Abstract
A reverse zymogram method for the detection of bacterial lysozyme inhibitors was developed. This method was validated by using a periplasmic protein extract of Escherichia coli containing a known inhibitor and subsequently led to the detection of a new proteinaceous hen egg white lysozyme inhibitor in Proteus mirabilis.
Lysozymes (EC 3.2.1.17) are hydrolytic enzymes that exert an antibacterial activity by cleaving the β(1,4)-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine in peptidoglycan, the major bacterial cell wall polymer. They are commonly found in animals, plants, microorganisms, and viruses and are divided into different families and types (8). In animals, there are three major lysozyme types; c- and g-type lysozymes typically occur in vertebrates, while invertebrates generally have i-type and sometimes also c-type lysozymes (7).
In 2001, the first proteinaceous lysozyme inhibitor was accidentally discovered in the periplasm of Escherichia coli and named Ivy, for inhibitor of vertebrate lysozyme (11). Ivy orthologs share a specific protruding five-residue loop that is essential for its inhibitory activity and are encountered in a number of gram-negative bacteria (1). Most recently, a second family of c-type lysozyme inhibitors was discovered in gram-negative bacteria (2). This novel and widely distributed family is characterized by a common conserved COG3895 domain (Cluster of Orthologous Groups, http://www.ncbi.nlm.nih.gov/COG/; 2, 14) and can be divided into periplasmic (PliC) or membrane-bound (MliC) lysozyme inhibitors of c-type lysozyme. Notably, Ivy (from E. coli), PliC of Salmonella enterica serovar Enteritidis, and MliC from E. coli, as well as from Pseudomonas aeruginosa, were all shown to contribute to the protection of the bacterial cell against challenge with hen egg white lysozyme (HEWL), the model c-type lysozyme (2, 4). Therefore, lysozyme inhibitors may have evolved to counter lysozyme as an important component of the immune system of animal hosts. In pathogenic bacteria, they may even contribute to virulence, which would make them an attractive novel target for antibacterial drug development (1, 2). In support of such a role, we recently found that the lysozyme inhibitor Ivy is required for the ability of E. coli to grow in saliva (5).
Our earlier described method to screen for lysozyme inhibitors is based on detecting inhibitory activity in bacterial extracts, and although it provides a good indication of the presence of inhibitory activity (2, 4), it inherently fails to reveal any information about the nature of the inhibitor (proteinaceous or not) or about the possible presence of multiple inhibitors in an extract. We have noticed that this technique is susceptible to false-positive results when searching for proteinaceous inhibitors due to the presence of substances that inhibit or influence HEWL activity (e.g., acidic polymers, salts, metal ions, lipopolysaccharides, peptidoglycan fragments, etc.). While some of these compounds may also have biological relevance, we were mainly interested in novel, highly specific proteinaceous HEWL inhibitors like the ones recently discovered in bacteria (2, 11). To detect these inhibitors in cell extracts or culture supernatant, a reverse zymogram method using HEWL and Micrococcus luteus cells as a substrate was developed. This method was validated for a periplasmic protein extract of E. coli containing Ivy and subsequently led to the detection of a proteinaceous HEWL inhibitor different from Ivy in a periplasmic extract of Proteus mirabilis which was further investigated.
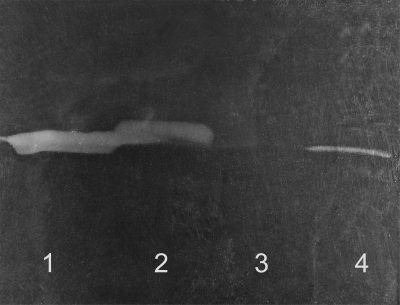
Periplasmic protein extracts isolated as described elsewhere (4) from wild-type E. coli MG1655, an isogenic ivy::Kan strain (Ivy knockout strain; 4), and a wild-type strain containing the plasmid pAA410 (Ivy overexpression strain; 4) were examined for HEWL inhibitory activity with the traditional assay described in reference 3. As expected, increasing amounts of inhibitory activity were measured for the knockout, wild-type, and overexpression extracts, in that order. The HEWL inhibitory activities of the periplasmic protein extracts were as follows: E. coli MG1655 ivy::Kan, 0 IU/ml; E. coli MG1655, 18 IU/ml; and E. coli MG1655(pAA410), 746 IU/ml. The reverse zymogram technique, already frequently used in the detection of protease inhibitors (13), was optimized for the detection of HEWL inhibitors with M. luteus as the substrate. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (10) with a 15% stacking and a 4% concentrating gel were supplemented with 1 μg/ml freeze-dried M. luteus ATCC 4698 cells (Sigma-Aldrich, Bornem, Belgium) and 0.25 μg/ml HEWL (66,000 U/mg protein; Fluka) just before polymerization. Samples of periplasmic protein extracts of the E. coli strains were prepared by boiling for 3 min in the presence of sample buffer (0.1 M Tris-HCl [pH 6.8], 1% SDS, 40% glycerol, 10 mM dithiothreitol, 0.0025% bromophenol blue) to denature the proteins. After electrophoresis with running buffer supplemented with 0.25 μg/ml HEWL, the gel was rinsed with deionized water for 30 min and transferred into renaturation buffer (20 mM sodium phosphate [pH 7.0] with 10 mM MgCl2 and 1% Triton X-100). Both the proteins from the periplasmic extracts and the HEWL (which might be partially denatured by the SDS present in the gel) will be allowed to renature in this buffer. Overnight incubation at 37°C with gentle agitation led to clarification of the gel due to lysis of the M. luteus cells, leaving opaque bands where presumed inhibitors were located in the gel. As expected, no inhibitory band was detected in the lane loaded with the periplasmic protein extract of the Ivy knockout strain (lane 3), while a weak and a more intense band with the same migration distance were seen for the wild type (lane 4) and the overexpression extract (lane 1), respectively (Fig. 1). This clearly indicates that proteinaceous HEWL inhibitors can be qualitatively and even semiquantitatively detected with this reverse zymogram technique.
FIG. 1.
Reverse zymogram analysis of periplasmic protein extracts of E. coli MG1655(pAA410) (lane 1), P. mirabilis LMM2010 (lane 2), E. coli MG1655 ivy::Kan (lane 3), and E. coli MG1655 (lane 4).
The novel method was then applied to look for inhibitors in P. mirabilis LMM2010 since the periplasmic extract of this organism showed HEWL inhibitory activity although its genome does not contain an ivy gene homolog (sequence determined by the P. mirabilis Sequencing Group at the Sanger Institute and available at http://www.sanger.ac.uk/cgi-bin/BLAST/submitblast/p_mirabilis). The reverse zymogram revealed an intense band of unlysed M. luteus, corresponding to a molecular mass slightly higher than that of Ivy, which has a molecular mass of 14.102 kDa (Fig. 1). Remarkably, the intensity of the P. mirabilis inhibitor band was comparable to that of the Ivy-overexpressing E. coli strain (compare lanes 2 and 1 in Fig. 1), while its inhibitory activity, as determined with the traditional assay (38 IU/ml), was about 20 times lower and on the order of magnitude of the inhibitory activity of wild-type E. coli extract (18 IU/ml). This may be due to the presence of other compounds in the periplasmic protein extract of P. mirabilis interfering with HEWL inhibitor binding and thereby affecting the values of inhibitory activity measured with the traditional assay. Other possible explanations are (i) the use of different buffers in the assays that might influence the activity of the inhibitor and (ii) incomplete renaturation of Ivy in our reverse zymogram method. The fact that both Ivy and PliC are renaturable is probably, at least in part, due to their low molecular masses. If high-molecular-mass lysozyme inhibitors also exist, it has yet to be determined if they will be renaturable and detectable by the reverse zymogram method described here. We demonstrated that the technique can also be used with goose egg white lysozyme, which belongs to a different lysozyme family than HEWL (data not shown). This opens perspectives for the identification of putative g-type lysozyme-specific inhibitors.
Interestingly, a BLAST search of the recently discovered HEWL inhibitor MliC from E. coli against the complete sequence of P. mirabilis delivered an open reading frame (ORF) with 35% identity over 57 amino acids. The ORF encodes a predicted periplasmic protein of 110 amino acids including a 23-amino-acid signal peptide, and just like MliC from E. coli and P. aeruginosa, it contains a conserved COG3895 domain. Anticipating its functionality as a lysozyme inhibitor, which will be demonstrated below, we designated this ORF PliC from P. mirabilis. A sequence alignment of this putative novel member of the PliC/MliC lysozyme inhibitor family with the three earlier described members is shown in Fig. 2. Two cysteine residues which, in the case of MliC from E. coli, have been shown to form a disulfide bridge (12) are conserved and might be important for conformational stability.
FIG. 2.
Amino acid sequence alignment (http://www.ebi.ac.uk/clustalw/; 15) of PliC from P. mirabilis LMM2010, PliC from S. enterica serovar Enteritidis ATCC 13076, MliC from E. coli MG1655, and MliC from P. aeruginosa PAO1. Residues that are identical in all of the sequences in the alignment are marked with asterisks in the bottom row, and conserved and semiconserved substitutions are marked with colons and periods, respectively. The lipobox of the lipoproteins is underlined, while conserved cysteine residues of the mature protein are highlighted in gray.
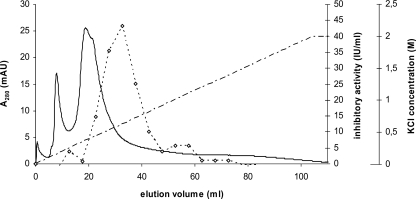
Subsequently, we isolated the putative HEWL inhibitor from P. mirabilis by loading 100 ml of a periplasmic protein extract with an inhibitory activity of 32 U/ml onto a HEWL affinity column and eluting it with a combined salt and high-pH gradient as described earlier (3). The resulting chromatogram and the inhibitory activities of the fractions after neutralization and dialysis are shown in Fig. 3. A single peak of HEWL inhibitory activity (43 U/ml) was detected at an elution volume of 33 ml. SDS-PAGE analysis of the fraction with strong HEWL inhibitory activity showed a single band with an estimated molecular mass of 18.5 kDa after silver staining according to reference 6 (data not shown). After lyophilization, the protein was visualized on a Coomassie-stained SDS-PAGE gel, and a small gel piece was subjected to trypsin digestion, followed by tandem mass spectrometry analysis. Two peptides, VSFPNDDLAVMDYNDELIVLK and YVGESFQLWGK, were identified with high confidence, leading to the same protein sequence found earlier by BLAST search with MliC from E. coli. There is, however, a remarkable discrepancy between the theoretically predicted molecular mass of this protein (9.6 kDa) and its molecular mass estimated from SDS-PAGE (18.5 kDa). The fact that the latter is almost twice the former could indicate that the protein occurs as a dimer. This seems unlikely since denaturing conditions were used for electrophoresis and the samples were boiled in the presence of SDS and dithiothreitol. However, deviations in molecular mass estimations from SDS-PAGE are not uncommon for acidic proteins (mature PliC from P. mirabilis has a pI of 4.33) and have been ascribed to poor binding of SDS (9). In view of its relatedness to the recently discovered family of HEWL inhibitors, the identified protein was renamed PliC (periplasmic lysozyme inhibitor of c-type lysozyme) of P. mirabilis. This protein constitutes the fourth member of the recently discovered family of lysozyme inhibitors.
FIG. 3.
Chromatogram showing the purification of HEWL-binding protein from P. mirabilis LMM 2010 periplasmic extract by affinity chromatography with immobilized HEWL. Protein concentration in eluate was monitored by A280 and expressed in milliabsorption units (mAU; solid line), and inhibitory activity of fractions against HEWL was monitored by traditional inhibitor assay (⋄). Elution was done with a gradient of 0 to 2.0 M KCl in 0.1 M Tris, pH 12.0 (dash-and-dot line).
In conclusion, reverse zymography, here optimized with the enzyme HEWL and Micrococcus as the substrate, is a useful technique for detecting bacterial lysozyme inhibitor proteins without nonproteinaceous compound interference with HEWL activity. Compared to the detection and isolation of lysozyme inhibitors by affinity chromatography, it has the advantage of allowing a higher throughput of samples, which will be useful in further screenings for novel lysozyme inhibitors. Extracts selected from this screening can then be subjected to affinity chromatography to isolate and further characterize the putative inhibitors. The reverse zymogram also allows differentiation between different inhibitors based on their electrophoretic mobilities and detection of multiple inhibitors with different mobilities in a single extract. Furthermore, the relative migration distance of the inhibitor provides an indication of the family to which the inhibitor belongs and whether only one or more inhibitors are present in an extract.
Acknowledgments
L.C. and D.D. have doctoral fellowships from the Flemish Institute for the Promotion of Scientific Technological Research (IWT), and A.A. has a postdoctoral fellowship from the Research Foundation-Flanders (FWO-Vlaanderen). This work was further financially supported by research grants from the Research Foundation-Flanders (FWO-Vlaanderen; G.0308.05 and G.0363.08) and by the Research Fund K.U.Leuven (research project GOA/03/10).
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Abergel, C., V. Monchois, D. Byrne, S. Chenivesse, F. Lembo, J.-C. Lazzaroni, and J.-M. Claverie. 2007. Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in gram-negative bacteria. Proc. Natl. Acad. Sci. USA 104:6394-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callewaert, L., A. Aertsen, D. Deckers, K. G. A. Vanoirbeek, L. Vanderkelen, J. M. Van Herreweghe, B. Masschalck, D. Nakimbugwe, J. Robben, and C. W. Michiels. 7 March 2008, posting date. A new family of lysozyme inhibitors contributing to lysozyme tolerance in gram-negative bacteria. PLoS Pathog. 4:e1000019. doi: 10.1371/journal.ppat.1000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callewaert, L., B. Masschalck, D. Deckers, D. Nakimbugwe, M. Atanassova, A. Aertsen, and C. W. Michiels. 2005. Purification of Ivy, a lysozyme inhibitor from Escherichia coli, and characterisation of its specificity for various lysozymes. Enzyme Microb. Technol. 37:205-211. [Google Scholar]
- 4.Deckers, D., B. Masschalck, A. Aertsen, L. Callewaert, C. G. M. Van Tiggelen, M. Atanassova, and C. W. Michiels. 2004. Periplasmic lysozyme inhibitor contributes to lysozyme resistance in Escherichia coli. Cell. Mol. Life Sci. 61:1229-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deckers, D., D. Vanlint, L. Callewaert, A. Aertsen, and C. W. Michiels. 2008. Role of the lysozyme inhibitor Ivy in growth or survival of Escherichia coli and Pseudomonas aeruginosa bacteria in hen egg white and in human saliva and breast milk. Appl. Environ. Microbiol. 74:4434-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heukeshoven, J., and R. Dernick. 1988. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis 9:28-32. [DOI] [PubMed] [Google Scholar]
- 7.Ito, Y., A. Yoshikawa, I. Hotani, S. Fukuda, K. Sugimura, and T. Imoto. 1999. Amino acid sequences of lysozymes newly purified from invertebrates imply wide distribution of a novel class in the lysozyme family. Eur. J. Biochem. 259:456-461. [DOI] [PubMed] [Google Scholar]
- 8.Jollès, P. 1996. Lysozymes: model enzymes in biochemistry and biology. Birkhäuser Verlag, Basel, Switzerland.
- 9.Kaufmann, E., N. Geisler, and K. Weber. 1984. SDS-PAGE strongly overestimates the molecular masses of the neurofilament proteins. FEBS Lett. 170:81-84. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli, M. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Monchois, V., C. Abergel, J. Sturgis, S. Jeudy, and J.-M. Claverie. 2001. Escherichia coli ykfE ORFan gene encodes a potent inhibitor of c-type lysozyme. J. Biol. Chem. 276:18437-18441. [DOI] [PubMed] [Google Scholar]
- 12.Revington, M., A. Semesi, A. Yee, C. H. Arrowsmith, and G. S. Shaw. 2006. The solution structure of the protein ydhA from Escherichia coli. J. Biomol. NMR 35:295-300. [DOI] [PubMed] [Google Scholar]
- 13.Snoek-van Beurden, P. A. M., and J. W. Von Den Hoff. 2005. Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. BioTechniques 38:73-83. [DOI] [PubMed] [Google Scholar]
- 14.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]