Abstract
In this study, 20 women with staphylococcal mastitis were randomly divided in two groups. Those in the probiotic group daily ingested 10 log10 CFU of Lactobacillus salivarius CECT5713 and the same quantity of Lactobacillus gasseri CECT5714 for 4 weeks, while those in the control one only ingested the excipient. Both lactobacillus strains were originally isolated from breast milk. On day 0, the mean staphylococcal counts in the probiotic and control groups were similar (4.74 and 4.81 log10 CFU/ml, respectively), but lactobacilli could not be detected. On day 30, the mean staphylococcal count in the probiotic group (2.96 log10 CFU/ml) was lower than that of the control group (4.79 log10 CFU/ml). L. salivarius CECT5713 and L. gasseri CECT5714 were isolated from the milk samples of 6 of the 10 women of the probiotic group. At day 14, no clinical signs of mastitis were observed in the women assigned to the probiotic group, but mastitis persisted throughout the study period in the control group women. In conclusion, L. salivarius CECT5713 and L. gasseri CECT5714 appear to be an efficient alternative for the treatment of lactational infectious mastitis during lactation.
Mastitis, an inflammation of one or more lobules of the mammary gland, is a common disease during lactation since its incidence oscillates between 3 and 33% of lactating mothers (5, 28). Although the disease may happen at any point during the lactation period, between 75 and 95% of cases occur within the first 12 weeks, and the frequency is particularly higher during the second and third weeks postpartum (21).
Lactational mastitis usually has an infectious origin (9). Traditionally, Staphylococcus aureus has been considered the main etiological agent, although Staphylococcus epidermidis is emerging as the leading cause in both human and veterinary medicine (3, 25, 29). Multidrug resistance to antibiotics and/or formation of biofilms is very common among clinical isolates of these two staphylococcal species; therefore, it is not strange that ca. 70 to 90% of the cases of staphylococcal mastitis in bovines (where this condition has been exhaustively studied) are refractory to antibiotherapy (27).
Breast milk is an important source of bacteria to the infant gut, where they play a key role in the initiation and development of the gut microbiota. Bacteria commonly isolated from this biological fluid include staphylococci, streptococci, lactococci, lactobacilli, and enterococci (6, 12). In previous studies, we isolated lactobacillus strains belonging to the species Lactobacillus gasseri, L. fermentum, and L. salivarius from milk of healthy mothers (10, 12) and showed that their probiotic potential is similar to that of the strains commonly used in commercial probiotic products (10, 14). It has also been suggested that commensal bacteria isolated from human milk have potential use as bacteriotherapeutic agents for the prevention of breast infections caused by S. aureus (6).
Parallel studies suggested that lactobacilli and other lactic acid bacteria present in human milk may have an endogenous origin (12) and, upon interactions with dendritic cells in the maternal gut, these bacteria would reach the mammary gland along the enteromammary pathway (13), a hypothesis that has been confirmed recently (19). In this context, the aim of the present study was to evaluate whether oral administration of two Lactobacillus strains isolated from breast milk, L. salivarius CECT5713 and L. gasseri CECT5714, may be an alternative or a complement for treating staphylococcal lactational mastitis in cases in which previous antibiotherapy was unsuccessful. In addition, a second objective was to investigate whether the oral administration of lactobacilli may actually lead to their presence in breast milk.
MATERIALS AND METHODS
Design of the study and collection of the milk samples.
A total of 20 women age 26 to 34 years with clinical symptoms of staphylococcal mastitis participated in the study. All of them met the following criteria: breast redness and pain, flu-like symptoms (including fever ≥ 38.5°C), a milk staphylococcal count higher than 4 log10 CFU/ml, and a milk leukocyte count higher than 6 log10 CFU/ml. Most of them (n = 14) presented with fissures in the mammary areola and/or nipple. All of them had received antibiotherapy (cloxacillin, clindamycin, amoxicillin-clavulanic acid, and/or erythromycin) for 2 to 4 weeks, but the respective treatments (which finished at least 2 weeks before the present study) had failed to improve their condition. None of them ingested commercial probiotic foods or supplements during the study. Women with mammary abscesses or any kind of parallel diseases were excluded. All volunteers gave written informed consent to the protocol, which was approved by the Ethical Committee of Hospital Clínico of Madrid (Spain). The volunteers were randomized into two groups (probiotic and control) by sealed envelope, and neither volunteers nor investigators knew the code during the investigation.
The study lasted 30 days and, during this period, the probiotic group (n = 10) daily consumed a capsule with 200 mg of a freeze-dried probiotic containing ∼10 log10 CFU each of L. salivarius CECT5713 and L. gasseri CECT5714 in a matrix of methylcellulose. Both strains were originally isolated from the breast milk of healthy women (10, 12). The entire process to obtain the probiotic strains and to prepare the capsules was performed in the industrial probiotic plant of Puleva Biotech S.A. (Granada, Spain). The capsules were kept at 4°C throughout the study. The viability of both strains was measured weekly by triplicate to guarantee that it was >99.99999% throughout the study. For this purpose, appropriate dilutions of the capsule content were spread onto plates of MRS agar (Oxoid, Basingstoke, United Kingdom), and identification of the colonies was carried out by species-specific PCR as described below. The placebo group daily received a capsule containing 200 mg of the same methylcellulose batch. Breast milk samples were obtained from the volunteers at the beginning of the study (before the ingestion of the first capsule [day 0]) and at the end of the trial period (day 30). One of the volunteers of the control group only provided milk samples from the right breast because the left one was not functional as a result of a previous carcinoma. To collect the breast milk samples, nipple and mammary areola were cleaned with soap and sterile water, and then chlorhexidine was applied. The breast milk sample was collected in a sterile tube after manual expression by using sterile gloves. The first drops (approximately 250 μl) were discarded to avoid chlorhexidine contamination. The evolution of the symptoms was evaluated weekly by midwives of the day care centers to which the volunteers were ascribed.
Count and identification of bacteria in the milk samples.
Proper dilutions of the fresh breast milk samples were spread onto Baird-Parker (BP) agar plates (bioMérieux, Marcy l'Etoile, France) for selective isolation and quantification of staphylococci and, in parallel, onto agar plates of MRS agar supplemented with l-cysteine (0.5 g/liter) (MRS-Cys) for the isolation of lactobacilli. All of the plates were incubated for 48 h at 37°C, the BP plates in aerobic conditions and those of MRS-Cys anaerobically (85% nitrogen, 10% hydrogen, 5% carbon dioxide) in a MACS-MG-1000-anaerobic workstation (DW Scientific, Shipley, United Kingdom). Although staphylococci can grow in MRS-Cys, they are easily differentiated from lactobacilli (gram-positive, catalase-negative rods and gram-positive, catalase-positive cocci, respectively). Lactobacilli do not grow in BP medium. As a consequence, all of the colony types growing on BP and MRS-Cys plates were subjected to microscope observation (shape, Gram staining) and assayed for catalase activity.
Staphylococci (10 colonies from each milk sample) were identified at the species level by classical morphological and biochemical tests and by a novel multiplex PCR method based on the dnaJ genes. Briefly, a single colony growing on solid medium was resuspended in 100 μl of sterile deionized water. Then, 100 μl of chloroform-isoamyl alcohol (24:1) was added to the suspension, which was stirred for 5 s and centrifuged at 16,000 × g for 5 min at 4°C. Subsequently, 5 to 10 μl of the upper aqueous phase was used as a source of DNA template for PCR with the primers J-StGen (5′-TGGCCAAAAGAGACTATTATGA-3′), J-StAur (5′-GGATCTCTTTGTCTGCCG-3′), and J-StEpi (5′-CCACCAAAGCCTTGACTT-3′) in an Icycler thermocycler (Bio-Rad Laboratories, Richmond, CA). The primer pairs J-StGen/J-StAur and J-StGen/J-StEpi result in a 337-bp S. aureus species-specific fragment and a 249-bp S. epidermidis species-specific fragment, respectively. The PCR conditions were as follows: 1 cycle of 94°C for 4 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, with a final extension of 72°C for 5 min. The identification of the staphylococcal isolates either as Staphylococcus epidermidis or S. aureus was confirmed by 16S rRNA sequencing by using the primers pbl16 (5′-AGAGTTTGATCCTGGCTCAG-3′) and mlb16 (5′-GGCTGCTGGCACGTAGTTAG-3′) (8). The PCR conditions were as follows: 96°C for 30 s, 48°C for 30 s, and 72°C for 45 s (40 cycles) and a final extension at 72°C for 4 min. The amplicons were purified by using a Nucleospin Extract II kit (Macherey-Nagel, Düren, Germany) and sequenced at the Genomics Unit of the Universidad Complutense de Madrid (Spain). The resulting sequences were used to search sequences deposited in the EMBL database using BLAST algorithm, and the identities of the isolates were determined on the basis of the highest scores (≥99%).
Microbiological data, recorded as CFU/ml of milk, were transformed to logarithmic values before statistical analysis. The reported values of bacterial counts are the mean values of duplicate or triplicate determinations ± the standard deviations (SD). The Kruskal-Wallis test was applied to determine whether the obtained mean values of log staphylococcal counts within each experimental group were identically distributed before starting the treatment. The Mann-Whitney (Wilcoxon) test was used to evaluate the differences between the probiotic and the control group. The significance level was established at P < 0.01. All analyses were performed by using the Statgraphics Plus 5.0 software (Manugistics, Inc., Rockville, MD).
Detection of L. salivarius and L. gasseri in the milk samples by colony hybridization, species-specific PCR, and 16S rRNA sequencing.
A DNA-DNA colony hybridization assay was developed to investigate whether the oral administration of the lactobacillus strains led to their presence in breast milk at day 30. For this purpose, two species-specific probes (for the detection of L. salivarius and L. gasseri, respectively) were designed on the basis of unique 16S rRNA sequences. In the case of L. salivarius, a fragment (210 bp) was amplified from L. salivarius CECT5713 genomic DNA using the primers SAL91F (5′-ATTCACCGTAAGAAGT-3′) and SAL285R (5′-TATCATCACCTTGGTAG-3′). Genomic DNA was isolated from 10 ml overnight MRS cultures by using the DNeasy tissue kit (Qiagen, Hilden, Germany) according to the protocol recommended by the supplier for isolation of genomic DNA from gram-positive bacteria. Separately, a fragment (200 bp) was amplified from L. gasseri CECT5714 genomic DNA using the primers Gas I (5′-GAGTGCGAGAGCACTAAAG-3′) and Gas II (5′-TATCATCACCTTGGTAG-3′). The PCR conditions were as follows: 95°C for 2 min (1 cycle); 95°C for 30 s, 46°C (L. salivarius) or 55°C (L. gasseri) for 30 s, and 72°C for 45 s (40 cycles); and a final extension at 72°C for 4 min. Both PCR fragments were purified by using the QIAquick spin PCR purification kit (Qiagen) and labeled by using the Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare, Little Chalfont, United Kingdom).
For colony hybridization, colonies (100 per mother) obtained on MRS-Cys plates from breast milk samples (day 30) were grown in MRS broth at 37°C overnight. These cultures were then spotted in a regular array on two sets of MRS-Cys replica plates. Once the colonies had grown, nylon Hybond-N+ discs (GE Healthcare) were laid directly on the culture surfaces and kept for at least 1 min. Then, both hybridization and detection were carried out according to the instructions of the Amersham ECL direct nucleic acid labeling and detection system with the modifications previously described (15) and a probe concentration of approximately 10 ng/ml. The identity of the isolates that gave a positive signal after colony hybridization was confirmed by 16S rRNA sequencing using the primers pbl16 and mlb16 as described above.
In parallel, identification of the isolates was also assessed by species-specific PCR using the primers lowlac (5′-CGACGACCATGAACCACCTGT-3′) and sal1 (5′-ATTCACTCGTAAGAAGT-3′) for L. salivarius, which results in a 993-bp fragment (2), and the primers Lgas-1 (5′-AGCGACCGAGAAGAGAGAGA-3′) and Lgas-2 (5′-TGCTATCGCTTCAAGTGCTT-3′) for L. gasseri, which generates a 360-bp fragment (23).
Identification of L. salivarius CECT5713 and L. gasseri CECT5714 in the milk samples by PFGE.
Later, and to check whether L. salivarius and L. gasseri isolates actually belonged to the strains CECT5713 and CECT5714, respectively, the samples were subjected to pulsed-field gel electrophoresis (PFGE) genotyping. Chromosomal DNA was extracted from the isolates and digested with the endonuclease SmaI (New England Biolabs, Ipswich, MA) at 25°C for 24 h. Electrophoresis was carried out in a CHEF DR II apparatus (Bio-Rad, Birmingham, United Kingdom) in 1% (wt/vol) SeaKem GTG agarose (FMC, Philadelphia, PA) with 0.5× TBE buffer (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA [pH 8.0]) at 15°C. A constant voltage of 200 V was applied to the system, and fragment separation was performed by using a two-phase program. The electrophoretic conditions for separating the SmaI fragments were a pulse from 0.5 to 5 s for 10 h and then another from 0.5 to 10 s for 6 h. LowRange PFG marker and MidRange PFG marker I (New England Biolabs) were used as molecular size standards. Agarose gels were stained with ethidium bromide (0.5 μg/ml), and images were digitized with a GelPrinter Plus System (TDI, Madrid, Spain).
RESULTS
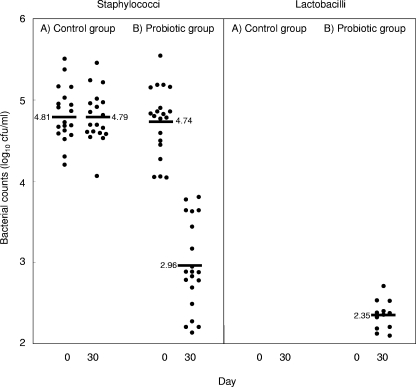
Counts of staphylococci and lactobacilli in the milk samples. At day 0, the total staphylococcal counts in the breast milk of all of the women ranged from 4.04 to 5.54 log10 CFU/ml (Fig. 1). Mean staphylococcal counts in the probiotic and control groups (Table 1) were similar (4.74 and 4.81 log10 CFU/ml, respectively). The Kruskal-Wallis test confirmed that the mean values of log staphylococcal counts were identically distributed in both groups before the trial (P = 0.806). On the other hand, lactobacilli could not be detected at that sampling time in any of these samples. By using species-specific PCR and 16S rRNA sequencing, the staphylococci isolated from milk of subjects 1, 2, 5, 8, 12, 14, 19, and 20 were identified as S. aureus, while those present in the rest of the women were identified as S. epidermidis. The partial 16S RNA gene sequences obtained from the S. aureus and the S. epidermidis isolates were deposited in the EMBL nucleotide sequence database under accession numbers EU280807 and EU280808, respectively. No other staphylococcal species were detected.
FIG. 1.
Staphylococcal counts in the milk samples obtained from women of the control (A) and probiotic (B) groups at days 0 and 30. The black bars (and the associated number) indicate the mean of the values.
TABLE 1.
Staphylococcus and lactobacillus counts in the breast milk samples
| Group | Subject | Breasta | Count (log10 CFU/ml ± SD)d
|
|||
|---|---|---|---|---|---|---|
| Staphylococci
|
Lactobacilli
|
|||||
| Day 0 | Day 30 | Day 0 | Day 30 | |||
| Probiotic | 1 | R | 4.86 ± 0.01 | 2.77 ± 0.10 | ND | 2.53 ± 0.05 |
| groupb | L | 4.82 ± 0.02 | 2.20 ± 0.01 | ND | 2.53 ± 0.02 | |
| 2 | R | 5.54 ± 0.04 | 3.77 ± 0.06 | ND | ND | |
| L | 4.05 ± 0.08 | 2.68 ± 0.01 | ND | ND | ||
| 3 | R | 5.18 ± 0.09 | 3.64 ± 0.01 | ND | ND | |
| L | 5.18 ± 0.13 | 3.64 ± 0.03 | ND | ND | ||
| 4 | R | 4.77 ± 0.03 | 2.88 ± 0.03 | ND | 2.12 ± 0.11 | |
| L | 4.50 ± 0.02 | 2.88 ± 0.03 | ND | 2.18 ± 0.12 | ||
| 5 | R | 4.04 ± 0.07 | 2.20 ± 0.03 | ND | 2.37 ± 0.07 | |
| L | 4.44 ± 0.06 | 2.82 ± 0.02 | ND | 2.38 ± 0.03 | ||
| 6 | R | 4.83 ± 0.04 | 3.44 ± 0.04 | ND | ND | |
| L | 4.80 ± 0.01 | 2.49 ± 0.10 | ND | ND | ||
| 7 | R | 4.06 ± 0.09 | 2.27 ± 0.12 | ND | 2.32 ± 0.09 | |
| L | 5.15 ± 0.09 | 3.17 ± 0.09 | ND | 2.09 ± 0.10 | ||
| 8 | R | 5.16 ± 0.12 | 3.62 ± 0.04 | ND | ND | |
| L | 4.90 ± 0.08 | 3.80 ± 0.11 | ND | ND | ||
| 9 | R | 4.59 ± 0.06 | 2.94 ± 0.05 | ND | 2.35 ± 0.06 | |
| L | 4.78 ± 0.02 | 2.78 ± 0.04 | ND | 2.70 ± 0.05 | ||
| 10 | R | 4.85 ± 0.02 | 2.13 ± 0.08 | ND | 2.40 ± 0.03 | |
| L | 4.27 ± 0.08 | 2.88 ± 0.05 | ND | 2.20 ± 0.20 | ||
| Control | 11 | R | 4.95 ± 0.02 | 4.60 ± 0.04 | ND | ND |
| groupc | L | NA | NA | NA | NA | |
| 12 | R | 5.16 ± 0.02 | 5.21 ± 0.03 | ND | ND | |
| L | 5.16 ± 0.06 | 5.24 ± 0.09 | ND | ND | ||
| 13 | R | 4.51 ± 0.08 | 4.54 ± 0.07 | ND | ND | |
| L | 4.57 ± 0.06 | 4.61 ± 0.03 | ND | ND | ||
| 14 | R | 4.66 ± 0.05 | 4.65 ± 0.09 | ND | ND | |
| L | 4.68 ± 0.04 | 4.69 ± 0.04 | ND | ND | ||
| 15 | R | 4.90 ± 0.02 | 4.91 ± 0.01 | ND | ND | |
| L | 4.68 ± 0.02 | 4.69 ± 0.02 | ND | ND | ||
| 16 | R | 5.50 ± 0.09 | 5.45 ± 0.03 | ND | ND | |
| L | 5.03 ± 0.05 | 4.97 ± 0.08 | ND | ND | ||
| 17 | R | 5.37 ± 0.01 | 4.84 ± 0.03 | ND | ND | |
| L | 4.58 ± 0.04 | 4.58 ± 0.07 | ND | ND | ||
| 18 | R | 4.72 ± 0.06 | 4.96 ± 0.02 | ND | ND | |
| L | 4.86 ± 0.02 | 4.81 ± 0.02 | ND | ND | ||
| 19 | R | 4.62 ± 0.12 | 4.59 ± 0.04 | ND | ND | |
| L | 4.94 ± 0.02 | 5.01 ± 0.05 | ND | ND | ||
| 20 | R | 4.20 ± 0.05 | 4.06 ± 0.04 | ND | ND | |
| L | 4.30 ± 0.04 | 4.53 ± 0.04 | ND | ND | ||
The milk sample was obtained from the right (R) or left (L) breast.
Mean values (log10 CFU/ml ± SD): 4.74 ± 0.41 (staphylococci at day 0), 2.96 ± 0.55 (staphylococci at day 30), and 2.35 ± 0.18 (lactobacilli at day 0).
Mean values (log10 CFU/ml ± SD): 4.81 ± 0.22 (staphylococci at day 0) and 4.79 ± 0.23 (staphylococci at day 30).
ND, not determined; NA, sample not available.
On day 30, the mean staphylococcal count in the probiotic group (2.96 log10 CFU/ml) was statistically lower than that corresponding to the control group (4.79 log10 CFU/ml). Reductions of approximately 1.5 to 2.0 log cycles in the staphylococcal count were observed among the milk samples of the probiotic group. In contrast, the concentration of staphylococci in the control group remained stable during the trial period (Fig. 1). Again, all of the staphylococcal isolates were identified as S. aureus (subjects 1, 2, 5, 8, 12, 14, 19, and 20) or S. epidermidis (the rest of the women). A Mann-Whitney test revealed that there were statistically significant differences between the probiotic and the control group concerning the staphylococcal values (Wilcoxon, P = 0.002). In relation to the lactobacilli, isolates belonging to this genus could not be detected in any of the control group samples, but they were isolated (2.09 to 2.70 log10 CFU/ml) in the samples obtained from 6 of the 10 women in the probiotic group (Table 1).
Evolution of the clinical symptoms.
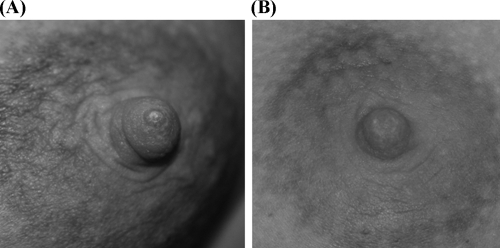
Clinical symptoms were evaluated weekly by a midwife. At day 7, symptoms had notably improved among women of the probiotic group since local inflammation and flu-like signs had disappeared. At day 14, no clinical signs of mastitis were observed in the women assigned to this group and, in the case of those that initially displayed fissures in the nipple and/or mammary areola (subjects 1, 3, 4, 5, 6, and 9), the fissures were completely healed (Fig. 2). In contrast, clinical signs persisted in control group women throughout the study period.
FIG. 2.
Mammary areola of one of the probiotic group women at day 0, in which redness and a nipple crack are clearly visible (A) and at day 14 show a normal appearance (B).
Detection of L. salivarius CECT5713 and L. gasseri CECT5714 in the milk samples.
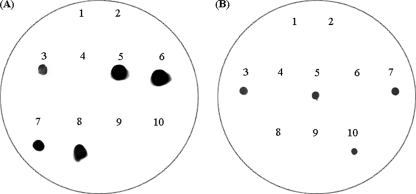
A total of 100 isolates (per mother) obtained on MRS-Cys plates from day 30 milk samples were spotted in a regular array onto two sets of MRS-Cys replica plates. One of the sets was submitted to colony hybridization using a L. gasseri-specific probe and, subsequently, an L. salivarius-specific probe. Both L. salivarius and L. gasseri isolates were detected in the milk samples of six of the women belonging to the probiotic group (Fig. 3). In the six cases, ca. 70% of the lactobacilli hybridized with the L. salivarius probe, while the remaining 30% hybridized with that of L. gasseri. The isolates that did not react with any of the probes were later identified as staphylococci, which is not strange since this bacterial group can grow on MRS-Cys plates. All of the potential lactobacillus isolates hybridized with any of these probes, which suggested that only these two species were present in the samples. These results were confirmed by L. salivarius and L. gasseri species-specific PCR using primers different from those used to generate the respective probes and by nucleotide sequencing of PCR fragments corresponding to the 16S rRNA gene. The sequences obtained from one of the L. gasseri and one of the L. salivarius isolates have been deposited in the GenBank database under accession numbers EU035754 and AM087452, respectively.
FIG. 3.
Colony hybridization analysis of isolates obtained from the milk sample of woman 1 (probiotic group) at day 30 using either an L. salivarius-specific (A) or an L. gasseri-specific probe (B). (A) Spots: 1, L. gasseri CECT5714; 3, L. salivarius CECT5713; 5, 6, 7, and 8, positive isolates belonging to the species L. salivarius; 2, 4, 9, and 10, negative isolates that were identified as Staphylococcus spp. (B) Spots: 1, L. salivarius CECT5713; 3, L. gasseri CECT5714; 5, 7, and 10, positive isolates belonging to the species L. gasseri; 2, 4, 6, 8, and 9, negative isolates (Staphylococcus spp.).
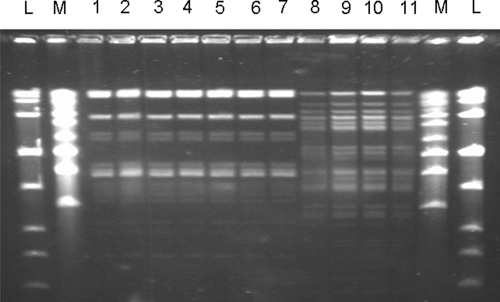
Finally, the lactobacillus isolates were genetically typified by the PFGE technique. The profiles revealed that all L. salivarius and L. gasseri isolates actually belonged to the strains CECT5713 and CECT5714, respectively (Fig. 4).
FIG. 4.
PFGE patterns of SmaI-digested genomic DNA from L. salivarius CECT5713 (lane 1), six milk isolates that hybridized with the L salivarius probe in the colony hybridization assay (lanes 2 to 7), L. gasseri CECT5714 (lane 8), and three milk isolates that hybridized with the L. gasseri probe in the hybridization assay (lanes 9 to 11). L and M represent the standards LowRange PFG and MidRange PFG, respectively.
DISCUSSION
Staphylococci are the main etiological agents of infectious mastitis during lactation. At the species level, S. aureus has been traditionally considered the most common agent; however, recent studies have shown the increasing importance of S. epidermidis in bovine mastitis and have revealed that its incidence could be even higher than that of S. aureus (3, 25, 29). In fact, in the present study, 40% of the women carried S. aureus in their milk, while 60% of them harbored S. epidermidis isolates. Previously, it had been suggested that coagulase-negative staphylococci should be considered as a possible etiologic agent of mastitis in nursing women since the inoculation of S. epidermidis strains isolated from human mastitis into the mammary glands of lactating mice led to clinical and histological signs of mastitis (24). Therefore, the results of the present study confirm that S. epidermidis may be an underrated cause of human lactational mastitis.
Independently of the species involved, mastitis-causing strains usually display two common properties: resistance to methicillin and other antibiotics and a high ability to form biofilms. This explains why this condition uses to be elusive to antibiotherapy and why it usually becomes a recurrent or chronic infection. In fact, ca. 25% of mothers cite such condition as their reason to cease breast-feeding (28). In this context, the development of new strategies based on bacteriotherapy, a practice that makes use of beneficial bacteria to prevent or treat colonization of the host by pathogens (7), as an alternative or complement to antibiotherapy is particularly attractive. In a previous study, we isolated a variety of lactobacillus strains from human milk, including L. salivarius CECT5713 and L. gasseri CECT5714 (10, 12). Subsequent studies revealed that both strains were good probiotic candidates since they reached high survival rates when exposed to the gastrointestinal tract conditions, showed a strong adherence to intestinal cells, stimulate the expression of mucin-encoding genes, produced antimicrobial compounds (lactate, acetate and hydrogen peroxide) in vitro, and displayed in vivo antibacterial properties against pathogenic bacteria (10, 14, 18). The presence of hydrogen peroxide-producing lactobacilli in breast milk seems especially interesting since it has been reported that lactobacilli with such an ability inhibit the growth of S. aureus (17). Because of their anti-infectious properties and breast milk origin, these strains are particularly appealing as a probiotic alternative for the treatment of infectious mastitis. In addition, it has already been shown that lactic acid bacteria isolated from human milk have potential use as bacteriotherapeutic agents in preventing neonatal and maternal breast infections caused by S. aureus (6).
In the present study, reductions of approximately 1.5- to 2-log cycles in the milk staphylococcal counts led to a rapid improvement of the mastitic condition. The final staphylococcal count was 2 to 3 log10 CFU/ml, and this number has been reported as a normal and acceptable staphylococcus load in milk of healthy women (6, 28). The fact that, after the probiotic treatment, L. salivarius CECT5713 and L. gasseri CECT5714 were the unique lactobacilli detected in milk is not surprising since the Lactobacillus composition of infant feces and breast milk is host specific and usually includes a low number of lactobacillus species (6, 11, 12); for example, the examination of Lactobacillus gut colonization in 112 breast-fed infants showed that during the first 6 months of life, 26% of them had no lactobacilli, 37% carried a single strain, 26% two strains, and only 11% three or more strains (1). L. salivarius CECT 5713 was the strain predominant in milk after the probiotic treatment. Studies with other L. salivarius strains in animal models and clinical trials have demonstrated their probiotic function and, particularly, their anti-inflammatory effects (4, 16, 22). The combination of anti-infectious and anti-inflammatory properties may explain the effect of the probiotic treatment in the present study.
Lactobacilli present in the maternal gut can cross the intestinal epithelium and reach the mammary gland through an endogenous route, the enteromammary pathway, which is responsible for the abundance of elements of the immunological system in human milk. It has been demonstrated that dendritic cells can penetrate the gut epithelium to take up noninvasive bacteria directly from the gut lumen (20). Once associated with gut-associated lymphoid tissue cells, live noninvasive bacteria can spread to other locations since there is a circulation of lymphocytes within the mucosal associated lymphoid system. Bacterium-stimulated cells move from the intestinal mucosa to colonize distant mucosal surfaces, such as those of the respiratory and genitourinary tracts, salivary and lachrymal glands and, most significantly, that of the lactating mammary gland. In fact, up to 16 lactobacillus species, including L. gasseri and L. salivarius, have been previously isolated from the blood of healthy people (26). Such enteromammary bacterial circulation has been confirmed recently (19) and would explain the beneficial effect observed in the present study since no lactobacilli could be isolated from breast skin of any women of the probiotic group (data not shown), which rules out the hypothesis of a direct fecal contamination of the breast.
The results obtained here suggest that L. salivarius CECT 5713 and L. gasseri CECT5714 can be used as an effective alternative to antibiotics for the treatment of infectious mastitis during lactation. Therefore, in the coming months we will begin a large multicentric trial to confirm the effect and, in addition, study a potential preventive effect in pregnant women with a previous history of lactational mastitis.
Acknowledgments
This study was supported by the FUN-C-FOOD (Consolider-Ingenio 2010) and AGL2007-62042 projects from the Ministerio de Educación y Ciencia (Spain).
We are grateful to S. Ferrer (Universidad de Valencia, Valencia, Spain) for assistance in PFGE analyses and to the Association “Amamantar” (Avilés, Asturias) for support in the collection of the samples.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Ahrné, S., E. Lönnermark, A. E. Wold, N. Aberg, B. Hesselmar, R. Saalman, I. L. Strannegård, G. Molin, and I. Adlerberth. 2005. Lactobacilli in the intestinal microbiota of Swedish infants. Microbes Infect. 7:1256-1262. [DOI] [PubMed] [Google Scholar]
- 2.Chagnaud, P., K. Machinis, L. A. Coutte, A. Marecat, and A. Mercenier. 2001. Rapid PCR-based procedure to identify lactic acid bacteria: application to six common Lactobacillus species. J. Microbiol. Methods 44:139-148. [DOI] [PubMed] [Google Scholar]
- 3.dos Santos Nascimento, J., P. C. Fagundes, M. A. de Paiva Brito, K. R. dos Santos, and M. do Carmo de Freire Bastos. 2005. Production of bacteriocins by coagulase-negative staphylococci involved in bovine mastitis. Vet. Microbiol. 106:61-71. [DOI] [PubMed] [Google Scholar]
- 4.Dunne, C., L. O'Mahony, L. Murphy, G. Thornton, D. Morrissey, S. O'Halloran, M. Feeney, S. Flynn, G. Fitzgerald, C. Daly, B. Kiely, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 2001. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr. 73(Suppl.):386S-392S. [DOI] [PubMed] [Google Scholar]
- 5.Foxman, B., H. D'Arcy, B. Gillespie, J. K. Bobo, and K. Schwartz. 2002. Lactation mastitis: occurrence and medical management among 946 breast-feeding women in the United States. Am. J. Epidemiol. 155:103-114. [DOI] [PubMed] [Google Scholar]
- 6.Heikkilä, M. P., and P. E. J. Saris. 2003. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 95:471-478. [DOI] [PubMed] [Google Scholar]
- 7.Huovinen, P. 2001. Bacteriotherapy: the time has come. BMJ 323:353-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kullen, M. J., R. B. Sanozky-Dawes, D. C. Crowell, and T. R. Klaenhammer. 2005. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 89:511-516. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence, R. A., and R. M. Lawrence. 2005. Breastfeeding: a guide for the medical profession, 6th ed. Mosby, St. Louis, MO.
- 10.Martín, R., E. Jiménez, M. Olivares, M. L. Marín, L. Fernández, J. Xaus, and J. M. Rodríguez. 2006. Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair. Int. J. Food Microbiol. 112:35-43. [DOI] [PubMed] [Google Scholar]
- 11.Martín, R., H. G. H. J. Heilig, E. G. Zoetendal, E. Jiménez, L. Fernández, H. Smidt, and J. M. Rodríguez. 2007. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res. Microbiol. 158:31-37. [DOI] [PubMed] [Google Scholar]
- 12.Martín, R., S. Langa, C. Reviriego, E. Jiménez, M. L. Marín, J. Xaus, and J. M. Rodríguez. 2003. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 143:754-758. [DOI] [PubMed] [Google Scholar]
- 13.Martín, R., S. Langa, C. Reviriego, E. Jiménez, M. L. Marín, M. Olivares, J. Boza, J. Jiménez, L. Fernández, J. Xaus, and J. M. Rodríguez. 2004. The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci. Technol. 15:121-127. [Google Scholar]
- 14.Martín, R., M. Olivares, M. L. Marín, L. Fernández, J. Xaus, and J. M. Rodríguez. 2005. Probiotic potential of 3 lactobacilli strains isolated from breast milk. J. Hum. Lact. 21:8-17. [DOI] [PubMed] [Google Scholar]
- 15.Martínez, M. I., E. Rodríguez, M. Medina, P. E. Hernández, and J. M. Rodríguez. 1998. Detection of specific bacteriocin-producing lactic acid bacteria by colony hybridization. J. Appl. Microbiol. 84:1099-1103. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy, J., L. O'Mahony, L. O'Callaghan, B. Sheil, E. E. Vaughan, N. Fitzsimons, J. Fitzgibbon, G. C. O'Sullivan, B. Kiely, J. K. Collins, and F. Shanahan. 2003. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut 52:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ocaña, V. S., A. A. Pesce de Ruiz Holgado, and M. E. Nader-Macías. 1999. Selection of H2O2-generating Lactobacillus species for probiotic use. Curr. Microbiol. 38:279-284. [DOI] [PubMed] [Google Scholar]
- 18.Olivares, M., M. P. Díaz-Ropero, R. Martín, J. M. Rodríguez, and J. Xaus. 2006. Antimicrobial potential of four Lactobacillus strains isolated from breast milk. J. Appl. Microbiol. 101:72-79. [DOI] [PubMed] [Google Scholar]
- 19.Perez, P. F., J. Doré, M. Leclerc, F. Levenez, J. Benyacoub, P. Serrant, I. Segura-Roggero, E. J. Schiffrin, and A. Donnet-Hughes. 2007. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119:e724-e732. [DOI] [PubMed] [Google Scholar]
- 20.Rescigno, M., M. Urbano, B. Valsazina, M. Francoloni, G. Rotta, R. Bonasio, F. Granucci, J. P. Kraehenbuhl, and P. Ricciardi-Castagnoli. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2:361-367. [DOI] [PubMed] [Google Scholar]
- 21.Riordan, J. M., and F. H. Nichols. 1990. A descriptive study of lactation mastitis in long-term breast-feeding women. J. Hum. Lact. 6:53-58. [DOI] [PubMed] [Google Scholar]
- 22.Sheil, B., J. McCarthy, L. O'Mahony, M. W. Bennett, P. Ryan, J. J. Fitzgibbon, B. Kiely, J. K. Collins, and F. Shanahan. 2004. Is the mucosal route of administration essential for probiotic function? Subcutaneous administration is associated with attenuation of murine colitis and arthritis. Gut 53:694-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song, Y., N. Kato, C. Liu, Y. Matsumiya, H. Kato, and K. Watanabe. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187:167-173. [DOI] [PubMed] [Google Scholar]
- 24.Thomsen, A. C., S. C. Mogensen, and F. Love Jepsen. 1985. Experimental mastitis in mice induced by coagulase-negative staphylococci isolated from cases of mastitis in nursing women. Acta Obstet. Gynecol. Scand. 64:163-166. [DOI] [PubMed] [Google Scholar]
- 25.Thorberg, B. M., I. Kuhn, F. M. Aarestrup, B. Brandstrom, P. Jonsson, and M. L. Danielsson-Tham. 2006. Pheno- and genotyping of Staphylococcus epidermidis isolated from bovine milk and human skin. Vet. Microbiol. 115:163-172. [DOI] [PubMed] [Google Scholar]
- 26.Vankerckhoven, V. V., T. A. Autgaerden, G. Huys, M. Vancanneyt, J. Swings, and H. Goossens. 2004. Establishment of the PROSAFE collection of probiotic and human lactic acid bacteria. Microbial Ecol. Health Dis. 16:131-136. [Google Scholar]
- 27.Wall, R. J., A. M. Powell, M. J. Paape, D. E. Kerr, D. D. Bannerman, V. G. Pursel, K. D. Wells, N. Talbot, and H. W. Hawk. 2005. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat. Biotechnol. 23:445-451. [DOI] [PubMed] [Google Scholar]
- 28.World Health Organization. 2000. Mastitis: causes and management. World Health Organization, Geneva, Switzerland.
- 29.Zhang, S., and C. W. Maddox. 2000. Cytotoxic activity of coagulase-negative staphylococci in bovine mastitis. Infect. Immun. 68:1102-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]