healthy individuals and animals, work began to investigate whether strains of L. reuteri could be used to modulate intestinal health. In 1986, a yogurt and milk fermented with a strain of L. reuteri reduced the intestinal coliform count in pigs (26), and it was later proposed by Talarico et al. (35) that the beneficial effects were due to the production of reuterin, more specifically 3-hydroxypropionaldehyde (3-HPA), a by-product of glycerol fermentation (9). A follow-up study characterized reuterin as a broad-spectrum antibiotic, effective against both gram-positive and -negative strains of bacteria, as well as several fungi and protozoa (36). Subsequent to this, L. reuteri strains have been used commercially as probiotic agents (“live microorganisms which when administered in adequate amounts confer a health benefit on the host”) (11), with the presumption that they produce reuterin.
However, it has become apparent that this antibiotic compound is not unique to L. reuteri species. Rather, it has been shown to be produced by potentially pathogenic Enterobacter agglomerans, Klebsiella pneumoniae, Citrobacter freundii, and Aerobacter aerogenes (3, 25, 34), as well as by an intestinal constituent, Clostridium butyricum (13). The L. reuteri are not the only Lactobacillus species to produce reuterin, with an L. coryniformis cheese isolate (21), as well as L. collinoides (33), also found to be a producer.
In addition, it has recently been shown that not all L. reuteri strains produce this antibiotic (22). This raises interesting questions. Just how common is reuterin production among the microbial world? Should non-reuterin-producing L. reuteri be given a different designation than those that produce the compound? Are there other important differences among L. reuteri strains that would support this reclassification?
In 1998, L. acidophilus RC-14, a probiotic isolated in 1985 and described for its anti-infective properties in 1987 (28), was reclassified as L. fermentum RC-14 based on ribotyping analysis (41). In 2006, the strain was tested by DNA-DNA hybridization and reclassified as L. reuteri RC-14. By then, it had been used in numerous clinical studies and shown to populate the vagina and reduce the risk of infection (2, 27, 29). In vitro studies led to the theory that this strain acted through its ability to produce biosurfactants, hydrogen peroxide, and lactic acid and to modulate the immune response (14, 19, 20). In order to rule out any involvement of reuterin in the activity of the strain, which would make this the first non-reuterin-producing L. reuteri probiotic strain, the following methods were used: testing for the presence of the gene encoding the large subunit of glycerol dehydratase (Gld), the enzyme required to produce 3-hydroxypropionaldehyde from glycerol (9), and the use of both high-performance liquid chromatography (HPLC) and a colorimetric assay to detect reuterin directly within culture supernatants after growth in glycerol.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The following strains were utilized in the study: L. reuteri SD2112 (ATCC 55730), L. reuteri 1063, L. reuteri 1068, L. reuteri ATCC 23272T (T-type strain), L. reuteri RC-14, L. reuteri 359, L. reuteri 656, L. amylovorus 20552, L. casei 393T, L. crispatus 33820, L. delbrueckii 20074T, L. fermentum 14 (Fuller strain) (4), L. fermentum 11739, L. gasseri 33323, L. johnsonii 20553, L. plantarum 14917T, L. rhamnosus GR-1, L. rhamnosus 7469T, and Escherichia coli C1214. For genomic DNA isolation, lactobacilli were cultured using MRS media (Becton Dickenson [BD], Oakville, Canada) for 48 h in anaerobic jars containing CO2-generating gas packs (BD). E. coli was cultured aerobically using brain-heart infusion media for 24 h with shaking (200 rpm). The growth conditions utilized for reuterin production and detection are described in their appropriate sections.
DNA isolation and PCR screening.
Genomic DNA (gDNA) was isolated from each bacterial strain by using InstaGene matrix (Bio-Rad, Mississauga, Ontario, Canada) according to the manufacturer's supplied protocol. Briefly, one to two bacterial colonies were isolated from each strain, washed once with sterile water, resuspended in 200 μl of InstaGene matrix, and incubated at 56°C for 30 min. Each suspension was then mixed, incubated in a boiling water bath for 8 min, mixed again, and centrifuged at 14,000 × g for 3 min. The resulting supernatant containing gDNA was removed and stored at −20°C until use.
All PCR primers and reagents were purchased from Invitrogen (Burlington, Ontario, Canada), and the gel electrophoresis reagents and equipment were from Bio-Rad. To amplify a portion of the gene encoding the large subunit of Gld (gldC), degenerate oligonucleotide primers were generated based upon regions of the gene highly conserved across currently published Lactobacillus gldC sequences (L. hilgardii [GenBank accession no. AY061969], L. diolivorans [AY061968], L. collinoides [AY061967], and L. reuteri 23272T [also termed F275, CP000705]). The primer sequences generated were: GDCRev-5′-GC[A/G]GC[C/T]TT[G/C]ATATCT[G/T][G/C]AACCAT-3′ (matching nucleotides 1800525 to 1800547 from CP000705) and GDCFor-5′-GC[A/C]TA[C/T]GC[A/T]GAAACCATTTCAGTTTA-3′ (matching nucleotides 1801252 to 1801227). As a positive DNA control, a portion of the 16S rRNA gene was also amplified from all samples by using the eubacterial primers 16SFor-5′-ACTCCTACGGGAGGCAGCAG-3′ and 16SRev-5′-GTATTACCGCGGCTGCTGGCAC-3′, highly conserved among bacteria (40). Each 50-μl PCR contained 100 pmol of each of the corresponding forward and reverse primers, 1× PCR buffer, 2 mM MgCl2, 200 μM deoxynucleoside triphosphates, 5 μl (∼0.5 μg) of gDNA, and 2.5 U of Taq polymerase. A total of 30 cycles of amplification were performed consisting of 1 min at 94°C, 30 s at 55°C, and 1 min at 72°C. A single initiation step of 5 min at 94°C prior to cycling and a final elongation step of 10 min at 72°C were also performed to optimize the accuracy and quantity of product produced. Products were separated on 1.2% agarose gels, stained with ethidium bromide, and visualized by using the GelDoc XR system. The expected product sizes were 728 bp for gldC and 201 bp for the 16S rRNA gene.
DNA probe generation and dot blot analysis.
A fragment of the expected 728-bp size was amplified from L. reuteri SD2112 gDNA by using GDC primers. This fragment was purified by using a PCR product purification spin column (Qiagen, Mississauga, Ontario, Canada) according to the manufacturer's supplied protocol, sequenced (Robarts Research Institute Sequencing Facility, London, Ontario, Canada), and used as a template in a PCR to generate a digoxigenin (DIG)-labeled probe. PCR was performed as already described except that the standard 200 μM dTTP used was reduced to 133 μM, and 67 μM DIG-dUTP added (Roche, Mississauga, Ontario, Canada). The probe was purified via spin column as described above, eluted in 50 μl of double-distilled H2O, and stored at −80°C until use. In a similar fashion, a 201-bp DIG-labeled probe corresponding to the 16S rRNA gene was generated from E. coli C1214 gDNA.
Two identical DNA dot blots were created by applying 2.5 μg of gDNA from selected bacterial strains in 15 μl of dilution buffer (10 mM Tris-HCl [pH 8.0], 50 μg of herring sperm DNA/ml) to positively charged nylon membranes (Roche) using a dot blot manifold (Schleicher and Schuell, Keene, NH) under vacuum. DNA was fixed to the membranes by a 1-min exposure to UV light on a transilluminator, and the blots were air dried. The blots were hybridized and detected in parallel (gldC and 16S rRNA gene) using the protocols and reagents outlined in Roche's DIG application manual for filter hybridization. Briefly, the blots were prehybridized for 30 min at 42°C in 10 ml of DIG-Easy Hyb solution in 15-ml conical tubes using a rotating hybridization oven (BioCan, Mississauga, Ontario, Canada). Fifteen microliters (∼250 ng) of each generated probe was separately added to 85 μl of double-distilled H2O and boiled for 5 min in a boiling water bath to denature the probes into single strands. The probes were then chilled for 2 min on ice and added to 5 ml of prewarmed (42°C) DIG-Easy Hyb. The prehybridization solution was poured off and replaced with the probe solution, and the blots were incubated for 10 h at 42°C with constant rotation. The probes were poured off, and the blots were incubated twice for 15 min each at room temperature in a low-stringency buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate), followed by three 10-min washes in a high-stringency buffer (0.5× SSC, 0.1% sodium dodecyl sulfate) at 65°C to remove nonspecifically bound probe. The blots were blocked for 30 min in DIG blocking solution (1% blocking reagent in 0.1 M maleic acid-0.15 M NaCl [pH 7.5]) and incubated with anti-DIG alkaline phosphatase antibody (75 mU/ml in blocking solution) for 30 min. Excess antibody was removed by two 15-min washes in washing buffer (0.1 M maleic acid, 0.15 M NaCl, 0.3% Tween 20) at room temperature, and the blots were equilibrated for 2 min in detection buffer (0.1 M Tris-HCl, 0.1 M NaCl [pH 9.5]. Colorimetric detection was performed in 340 μg of NBT (4-nitroblue tetrazolium chloride)/ml and 175 μg of BCIP (5-bromo-4-chloro-3-indolylphosphate)/ml in detection buffer until the desired color achieved. NBT and BCIP react with the alkaline phosphatase conjugated to the anti-DIG antibody, resulting in the formation of blue dye, which precipitates on to the membrane. The blots were scanned and stored in TE buffer (pH 8.0) to stop the colorimetric reaction.
HPLC analysis.
L. reuteri 1063 and L. reuteri 1068, L. reuteri SD2112, and L. reuteri RC-14 were cultured and harvested by the homologous method as described Dobrogosz and Lindgren (8). In short, the strains were inoculated in 1 liter of LCM medium (Trypticase, 10 g/liter; yeast extract, 5 g/liter; tryptose, 3 g/liter; KH2PO4, 3 g/liter; ammonium citrate, 1.5 g/liter; sodium acetate [trihydrate], 1 g/liter; MgSO4·7H2O, 1.2 g/liter; MnSO4·H2O, 0.13 g/liter; FeSO4·7H2O, 60 mg/liter; cysteine-HCl, 0.2 g/liter; Tween 80, 1 ml/liter). The pH was adjusted to 7.0. Sterile glucose was added to 20 mM after sterilization, followed by incubation for 48 h at 37°C. The cells were harvested by centrifugation and washed twice in sterile sodium phosphate buffer (pH 7.5). The cells were then suspended in 10 ml of sterile 250 mM glycerol and incubated for 6 h at 37°C. The suspension was centrifuged, and the supernatant was recovered, sterile filtered through a 0.45-μm-pore-size filter, and stored aseptically at 2°C until HPLC analysis. HPLC was performed as described Dobrogosz and Lindgren (8), except that a Shodex Sugar SH1011 (Phenomenex) column was used. The substance eluting at the expected elution time for reuterin was not further analyzed.
Colorimetric assay for reuterin detection.
The detection of reuterin was performed by using the method of Circle et al. (6). Overnight L. reuteri cultures were centrifuged at 4,000 rpm for 10 min at room temperature to pellet the cells. The cells were then washed twice with 50 mM potassium phosphate buffer (pH 7.5), and 100-mg portions were resuspended in 14 ml of 250 mM glycerol, followed by incubation at 37°C for 1 to 2 h. After the cells were pelleted as described above, the supernatant was passed through a 0.45-μm-pore-size syringe filter and stored at 4°C. A total of 300 μl of each supernatant was added to 225 μl of the 10 mM tryptophan solution, 900 μl of concentrated HCl was added, and the solution was incubated for 20 min at 37°C. The absorbance was measured at 560 nm by using a SpectraMax M5 microplate reader.
RESULTS
GldC gene screening.
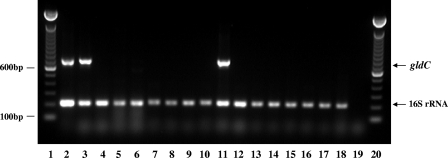
The conversion of glycerol to reuterin is known to be dependent upon the activity of Gld, an enzyme that dehydrates glycerol into the aldehyde through the removal of one molecule of H2O. Thus, we initially wanted to determine whether L. reuteri RC-14 possessed at least one of the genes necessary for its expression. Since gldC encodes the largest subunit of the three subunit enzyme (gldD and gldE encode the other two), we selected it for screening to provide the largest sequence in which to search for highly homologous regions to use in designing oligonucleotide primers and a DNA probe. Based upon the alignment of published gldC sequences taken from four species of Lactobacillus (L. hilgardii, L. coryniformis, L. diolovorans, and L. reuteri), degenerate primers were synthesized. Primers GDCRev and GDCFor were 23 nucleotides in length with five sites of degeneracy and 26 nucleotides long with three degenerate bases, respectively, and represented 100% matches for all four Lactobacillus sequences. PCR screening was performed on 16 Lactobacillus strains comprising 10 different species and included both positive (L. reuteri 23272T) and negative (E. coli C1214) control strains (Fig. 1). Three strains were positive for the expected 728-bp product, namely, L. reuteri 23272T, L. reuteri SD2112, and L. fermentum 14. Strain L. reuteri RC-14, as well as the 12 other lactobacilli tested, including two L. reuteri clinical isolates, were negative for gldC using this method. Based upon the Tm of the primer sequences and preliminary experiments involving the positive and negative control strains, an annealing temperature of 55°C was used for all PCRs. However, to ensure that the selected annealing temperature was not too high for the degenerate primers to cause false negatives in the reaction, identical PCRs were also carried out with an annealing temperature of 50°C. Similar results were obtained in terms of the amplification of the 728-bp gldC product (data not shown).
FIG. 1.
PCR screening of Lactobacillus strains for gldC sequence. Upper band at 728 bp represents the expected product using the gldC-specific primers, while the lower band at 201 bp represents that expected using the 16SrRNA gene primers (positive DNA control). Lanes:1 and 20, 100-bp ladder; 2 to 6, L. reuteri strains 23272T (positive strain control), SD2112, RC-14, 359, and 656, respectively; 7, L. amylovorus 20552; 8, L. casei 393T; 9, L. crispatus 33820; 10, L. delbrueckii 20074T; 11, L. fermentum 14; 12, L. fermentum 11739; 13, L. gasseri 33323; 14, L. johnsonii 20553; 15, L. plantarum 14917T; 16, L. rhamnosus GR-1; 17, L. rhamnosus 7469T; 18, E. coli C1214 (negative strain control); 19, negative DNA control.
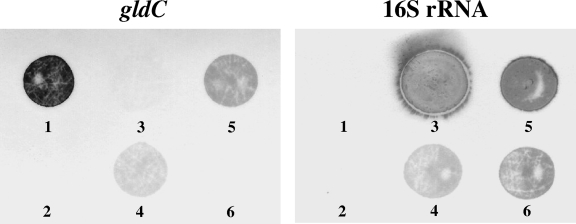
To confirm the PCR screening results, we generated two identical dot blots containing gDNA from three lactobacilli, namely, L. reuteri SD2112 (PCR positive), L. fermentum 14 (PCR positive), and L. reuteri RC-14 (PCR negative), as well as several controls. Blots were hybridized in parallel with DIG-labeled 728-bp gldC-specific and 201-bp 16S rRNA gene-specific DNA probes, respectively (Fig. 2). Probes were generated by PCR similarly to PCR screening but with the addition of DIG-labeled dUTP to the nucleotide mix and were evaluated via gel electrophoresis, showing a single band for each migrating ∼1.3-fold greater in size from their corresponding unlabeled products due to DIG incorporation (data not shown). The blotting results for the three lactobacilli were shown to be identical to those observed via PCR, supporting that L. reuteri RC-14 does not possess gldC.
FIG. 2.
DNA hybridization screening for gldC. Two identical DNA dot blots were prepared and separately screened for gldC (left) and 16S rRNA (right) gene sequences using DIG-labeled DNA probes. Spot 1, 5 ng of gldC PCR product (positive control); spot 2, DNA dilution buffer only (negative probe control). Spots 3 to 6 each contained 2.5 μg of gDNA from the following strains: spot 3, E. coli C1214 (negative strain control); spot 4, L. fermentum 14; spot 5, L. reuteri SD2112; and spot 6, L. reuteri RC-14.
HPLC and colorimetric assay.
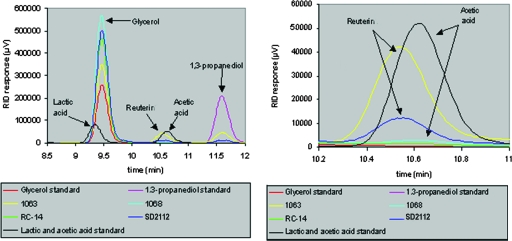
To augment the genetic screening results, L. reuteri RC-14 was cultured in glycerol, known to stimulate reuterin production in other L. reuteri strains, to examine whether or not it could produce the compound under these conditions. The controls L. reuteri 1063 and L. reuteri SD2112 are known to produce reuterin (18, 39), while L. reuteri 1068 is known not to produce it with glycerol in the medium. After a 6-h glycerol incubation, culture supernatants from all four strains were examined via HPLC (Fig. 3). As expected, both L. reuteri 1063 and L. reuteri SD2112 showed a peak at the expected elution time for reuterin between glycerol and 1,3-propanediol, whereas L. reuteri 1068 and L. reuteri RC-14 did not. Thus, RC-14 cannot be induced to produce reuterin in the presence of glycerol, conditions known to stimulate its production in other L. reuteri strains.
FIG. 3.
HPLC analysis of culture supernatants collected from L. reuteri strains. The left panel shows the time interval from 8.5 to 12 min of chromatograms of the four samples and standard compounds. The right panel shows an enlarged section of the chromatogram from 10.2 to 11 min. The data clearly demonstrate that there a is real-time difference between the reuterin peak (10.55 min) eluting 6/100 min before acetic acid (10.61 min) and the absence of reuterin in strain L. reuteri RC-14. Glycerol was the test substrate, and lactic and acetic acids are normal by-products of Lactobacillus glycerol fermentation. 1,3-Propanediol is a compound produced via the reduction of reuterin and therefore should not be present in the supernatants of non-reuterin-producing strains.
To support the HPLC results, we also monitored the production of reuterin by using a colorimetric assay. Two independent isolates of L. reuteri RC-14 were grown to stationary phase in MRS medium at 37°C. Cells were pelleted, washed once with phosphate-buffered saline, and resuspended in 15 ml of 250 mM glycerol. As a positive control, a reuterin-producing L. reuteri strain (ATCC 6475) was used. As a negative control, we utilized a L. reuteri strain in which the gldC gene was disrupted. As expected, only the positive control L. reuteri strain produced any colorimetric change, indicating the presence of reuterin (optical density at 560 nm [OD560] = 0.628). The two independent isolates of L. reuteri RC-14 (OD560 = 0.001,0.000) and the gldC mutant (OD560 = 0.001) did not yield any colorimetric change, indicating a lack of production of reuterin.
DISCUSSION
This study has shown that probiotic L. reuteri RC-14 does not appear to contain a gld gene, and reuterin production was not detected in this strain. This is important for a number of reasons. First, it indicates that reuterin production is not a mechanism of action of L. reuteri RC-14 to confer health benefits on the intestine and vagina, as this strain has been shown to do (1, 27). Thus, the long-held view that specific antimicrobial substances are required for probiotic strains to function and to displace pathogens is not supported by the present findings. Rather, probiotic strains such as L. reuteri RC-14 can function in other ways that are antagonistic to pathogen colonization, including modulating host immunity (17, 20) and producing biosurfactants and other antiadhesive factors (19, 38). This emphasizes that selection of a strain intended for probiotic use cannot be solely made based upon the presence of an antimicrobial compound. This supports the position of the Food and Agriculture Organization of the United Nations and World Health Organization, whose report (12) states this very point. However, the production of antibiotic-type substances is still being used as a means to identify and propagate new probiotic strains (7, 10).
Second, it reemphasizes the diversity of probiotic strains, even within the same species, and shows that species possess different functional properties. Such phenotypic differences between strains have been seen previously, for example, with L. rhamnosus GR-1 better functioning in the vagina than L. rhamnosus GG (5), which is better known for its intestinal modalities (15).
In the case of L. reuteri RC-14 and similar strains that do not produce reuterin, several questions arise. Should they become members of a nonreuterin subspecies of L. reuteri and studied further to determine whether additional characteristics warrant their designation as a novel species of Lactobacillus? Or, given that reuterin is produced by strains other than L. reuteri, should this compound be renamed? The reclassification of bacterial species is generally based on genotypic profiles, even though these, too, can differ between isolates. However, since strain L. reuteri RC-14 is also genetically closely related to L. fermentum, as shown by ribotying (41), and does not possess the genes required to produce a factor key to the identification and designation of L. reuteri strains, should it remain under this classification?
The factors that influence the evolution of bacterial species evolve with new techniques. However, the strain itself remains the same. Thus, L. reuteri RC-14 was formerly named L. acidophilus RC-14 (28) and L. fermentum RC-14 (27), and its probiotic properties predate the discovery of reuterin-producing L. reuteri strains. Since the naming and speciation of strains has more and more recently had commercial implications with probiotic products appearing in various forms around the world, it behooves microbiologists to carefully outline the exact criteria for strain speciation, beyond simple genomic profiles.
Acknowledgments
This study was supported by a grant from NSERC.
The provision of strain L. fermentum 14 by Roy Fuller is greatly appreciated.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Anukam, K. C., E. O. Osazuwa, B. E. Osadolor, A. W. Bruce, and G. Reid. 2008. Yogurt containing probiotic Lactobacillus rhamnosus GR-1 and L. reuteri RC-14 helps resolve moderate diarrhea and increases CD4 count in HIV/AIDS patients. J. Clin. Gastroenterol. 42:239-243. [DOI] [PubMed] [Google Scholar]
- 2.Anukam, K. C., E. Osazuwa, G. I. Osemene, F. Ehigiagbe, A. W. Bruce, and G. Reid. 2006. Clinical study comparing probiotic Lactobacillus GR-1 and RC-14 with metronidazole vaginal gel to treat symptomatic bacterial vaginosis. Microbes Infect. 8:2772-2776. [DOI] [PubMed] [Google Scholar]
- 3.Barbirato, F., J. P. Grivet, P. Soucaille, and A. Bories. 1996. 3-Hydroxypropionaldehyde, an inhibitory metabolite of glycerol fermentation to 1,3-propanediol by enterobacterial species. Appl. Environ. Microbiol. 62:1448-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrow, P. A., B. E. Brooker, R. Fuller, and M. J. Newport. 1980. The attachment of bacteria to the gastric epithelium of the pig and its importance in the microecology of the intestine. J. Appl. Bacteriol. 48:147-154. [DOI] [PubMed] [Google Scholar]
- 5.Cadieux, P., J. Burton, G. Gardiner, I. Braunstein, A. W. Bruce, C. Y. Kang, and G. Reid. 2002. Lactobacillus strains and vaginal ecology. JAMA 287:1940-1941. [DOI] [PubMed] [Google Scholar]
- 6.Circle, S. J., L. Stone, and C. S. Boruff. 1945. Acrolein determination by means of tryptophan: a colorimetric micro method. Ind. Eng. Chem. Anal. 17:259-262. [Google Scholar]
- 7.Deraz, S. F., E. N. Karlsson, A. A. Khalil, and B. Mattiasson. 2007. Mode of action of acidocin D20079, a bacteriocin produced by the potential probiotic strain, Lactobacillus acidophilus DSM 20079. J. Ind. Microbiol. Biotechnol. 34:373-379. [DOI] [PubMed] [Google Scholar]
- 8.Dobrogosz, W. J., and S. E. Lindgren. 15 December 1998. Method for inhibiting microorganism growth. U.S. patent 5,849,289.
- 9.Doleyres, Y., P. Beck, S. Vollenweider, and C. Lacroix. 2005. Production of 3-hydroxypropionaldehyde using a two-step process with Lactobacillus reuteri. Appl. Microbiol. Biotechnol. 68:467-474. [DOI] [PubMed] [Google Scholar]
- 10.Ermolenko, E. I., A. I. Chernysh, I. V. Martsinkovskaia, and A. N. Suvorov. 2007. Influence of probiotic enterococci on the growth of Streptococcus agalactiae. Zh. Mikrobiol. Epidemiol. Immunobiol. Sep-Oct:73-77. (In Russian.) [PubMed]
- 11.FAO/WHO. 2001. Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. Food and Agriculture Organization of the United Nations and World Health Organization Expert Consultation Report. http://www.fao.org/es/ESN/Probio/probio.htm.
- 12.FAO/WHO. 2002. Guidelines for the evaluation of probiotics in food. Food and Agriculture Organization of the United Nations and World Health Organization Working Group Report. ftp://ftp.fao.org/es/esn/food/wgreport2.pdf.
- 13.González-Pajuelo, M., J. C. Andrade, and I. Vasconcelos. 2005. Production of 1,3-propanediol by Clostridium butyricum VPI 3266 in continuous cultures with high yield and productivity. J. Ind. Microbiol. Biotechnol. 32:391-396. [DOI] [PubMed] [Google Scholar]
- 14.Heinemann, C., J. E. van Hylckama Vlieg, D. B. Janssen, H. J. Busscher, H. C. van der Mei, and G. Reid. 2000. Purification and characterization of a surface-binding protein from Lactobacillus fermentum RC-14 inhibiting Enterococcus faecalis 1131 adhesion. FEMS Microbiol. Lett. 190:177-180. [DOI] [PubMed] [Google Scholar]
- 15.Huebner, E. S., and C. M. Surawicz. 2006. Probiotics in the prevention and treatment of gastrointestinal infections. Gastroenterol. Clin. N. Am. 35:355-365. [DOI] [PubMed] [Google Scholar]
- 16.Kandler, O., K. O. Stetter, and R. Kohl. 1980. Lactobacillus reuteri sp. nov., a new species of heterofermentative lactobacilli. Zentralbl. Mikrobiol. Parasitenkd. Infektionskr. Hyg. Abt. 1 Orig. C 1:264-269. [Google Scholar]
- 17.Kim, S. O., H. I. Sheik, S.-D. Ha, A. Martins, and G. Reid. 2006. G-CSF mediated inhibition of JNK is a key mechanism for Lactobacillus rhamnosus-induced anti-inflammatory effects in macrophages. Cell. Microbiol. 8:1958-1971. [DOI] [PubMed] [Google Scholar]
- 18.Kwon, N. H., S. H. Kim, J. Y. Kim, J. Y. Lim, J. S. Ahn, B. W. Yoo, H. J. Kang, D. S. Lee, I. B. Kwon, and Y. H. Park. 2002. Antimicrobial activity of Lactobacillus reuteri SD 2112 against bovine pathogens and Escherichia coli O157:H7, abstr. J. Dairy Sci. 85(Suppl. 1):1304A. [Google Scholar]
- 19.Laughton, J., E. Devillard, D. Heinrichs, G. Reid, and J. McCormick. 2006. Inhibition of expression of a staphylococcal superantigen-like protein by a secreted signaling factor from Lactobacillus reuteri. Microbiology 152:1155-1167. [DOI] [PubMed] [Google Scholar]
- 20.Lorea Baroja, M., P. V. Kirjavainen, S. Hekmat, and G. Reid. 2007. Anti-inflammatory effects of probiotic yogurt in inflammatory bowel disease patients. Clin. Exp. Immunol. 149:470-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, R., M. Olivares, M. L. Marin, J. Xaus, L. Fernandez, and J. M. Rodriguez. 2005. Characterization of a reuterin-producing Lactobacillus coryniformis strain isolated from a goat's milk cheese. Int. J. Food Microbiol. 104:267-277. [DOI] [PubMed] [Google Scholar]
- 22.McCoy, S., and S. E. Gilliland. 2007. Isolation and characterization of Lactobacillus species having potential for use as probiotic cultures for dogs. J. Food Sci. 72:M94-M97. [DOI] [PubMed] [Google Scholar]
- 23.Molin, G., M.-L. Johansson, M. Stahl, S. Ahrné, R. Andersson, B. Jeppsson, and S. Bengmark. 1992. Systematics of the Lactobacillus population on rat mucosa with special reference to Lactobacillus reuteri. Antonie van Leeuwenhoek 61:175-183. [DOI] [PubMed] [Google Scholar]
- 24.Naito, S., H. Hayashidani, K. Kaneko, M. Ogawa, and Y. Benno. 1995. Development of intestinal lactobacilli in normal piglets. J. Appl. Bacteriol. 79:230-236. [DOI] [PubMed] [Google Scholar]
- 25.Pawelkiewicz, J., and B. Zagalak. 1965. Enzymic conversion of glycerol into β-hydroxy-propionaldehyde in a cell-free extract from Aerobacter aerogenes. Acta Biochim. Pol. 12:207-218. [PubMed] [Google Scholar]
- 26.Ratcliffe, B., C. B. Cole, R. Fuller, and M. J. Newport. 1986. The effect of yoghurt and milk fermented with a porcine intestinal strain of Lactobacillus reuteri on the performance and gastrointestinal flora of pigs weaned at 2 days of age. Food Microbiol. 3:203-211. [Google Scholar]
- 27.Reid, G., A. W. Bruce, N. Fraser, C. Heinemann, J. Owen, and B. Henning. 2001. Oral probiotics can resolve urogenital infections. FEMS Immunol. Med. Microbiol. 30:49-52. [DOI] [PubMed] [Google Scholar]
- 28.Reid, G., R. L. Cook, and A. W. Bruce. 1987. Examination of strains of lactobacilli for properties which may influence bacterial interference in the urinary tract. J. Urol. 138:330-335. [DOI] [PubMed] [Google Scholar]
- 29.Reid, G., D. Charbonneau, J. Erb, B. Kochanowski, D. Beuerman, R. Poehner, and A. W. Bruce. 2003. Oral use of Lactobacillus rhamnosus GR-1 and L. fermentum RC-14 significantly alters vaginal flora: randomized, placebo-controlled trial in 64 healthy women. FEMS Immunol. Med. Microbiol. 35:131-134. [DOI] [PubMed] [Google Scholar]
- 30.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 31.Ross, R. P., S. Morgan, and C. Hill. 2002. Preservation and fermentation: past, present and future. Int. J. Food Microbiol. 79:3-16. [DOI] [PubMed] [Google Scholar]
- 32.Sarra, P. G., F. Dellaglio, and V. Bottazzi. 1985. Taxonomy of lactobacilli isolated from the alimentary tract of chickens. Syst. Appl. Microbiol. 6:86-89. [Google Scholar]
- 33.Sauvageot, N., K. Gouffi, J. M. Laplace, and Y. Auffray. 2000. Glycerol metabolism in Lactobacillus collinoides: production of 3-hydroxypropionaldehyde, a precursor of acrolein. Int. J. Food Microbiol. 55:167-170. [DOI] [PubMed] [Google Scholar]
- 34.Slininger, P. J., and R. J. Bothast. 1985. Optimizing aerobic conversion of glycerol to 3-hydroxypropionaldehyde. Appl. Environ. Microbiol. 50:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talarico, T. L., I. A. Casas, T. C. Chung, and W. J. Dobrogosz. 1988. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 32:1854-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talarico, T. L., and W. J. Dobrogosz. 1989. Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 33:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tungjaroenchai, W., C. H. White, W. E. Holmes, and M. A. Drake. 2004. Influence of adjunct cultures on volatile free fatty acids in reduced-fat Edam cheeses. J. Dairy Sci. 87:3224-3234. [DOI] [PubMed] [Google Scholar]
- 38.Velraeds, M. M., B. van de Belt-Gritter, H. C. van der Mei, G. Reid, and H. J. Busscher. 1998. Interference in initial adhesion of uropathogenic bacteria and yeasts to silicone rubber by a Lactobacillus acidophilus biosurfactant. J. Med. Microbiol. 47:1081-1085. [DOI] [PubMed] [Google Scholar]
- 39.Wadström, T., K. Andersson, M. Sydow, L. Axelsson, S. Lindgren, and B. Gullmar. 1987. Surface properties of lactobacilli isolated from the small intestine of pigs. J. Appl. Bacteriol. 62:513-520. [DOI] [PubMed] [Google Scholar]
- 40.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong, W., K. Millsap, H. Bialkowska-Hobrzanska, and G. Reid. 1998. Differentiation of Lactobacillus species by molecular typing. Appl. Environ. Microbiol. 64:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]