Abstract
Bacterial vaginosis (BV) is a common syndrome associated with numerous adverse health outcomes in women. Despite its medical importance, the etiology and microbial ecology of BV remain poorly understood. We used broad-range PCR to census the community structure of the healthy and BV-affected vaginal microbial ecosystems and synthesized current publicly available bacterial 16S rRNA gene sequence data from this environment. The community of vaginal bacteria detected in subjects with BV was much more taxon rich and diverse than in subjects without BV. At a 97% sequence similarity cutoff, the number of operational taxonomic units (OTUs) per patient in 28 subjects with BV was nearly three times greater than in 13 subjects without BV: 14.8 ± 0.7 versus 5.2 ± 0.75 (mean ± standard error). OTU-based analyses revealed previously hidden diversity for many vaginal bacteria that are currently poorly represented in GenBank. Our sequencing efforts yielded many novel phylotypes (123 of our sequences represented 38 OTUs not previously found in the vaginal ecosystem), including several novel BV-associated OTUs, such as those belonging to the Prevotella species complex, which remain severely underrepresented in the current NCBI database. Community composition was highly variable among subjects at a fine taxonomic scale, but at the phylum level, Actinobacteria and Bacteroidetes were strongly associated with BV. Our data describe a previously unrecognized extent of bacterial diversity in the vaginal ecosystem. The human vagina hosts many bacteria that are only distantly related to known species, and subjects with BV harbor particularly taxon-rich and diverse bacterial communities.
A growing body of evidence suggests that human health is fundamentally affected by the composition and function of the vast human microbial ecosystem. Microbial cells in and on our bodies are approximately 10 times more numerous than our own cells and contain, in aggregate, about 100 times more genes, leading to the suggestion that humans and our microbial symbionts be considered superorganisms (12). There is increasing evidence that microbial communities strongly influence human health and quality of life. For example, in the gastrointestinal tract, the taxonomic composition of the microbial community may affect the propensity to develop obesity (30). In the vagina, bacterial vaginosis (BV) is marked by dramatic shifts in the types and relative proportions of a diverse community of bacteria as the vaginal ecosystem changes from a healthy to a diseased state (7, 9).
BV is strongly associated with several adverse health outcomes, including preterm labor and delivery (13, 14), pelvic inflammatory disease (29), and the acquisition and transmission of sexually transmitted diseases, such as human immunodeficiency virus (23-25). BV is the most common cause of vaginal discharge among women of reproductive age and results in millions of health care visits annually in the United States (26). Because BV is fundamentally a result of changes to the vaginal microbial community, full understanding and successful treatment will require knowledge of how the healthy community is altered in its taxonomic composition, community structure, and, ultimately, function.
Historically, a search for single etiologic agents in BV and the constraints of traditional cultivation techniques limited appreciation of the full extent of vaginal microbial diversity in this syndrome. BV has long been recognized as a complex condition, but until recently, lactobacilli found in the healthy vagina were thought to be replaced by a relatively small suite of taxa typically including Gardnerella vaginalis, Prevotella spp., Porphyromonas spp., Mobiluncus spp., and Mycoplasma hominis (7, 27). The advent of molecular tools, such as broad-range bacterial 16S rRNA gene PCR, has enabled community assessments independent of cultivation-based techniques and has greatly expanded our knowledge of the phylogenetic range and taxonomic diversity of bacteria found in the vagina. For example, intensive sequencing has revealed extensive diversity, not only within the Lactobacillus species complex (15), but also within other taxa most closely related to Atopobium, Clostridium, Leptotrichia, Prevotella, Peptostreptococcus, and Peptoniphilus, species not previously thought to be common in the human vagina (3, 9, 15, 33, 34).
Although there is agreement in the literature that no single agent likely causes BV, there is no consensus about what constitutes a pathogenic bacterial community in this syndrome. Comprehensive cultivation-independent comparisons of the vaginal bacterial communities from subjects with and without clinically defined BV have been rare. Without such fundamental data, our understanding of the pathogenesis of BV is constrained. In one study that compared broad-range bacterial community composition among patients with objectively defined BV status, subjects with BV had nearly four times as many taxa as subjects without BV, including uncultivated Clostridia-like bacteria, which were shown to be highly specific for BV (9).
In this paper, we extend this previous work to present a systematic phylogenetic analysis of vaginal bacterial community composition from subjects with defined BV status as determined by commonly accepted clinical criteria. We also synthesize current public 16S rRNA gene sequence data from bacteria found in the vaginal ecosystem (with inclusion of studies where BV status was not assessed) and place these data in a phylogenetic context. Our goal was to provide a comprehensive picture of our current knowledge of the community structure of the vaginal bacterial ecosystem.
MATERIALS AND METHODS
Sample collection and clinical assessment of BV status.
Vaginal swabs were collected from subjects attending either the Public Health-Seattle and King County Sexually Transmitted Diseases Clinic or the Harborview Medical Center Women's Research Clinic and immediately frozen for later analysis, as previously described (9). BV status was assessed using Amsel clinical criteria (1) for all subjects and confirmed using Gram stain criteria (Nugent [18] scores) for subjects attending the Women's Research Clinic. In this paper, we analyze data collected from 34 vaginal samples, including 21 collected from subjects with clinically defined BV and 13 samples collected from subjects without BV. This analysis includes data on six vaginal samples not previously reported (9) and provides a more exhaustive phylogenetic analysis based on the presence of high-quality sequence data (detailed below) representative of the two patient populations.
Written informed consent was obtained from all participants in the study, which was also approved by the Institutional Review Boards at the Fred Hutchinson Cancer Research Center and the University of Washington.
Molecular methods.
DNA was obtained from swabs using the UltraClean Soil DNA extraction kit (MoBio, Carlsbad, CA), chosen after testing guanidinium-based lysis protocols with and without bead beating. Out of the 969 sequences generated, 3 were detected only in the protocol without bead beating and 7 were detected only in the bead-beating protocol. Accordingly, we did not find that bead beating substantially affected our results. For broad-range PCR, the primers 338F (5′-ACTCCTRCGGGAGGCAGCAG-3′) and 1407R (5′-GACGGGCGGTGTGTRCA-3′) were used with a thermal-cycling protocol of denaturation at 95°C, followed by 21 to 25 cycles of 95°C for 30 s, 55°C for 30s, and 72°C for 90s, with a final extension at 72°C for 7 min.
Successfully amplified products were cloned using the Topo blunt cloning kit (Invitrogen, Carlsbad, CA), and PCR-amplified inserts were sequenced using vector primers and BigDye version 3 (Applied Biosystems, Foster City, CA). Single reads were generated for 736 sequences, with an average length of 811 base pairs. Two reads were obtained for 233 sequences, resulting in about 1-kb sequence lengths.
Data analysis.
Raw sequence data were edited, and contigs were assembled for each clone using Sequencher (Gene Codes, Ann Arbor, MI). The 969 sequences obtained from our sequencing efforts and all publicly available sequences from vaginal bacteria (554 sequences, listed below) were then submitted to the GreenGenes 16S rRNA gene database (5) for alignment using the NAST algorithm (4) and taxonomic classification of each sequence. All sequences were checked for chimeric anomalies using the Mallard program (2).
To classify sequences based on self-similarities rather than matches to an external database, sequences were grouped into operational taxonomic units (OTUs) with cutoffs of 99%, 97%, and 95% sequence similarity using the DOTUR software package implemented with the furthest-neighbor option, in which all of the sequences within an OTU are at least x% similar to all of the other sequences within the OTU (22). The commonly accepted phylogenetic species definition of 97% 16S sequence similarity (32) was used to define a core data set of representative sequences that were used for phylogenetic analyses.
Sequences were incorporated into ARB (17) for phylogenetic analysis and tree construction. Sequences from a nonredundant list of three nearest-neighbor isolates for each sequence were also added to the database. Alignments were manually corrected based on the secondary structure of the 16S rRNA and tree construction performed using the maximum-likelihood method in ARB (FastdnaML) with a 75% similarity filter constructed for each phylum. Bootstrap values (100 resamplings) were generated for maximum-likelihood trees created in PHYLIP (8) using the DNAML algorithm and neighbor-joining trees created in ARB and superimposed on nodes supported by all three methods.
Publicly available sequences were collected from previous surveys of vaginal bacterial diversity, which included 439 sequences from Hyman et al. (15) (AY958774 to AY959212), 7 unpublished sequences from Zozaya-Hinchliffe et al. (EF120360 to EF120366), 25 sequences from Verhelst et al. (31) (AJ585206 to AJ619714), 15 unpublished sequences from Zhang et al. (DQ666091 to DQ666105), and 68 sequences from Zhou at al (33) (AY267541 and -2, AY269020 to -34, AY271931 to -53, AY283264 to -75, AY335493 to -504, and DQ987868 to -9). Because subject BV status was not available for most of these studies, the corresponding sequences were excluded from comparisons of bacterial community structure in subjects with and without BV but were included in phylogenetic analyses of vaginal bacterial diversity.
Nucleotide sequence accession numbers.
Sequences from the 97 OTUs generated in this study were deposited in GenBank (AY738660, AY738684, AY738687, AY738691, AY738694, AY738697, AY738701 to -5, EF428974, and EU188937 to EU189021) and are listed in Table S1 in the supplemental material.
RESULTS
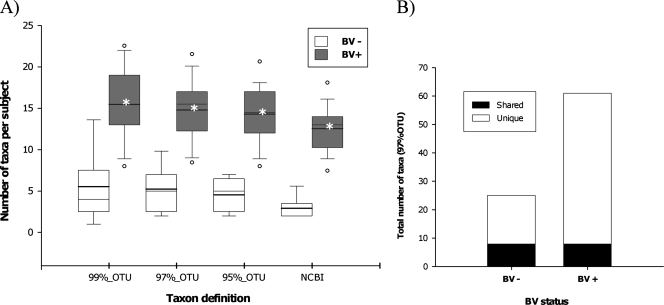
The structures of the bacterial communities were dramatically different in samples from women with and without BV. Increased taxonomic richness and diversity of BV-associated flora relative to normal vaginal flora was evident in a variety of analyses. The mean number of taxa detected per patient was significantly higher (P < 0.001) in subjects with BV than in subjects without BV for all phylotype definitions (Fig. 1A). Similarly, diversity was higher in subjects with BV; the Shannon diversity index was 1.4 to 4.1 times greater for subjects with BV than for subjects without BV, depending on the taxonomic definition (Table 1).
FIG. 1.
(A) Numbers of taxa per subject using four different taxon definitions. Taxon definitions of 99%, 97%, and 95% OTUs were assigned using the DOTUR package, and NCBI definitions were based on taxonomic classifications using the GreenGenes 16S rRNA database, as described in the text. The data represent 13 subjects without BV and 28 subjects with BV. The shaded boxes encompass the 25th to 75th percentiles of the data, the boldface lines indicate means, the lightface lines indicate median values, and the whiskers span the 5th to 95th percentiles. The asterisks indicate significant (P < 0.001) differences in mean values for subjects with BV versus subjects without BV, as determined by t tests. (B) Total number of taxa found across all subjects that are unique (e.g., found only in subjects with BV) and the number that are shared for each clinical state.
TABLE 1.
Richness and diversity values and best matches to taxon designations in NCBI
| Taxon definition | Total no. of taxa
|
Shannon diversity index
|
|||
|---|---|---|---|---|---|
| BV+ (n = 829) | BV− (n = 140) | BV+ (n = 829) | BV− (n = 140) | Ratio (BV+/BV−) | |
| 99% OTU | 85 (101) | 39 (67) | 1.54 | 1.12 | 1.4 |
| 97% OTU | 61 (75) | 25 (21) | 1.37 | 0.93 | 1.5 |
| 95% OTU | 49 (48) | 17 (16) | 1.33 | 0.81 | 1.6 |
| NCBI | 36 | 9 | 1.20 | 0.29 | 4.1 |
Richness and diversity values for taxon definitions of sequence similarity cutoffs of 99%, 97%, and 95% and best matches to taxon designations in NCBI for all 16S rRNA gene sequences generated from subjects with (BV+) and without (BV−) BV. Richness values in parentheses represent average Chao1 richness estimates calculated as described in the text.
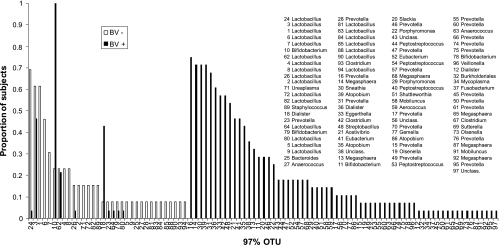
High interpatient variability of vaginal bacterial community structure was evident in several analyses, particularly for subjects with BV. First, the total number of phylotypes found across all subjects in each clinical group was dramatically higher than the per-patient mean (Fig. 1). Second, many taxa were relatively rare; for subjects with BV, 47 of the 97 OTUs identified were found in fewer than 25% of subjects, while only a single taxon, most closely related to Bifidobacterium/Gardnerella, was found in all subjects with BV (Fig. 2).
FIG. 2.
Proportions of subjects for which each of the 97 OTUs classified at a 97% sequence similarity were encountered. The list indicates genus-level identification of each OTU based on the NCBI taxonomy. Note that sequences designated Bifidobacterium by NCBI correspond to sequences classified as Gardnerella by the RDP. The accession number for each OTU is listed in Table S1 in the supplemental material.
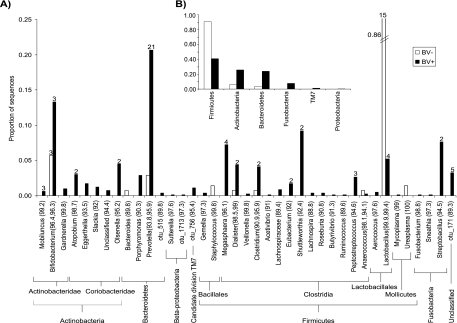
There were dramatic differences in the relative proportions of major groups of bacteria between subjects with and without BV. Relative to subjects without BV, those with BV had much higher proportions of Actinobacteria (4.5 times) and Bacteroidetes (6.7 times) and a much lower proportion of Firmicutes (0.45 times) (Fig. 3B). Several phylogenetic groups were found only in samples from subjects with BV; Fusobacteria, candidate division TM7, and Betaproteobacteria together accounted for 9% of clones from subjects with BV but were not detected in clone libraries from subjects without BV (Fig. 3B).
FIG. 3.
Taxonomic affiliations and relative proportions of sequences summarized at the genus (A) and phylum (B) levels. The taxa in panel A represent the closest matches from the NCBI database, with the numbers in parentheses representing the mean percent match for each group of sequences for subjects with BV (829 sequences) and subjects without BV (140 sequences). Note that some sequences designated Bifidobacterium by NCBI correspond to sequences consistent with G. vaginalis; there is significant sequence heterogeneity within the G. vaginalis species complex. The y axes in both panels show the proportion of sequences from subjects with and without BV calculated separately. The numbers above the bars represent the number of OTUs within each group defined at a minimum sequence similarity of 97%; only numbers >1 are shown. Note the break in the axis for Lactobacillus-like sequences from subjects without BV, where lactobacilli comprise 86% of the sequences.
Subjects without BV had bacterial communities dominated by Lactobacillus species, accounting for 86% of all sequences from subjects without BV (Fig. 3). In contrast, subjects with BV did not possess a single dominant taxon but rather harbored a diverse array of vaginal bacteria, many present at low relative abundance. Of phylotypes unique to subjects with BV, nine were singletons (encountered only once), and the four most common (sequences most closely related to Olsenella, Megasphaera, Streptobacillus, and Shuttleworthia) accounted for 4%, 7%, 8%, and 9%, respectively, of all sequences retrieved from subjects with BV (Fig. 3).
For several taxonomic groups in particular, the true extent of diversity is not reflected in the current GenBank database. For example, even though about 20% of sequences from subjects with BV were classified as Prevotella according to the best match in GenBank, this group of sequences actually contained 21 different taxa based on a species definition of 97% sequence similarity (Fig. 3A). Similarly, within the group of sequences most closely related to Lactobacillus in GenBank, there were actually 15 different phylotypes (Fig. 3A). Estimates of taxonomic richness and diversity based on best matches to named taxa may severely underestimate the true extent of bacterial diversity in the vagina.
Taxonomic designations of sequences using two different classification schemes, NCBI and the Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu), were largely congruent (Tables 2, 3, and 4) but also demonstrated important discrepancies. For example, sequences classified as Bifidobacterium by NCBI belonged to Gardnerella in the RDP scheme (Tables 2 and 4). Similarly, sequences classified as Clostridium by NCBI were called Acetivibrio by RDP (Table 2). In general, NCBI designations split sequences into more groups, while the RDP scheme lumped sequences together: the 60 sequences classified as Acetivibrio by RDP belonged to four different taxonomic groups in the NCBI scheme, and 47 sequences classified as Coriobacteriaceae by RDP represented five taxonomic groups according to NCBI (Table 2).
TABLE 2.
Comparison of NCBI and RDP taxonomic designations for Actinobacteria, Bacteroidetes, and Firmicutes sequences obtained from subjects with BV (n = 580)
| NCBI designation | No. of sequences for RDP designation:
|
Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Actinobacteria
|
Bacteroidetes
|
Firmicutes
|
||||||||||||
| Atopobium | Coriobacteriaceae | Eggerthella | Gardnerella | Mobiluncus | Olsenella | Slackia | Porphyromonas | Tannerella | Prevotella | Acetivibrio | Lachnospiraceae | Shuttleworthia | ||
| Actinobacteria | ||||||||||||||
| Atopobium | 4 | 9 | 12 | 25 | ||||||||||
| Bifidobacterium | 110 | 110 | ||||||||||||
| Coriobacteriaceae | 4 | 2 | 6 | |||||||||||
| Eggerthella | 10 | 4 | 14 | |||||||||||
| Gardnerella | 8 | 8 | ||||||||||||
| Mobiluncus | 5 | 5 | ||||||||||||
| Olsenella | 20 | 17 | 37 | |||||||||||
| Slackia | 2 | 4 | 4 | 10 | ||||||||||
| Bacteroidetes | ||||||||||||||
| Bacteroidales | 3 | 3 | ||||||||||||
| Porphyromonas | 24 | 24 | ||||||||||||
| Prevotella | 171 | 171 | ||||||||||||
| Firmicutes | ||||||||||||||
| Acetivibrio | 1 | 1 | ||||||||||||
| Butyvibrio | 1 | 1 | ||||||||||||
| Clostridium | 34 | 34 | ||||||||||||
| Eubacterium | 14 | 14 | ||||||||||||
| Lachnospira | 1 | 2 | 3 | |||||||||||
| Lachnospiraceae | 7 | 7 | ||||||||||||
| Roseburia | 3 | 3 | ||||||||||||
| Ruminococcus | 1 | 1 | ||||||||||||
| Shuttleworthia | 76 | 76 | ||||||||||||
| OTU_171 | 3 | 1 | 18 | 5 | 27 | |||||||||
| Total | 6 | 47 | 6 | 118 | 5 | 29 | 4 | 24 | 3 | 175 | 60 | 25 | 78 | 580 |
TABLE 3.
Comparison of NCBI and RDP taxonomic designations for Firmicutes and Fusobacteria sequences obtained from subjects with BV (n = 249)
| NCBI designation | No. of sequences for RDP designation:
|
Total | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Firmicutes
|
Fusoba
|
|||||||||||||||
| Aerococcus | Anaerococcus | Dialister | Gemella | Lactobacillus | Megasphaera | Micromonas | Mycoplasma | Peptoniphilus | Veillonella | Fusobacterium | Fusobacteriaceae | Streptobacillus | Sutterella | TM7 | ||
| Firmicutes | ||||||||||||||||
| Aerococcus | 4 | 4 | ||||||||||||||
| Anaerococcus | 2 | 2 | ||||||||||||||
| Dialister | 36 | 36 | ||||||||||||||
| Gemella | 3 | 3 | ||||||||||||||
| Lactobacillus | 43 | 43 | ||||||||||||||
| Megasphaera | 60 | 60 | ||||||||||||||
| Mycoplasma | 1 | 1 | ||||||||||||||
| Peptostreptococcus | 12 | 10 | 22 | |||||||||||||
| Veillonella | 2 | 2 | ||||||||||||||
| Fusobacteria | ||||||||||||||||
| Fusobacterium | 1 | 1 | ||||||||||||||
| Sneathia | 1 | 1 | ||||||||||||||
| Streptobacillus | 63 | 63 | ||||||||||||||
| Burkholderiales | 1 | 1 | ||||||||||||||
| Sutterella | 1 | 1 | ||||||||||||||
| TM7 | 9 | 9 | ||||||||||||||
| Total | 4 | 2 | 36 | 3 | 43 | 60 | 12 | 1 | 10 | 2 | 1 | 1 | 63 | 2 | 9 | 249 |
TABLE 4.
Comparison of NCBI and RDP taxonomic designations for sequences obtained from subjects without BV (n = 140)
| NCBI designation | No. of sequences for RDP designation:
|
Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Actinobacteria |
Bacteroidetes
|
Firmicutes
|
||||||||
| Gardnerella | Prevotella | Bacteroidales | Anaerococcus | Clostridiaceae | Dialister | Lactobacillus | Staphylococcus | Ureaplasma | ||
| Actinobacteria | ||||||||||
| Bifidobacterium | 8 | 8 | ||||||||
| Bacteroidetes | ||||||||||
| Bacteroides | 1 | 1 | ||||||||
| Prevotella | 4 | 4 | ||||||||
| Firmicutes | ||||||||||
| Anaerococcus | 1 | 1 | ||||||||
| Clostridium | 1 | 1 | ||||||||
| Dialister | 1 | 1 | ||||||||
| Lactobacillus | 120 | 120 | ||||||||
| Staphylococcus | 2 | 2 | ||||||||
| Ureaplasma | 2 | 2 | ||||||||
| Total | 8 | 4 | 1 | 1 | 1 | 1 | 120 | 2 | 2 | 140 |
In the entire data set of 1,523 publicly available sequences, we found a total of 195 OTUs using the bacterial species definition of 97% 16S rRNA gene sequence similarity, including 38 novel OTUs representing 123 sequences unique to the data presented here. Of these, 31 OTUs (115 sequences) were found only in subjects with BV. We encountered 59 OTUs in our sequencing set that were also reported elsewhere in the data set of publicly available sequences, while 98 OTUs were detected in other studies (all but 5 of these by Hyman et al. [15]) but not in our patient populations.
Six sequences deposited in GenBank (AY959158, AY958981, AY959016, AY959073, AY958933, and AY959044) were found to be consistent with chimeric data (P < 0.001) and so were excluded from our analyses.
Phylogenetic analyses of BV-associated bacteria.
The true diversity and novelty of BV-associated bacteria were apparent when sequences generated in this study were compared to publicly available sequences from previous surveys of vaginal bacterial diversity and their nearest cultivated and uncultivated matches.
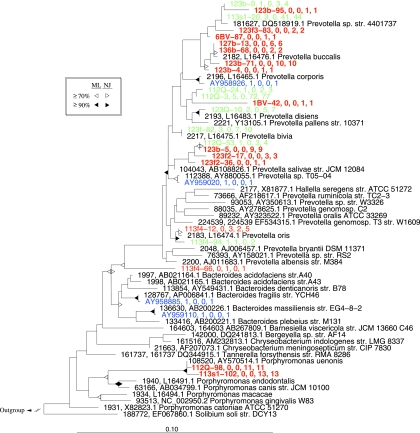
Within the phylum Bacteroidetes, sequences in our data set were most closely related to one of three genera: Prevotella, Bacteroides, and Porphyromonas (Fig. 4). Most (196/222) belonged to the Prevotella group but were only distantly related to their nearest relative and represented many novel sequence types not found in previous studies of vaginal bacteria. In the Prevotella species complex, we found 13 novel OTUs (Fig. 4). Two novel OTUs representing 24 sequences were most closely related to Porphyromonas uenonis (Fig. 4) and were found only in subjects with BV.
FIG. 4.
Phylogenetic relationships among representative taxa belonging to the phylum Bacteroidetes. The tree was reconstructed using the maximum-likelihood method in ARB with a 75% similarity column filter. Representative taxa were defined using a 97% OTU definition, as described in the text. Additional taxa represent a nonredundant list of nearest neighbors current as of April 2008. Taxa in red represent OTUs unique to our sequencing efforts, OTUs in green were common to our study and at least one other, and OTUs shown in blue were detected in other studies but not encountered in our sequencing. The numbers after each OTU indicate the number of sequences with unknown BV status, the number of sequences from subjects without BV, the number of sequences from subjects with BV, and the total number of sequences within each OTU. OTUs shown in boldface were found only in subjects with BV. Triangles at nodes represent bootstrap values from 100 resamplings of maximum-likelihood (ML) trees built in PHYLIP and neighbor-joining (NJ) trees, as shown in the legend and described in the text. str., strain.
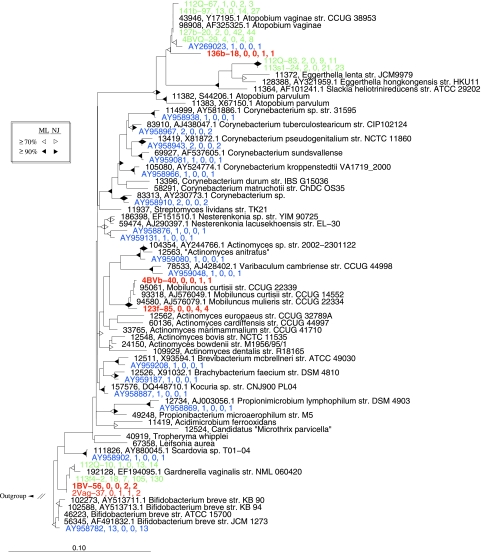
Within the Actinobacteria, most sequences (276/302) belonged to clades affiliated with Atopobium, Eggerthella, or Gardnerella, and all of the shared OTUs across studies belonged to one of these three groups (Fig. 5). Of these 306 sequences, only eight representing two OTUs in the Gardnerella group were from subjects confirmed not to have BV (Fig. 5). Our sequencing also revealed several novel OTUs in this phylum: one related to Atopobium, two most similar to Mobiluncus, and two Gardnerella-like OTUs (Fig. 5). Of the 17 OTUs not found in our sequences, 13 were singletons representing a wider phylogenetic range outside of the three main BV-associated genera of the Actinobacteria, and all were from patients with unknown BV status (Fig. 5).
FIG. 5.
Phylogenetic relationships among representative taxa belonging to the phylum Actinobacteria. The tree reconstruction methods, color scheme, and bootstrap representations are as described in the legend to Fig. 4.
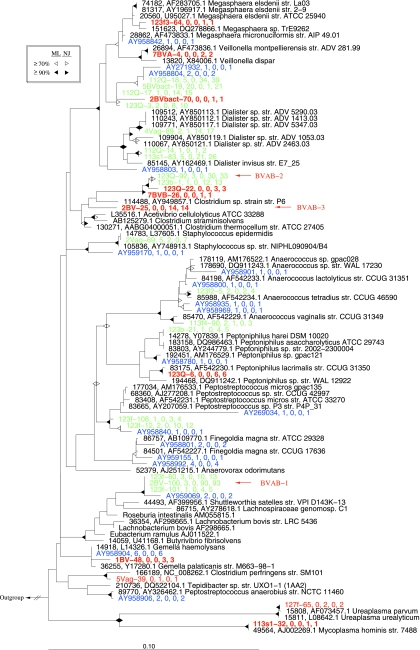
Within the Firmicutes, the majority of sequences (313/387) belonged to one of four main groups related to known taxa: Shuttleworthia, Megasphaera/Veillonella, Dialister, and Clostridium (Fig. 6). Of these, only a single sequence was from a subject confirmed not to have BV. We identified 11 novel OTUs representing 35 sequences within the Firmicutes; of these, only three sequences from two OTUs related to Clostridium perfringens and Ureaplasma were from subjects without BV (Fig. 6).
FIG. 6.
Phylogenetic relationships among representative taxa belonging to the phylum Firmicutes. The tree reconstruction methods, color scheme, and bootstrap representations are as described in the legend to Fig. 4.
Previously identified (9) BV-associated bacteria appear to be truly novel taxa that are commonly encountered in subjects with BV. Ninety-three sequences most closely related to Shuttleworthia encompass those previously designated (9) as BV-associated bacterium 1 (BVAB-1) (Fig. 6). Three additional OTUs closely related to BVAB-1 were also identified (Fig. 6). Sequence types designated BVAB-2 and BVAB-3 (9) were most closely related to Acetovibrio cellulolyticus and several Clostridium strains and together accounted for 47 sequences (Fig. 6). Three additional OTUs (two of them novel) closely related to BVAB-2 represented an additional 27 sequences (Fig. 6). All three of the BVAB phylotypes had low similarities to their nearest known cultivated neighbors and together accounted for 14% of all sequences from subjects with BV.
DISCUSSION
The microbial ecosystem resident on the human vaginal epithelium remains poorly understood in terms of census, microbe-microbe interactions, and microbial interactions with the host. In particular, details about the relationship between newly described fastidious BV-associated bacteria and the manifestations of BV are just starting to emerge. In this study, we compared vaginal bacterial community structures between subjects with and without clinically defined BV and synthesized publicly available data to better understand the diversity of healthy and diseased vaginal bacterial communities.
For patients afflicted with BV, there is a profound shift in the types of bacteria present in the vagina and their absolute and relative abundances. Data presented here clearly demonstrate that BV is associated with a dramatic increase in the taxonomic richness and diversity of the vaginal bacterial community. Our analyses highlight three main points.
First, the true extent of vaginal bacterial diversity may be severely underestimated by attributing taxonomic identities to sequences simply according to their best match to currently known taxa. Although the use of the NCBI taxonomic classification scheme gives a familiar nomenclature for sequences obtained from any environment, in the case of the vaginal ecosystem, it also clearly hides a significant amount of diversity. Multiple OTUs within sequence groups matching a single taxon as defined by NCBI and low similarity scores to these taxa demonstrate that we are just beginning to reveal the true extent of the diversity of the vaginal flora.
Vaginal bacteria are generally underrepresented in the NCBI database, but for some groups in particular, the true extent of diversity is hidden by their poor representation in GenBank. For example, sequences classified as Prevotella actually contained 21 different OTUs using a 97% definition. Similarly, within sequences classified as Lactobacillus, there were 15 different OTUs. The diversity contained within the Lactobacillus species complex of the vagina has been noted previously by other researchers (15, 34), but the functional importance of this level of diversity remains unknown. Ecological theory suggests that each phylotype occupies its own niche in its environment and thus must be somehow unique, but the details of how BV-associated microbes partition niche space in the vaginal ecosystem and the correspondence between 16S rRNA-defined phylotypes and various functional capabilities remain relatively unexplored.
It is likely that additional sampling will reveal even greater richness and diversity in subjects with BV. When a taxon was defined using a 97% OTU definition, we observed a total of 61 taxa across all subjects with BV, but richness estimates of the true number of taxa present ranged from 63 to 111 (95% confidence interval), with an average estimate of 75 taxa (Table 1). For subjects without BV, our sampling effort was more complete; we encountered 25 taxa with a 97% OTU definition, and estimates of the total ranged from 20 to 30 (95% confidence interval), with an average of 21 (Table 1).
The loss of information caused by lumping multiple OTUs into a single taxon and the subsequent underestimation of the true richness and diversity of the vaginal ecosystem highlight the second important result of our analyses. The application of an OTU-based analysis to the full data set of our sequences and those from previous surveys of the vaginal ecosystem revealed many novel phylotypes associated with BV. We found 38 OTUs not previously encountered in the vaginal ecosystem, including 31 OTUs that were found only in subjects with BV. These OTUs were found across all of the major groups of BV-related taxa (Fig. 4, 5, and 6), providing additional evidence that many more taxa are associated with BV than was previously thought.
Third, although the vaginal bacterial communities found in subjects with and without BV are distinctly different, the structures of these vaginal bacterial communities have high intersubject variability within each clinical group. For both groups, the total number of taxa encountered across all subjects was at least four times greater than the per-patient mean (Fig. 1), and many taxa were relatively uncommon (Fig. 2). This variability in community membership and structure has important implications for understanding the etiology of BV and for developing diagnostic tools. Why is the taxonomic composition of BV-associated bacteria so different for each patient? High interpatient variability has been observed previously for microbial communities in the vagina (3) and other areas of the human body (6, 11). Stochastic differences in colonization could generate this pattern, but so could other factors, such as differences in host immune response, expression of ligands for bacterial attachment to epithelial cells, the chemical and physical environments of the host, or intra- and interspecific microbial competition.
Although no single bacterium can be identified as uniquely associated with BV, aggregating our data at higher taxonomic levels made it clear that several taxonomic groups do have a strong association. Common practice has generally been to describe sampled communities using the finest level of taxonomic discrimination possible, but aggregating data into higher taxonomic groups could be more informative and may avoid currently unresolved problems in classifying many BV-related bacteria at the genus or species level. For example, when treated at the genus level, taxa such as Eggerthella, Mobiluncus, and Slackia were relatively rare in subjects with BV, but when considered as a phylum, Actinobacteria accounted for 26% of all sequences from subjects with BV versus 6% from those without. Actinobacteria and Bacteroidetes in particular were much more common in subjects with BV than in those without BV (4.5 times and 6.7 times, respectively). The ability to identify a set of signature taxa common to BV could have important clinical and diagnostic implications.
Aggregating taxonomic data may also reduce problems associated with the current poorly resolved systematics of many BV-associated bacteria. For example, under the NCBI naming scheme, sequences classified as Gardnerella were relatively rare in subjects with BV, which contradicts generally held notions of a strong association between BV and Gardnerella. However, the 110 sequences classified as Bifidobacterium by NCBI were all classified as Gardnerella by the RDP (Table 2). The discrepancies among different taxonomic classification schemes highlight the current state of systematics for many BV-associated taxa and emphasize the importance of OTU-based analyses to capture the true diversity within a pool of sequences.
The polymicrobial nature of BV clearly raises the possibility of interactions among vaginal microbes, including syntrophies, which might contribute to the pathogenesis of BV (see reference 20 for a review). Ammonia transfer from Prevotella bivia to G. vaginalis has been demonstrated (19), but much remains to be learned about interspecific interactions of vaginal microbes and their contribution to the etiology of BV. Subjects with BV have volatile amines in vaginal fluid (the basis for the “whiff test” used in the clinical diagnosis of BV) with elevated levels of trimethylamine, putrescine, cadaverine, and tyramine, but the microbes responsible for generating these metabolic products are not clearly identified. An interesting recent paper has shown the importance of direct competition between lactobacilli and G. vaginalis, apparently independently of pH and H2O2 production (21). Clearly, much work remains to be done to tease apart the microbial interactions, metabolic processes, and host factors that lead to BV; it is our hope that increased knowledge about the composition and structure of BV-associated bacterial communities will facilitate these studies.
Limitations.
The data presented here reflect different sample sizes for subjects with and without BV. However, initial screening of samples using restriction fragment length polymorphisms indicated that the extreme differences in taxonomic richness between the two patient populations justified the different sample sizes in order to achieve similar sampling depths. It is axiomatic that representative sampling of any community requires that the sampling effort be tied to the structure of the community; depauperate communities naturally require less sampling effort than taxon-rich and diverse communities to achieve equivalent sampling saturation. In the case of subjects without BV, fewer sequences were obtained due to many fewer restriction fragment length polymorphism patterns because of the dominance by Lactobacillus. Richness estimators (Table 1) indicated subjects without BV were adequately sampled, and additional diversity will be discovered in subjects with BV. Future work utilizing techniques such as pyrosequencing could provide greatly improved coverage of BV-associated genetic diversity but with more limited phylogenetic resolution due to limited sequence lengths.
Although the use of cloning to identify bacteria from complex samples has been criticized for potentially biased amplification (28), the clone library approach has also compared favorably to PCR-independent methods, such as fluorescence in situ hybridization, in its estimation of the relative proportions of various taxonomic groups (16). The partial 16S rRNA gene sequences obtained in our study (∼800 to 1,000 bp) may contribute to some of the discrepancies in taxonomic assignments between the NCBI and RDP classifications. Full-length sequences (∼1.5 kb) would likely improve phylogenetic resolution for some taxa, but in this study, we opted for a relatively high-throughput approach to compare a large number of sequences from subjects with and without BV.
Comparison to previous results.
In our phylogenetic analysis, it is apparent that novel OTUs found in our study generally belong to different phylogenetic clades than OTUs not encountered in our study. This is perhaps most striking for the Bacteroidetes (Fig. 4), for which we found 13 OTUs that had not been discovered in previous studies of vaginal bacteria. Similarly, for the Actinobacteria (Fig. 5), we found only two OTUs representing five sequences that did not group with either Atopobium, Eggerthella, or Gardnerella/Bifidobacterium, while Hyman et al. found 15 OTUs outside of this main group. This pattern could reflect different study populations, the inherent variability of BV, PCR primer bias, or different sampling intensities across studies. Most (82/98) of the OTUs not found by us have two or fewer members represented, and all but six of these were found only by Hyman et al. (15), suggesting that their intensive sequencing effort did find uncommon members of the community. However, it is also possible that these sequences could originate from low-level PCR contaminants, which are more likely to become evident with more intensive sampling of clone libraries (10). Taq polymerase, used for PCR, is known to be contaminated with low levels of bacterial 16S rRNA genes. Demographic differences among patient populations have also been demonstrated (34) and may explain these differences.
Conclusions.
The structures of the vaginal bacterial communities differ dramatically between subjects with and without BV. BV is associated with increased taxonomic richness and diversity. At a species or genus level, the composition of the vaginal bacterial community has high interpatient variability, yet at higher taxonomic levels, several bacterial groups are strongly associated with BV, most notably Actinobacteria and Bacteroidetes. Our data describe a previously unrecognized extent of diversity in the vaginal ecosystem in general and of BV-associated bacteria in particular. The true extent of diversity within several key taxonomic groups is grossly underrepresented in the current NCBI database. The most prominent of these are Prevotella-like sequences, commonly found in subjects with BV, and Lactobacillus-like sequences, common in subjects without BV.
Using Web-based tools freely available to the research community, our analysis provides a comprehensive census of vaginal bacterial communities and their association with BV. It is our hope that the data presented here will stimulate the formulation of new hypotheses about the metabolic functions, syntrophic interactions, and niche partitioning of bacteria colonizing the vaginal ecosystem. Continuing investigations of BV will almost certainly reveal complex syntrophies, cell-to-cell signaling, and bacterial-host interactions that will shed light on how consortia of bacteria interact to form pathogenic communities in the human host.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases with grants to D.N.F. (RO1 AI061628) and to J.M.M. (AI052228).
We thank Andrew Millard for assistance with Perl scripting.
Footnotes
Published ahead of print on 16 May 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amsel, R., C. A. Spiegel, K. C. Chen, D. Eschenbach, and K. K. Holmes. 1983. Nonspecific vaginitis: diagnostic criteria and microbial and epidemiologic associations. Am. J. Med. 74:14-22. [DOI] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton, J. P., and G. Reid. 2002. Evaluation of the bacterial vaginal flora of 20 postmenopausal women by direct (Nugent score) and molecular (polymerase chain reaction and denaturing gradient gel electrophoresis) techniques. J. Infect. Dis. 186:1770-1780. [DOI] [PubMed] [Google Scholar]
- 4.DeSantis, T. Z., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, D. A. Relman, Z. Gao, C. H. Tseng, Z. Pei, and M. J. Blaser. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eschenbach, D. A. 1993. History and review of bacterial vaginosis. Am. J. Obstet. Gynecol. 169:441-445. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1989. PHYLIP: Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 9.Fredricks, D. N., T. L. Fiedler, and J. M. Marrazzo. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353:1899-1911. [DOI] [PubMed] [Google Scholar]
- 10.Fredricks, D. N., and J. M. Marrazzo. 2005. Molecular methodology in determining vaginal flora in health and disease: its time has come. Curr. Infect. Dis. Rep. 7:463-470. [DOI] [PubMed] [Google Scholar]
- 11.Gao, Z., C. H. Tseng, Z. H. Pei, and M. J. Blaser. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA 104:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gravett, M. G., D. Hummel, D. A. Eschenbach, and K. K. Holmes. 1986. Preterm labor associated with subclinical amniotic-fluid infection and with bacterial vaginosis. Obstet. Gynecol. 67:229-237. [DOI] [PubMed] [Google Scholar]
- 14.Hillier, S. L., R. P. Nugent, D. A. Eschenbach, M. A. Krohn, R. S. Gibbs, D. H. Martin, M. F. Cotch, R. Edelman, J. G. Pastorek, A. V. Rao, D. McNellis, J. A. Regan, J. C. Carey, and M. A. Klebanoff. 1995. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. N. Engl. J. Med. 333:1737-1742. [DOI] [PubMed] [Google Scholar]
- 15.Hyman, R. W., M. Fukushima, L. Diamond, J. Kumm, L. C. Giudice, and R. W. Davis. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. USA 102:7952-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 17.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nugent, R. P., M. A. Krohn, and S. L. Hillier. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J. Clin. Microbiol. 29:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pybus, V., and A. B. Onderdonk. 1997. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J. Infect. Dis. 175:406-413. [DOI] [PubMed] [Google Scholar]
- 20.Pybus, V., and A. B. Onderdonk. 1999. Microbial interactions in the vaginal ecosystem, with emphasis on the pathogenesis of bacterial vaginosis. Microbes Infect. 1:285-292. [DOI] [PubMed] [Google Scholar]
- 21.Saunders, S., A. Bocking, J. Challis, and G. Reid. 2007. Effect of Lactobacillus challenge on Gardnerella vaginalis biofilms. Colloids Surf. B 55:138-142. [DOI] [PubMed] [Google Scholar]
- 22.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid, G., L. Markowitz, R. Joesoef, and E. Koumans. 2000. Bacterial vaginosis and HIV infection. Sex. Transm. Infect. 76:3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sewankambo, N., R. H. Gray, M. J. Wawer, L. Paxton, D. McNairn, F. WabwireMangen, D. Serwadda, C. J. Li, N. Kiwanuka, S. L. Hillier, L. Rabe, C. A. Gaydos, T. C. Quinn, and J. KondeLule. 1997. HIV-1 infection associated with abnormal vaginal flora morphology and bacterial vaginosis. Lancet 350:546-550. [DOI] [PubMed] [Google Scholar]
- 25.Sha, B. E., M. R. Zariffard, Q. J. Wang, H. Y. Chen, J. Bremer, M. H. Cohen, and G. T. Spear. 2005. Female genital-tract HIV load correlates inversely with Lactobacillus species but positively with bacterial vaginosis and Mycoplasma hominis. J. Infect. Dis. 191:25-32. [DOI] [PubMed] [Google Scholar]
- 26.Sobel, J. D. 2005. What's new in bacterial vaginosis and trichomoniasis? Infect. Dis. Clin. N. Am. 19:387-406. [DOI] [PubMed] [Google Scholar]
- 27.Spiegel, C. A. 2002. Bacterial vaginosis. Rev. Med. Microbiol. 13:43-51. [Google Scholar]
- 28.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweet, R. L. 1995. Role of bacterial vaginosis in pelvic inflammatory disease. Clin. Infect. Dis. 20:S271-S275. [DOI] [PubMed] [Google Scholar]
- 30.Turnbaugh, P. J., R. E. Ley, M. A. Mahowald, V. Magrini, E. R. Mardis, and J. I. Gordon. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027-1031. [DOI] [PubMed] [Google Scholar]
- 31.Verhelst, R., H. Verstraelen, G. Claeys, G. Verschraegen, J. Delanghe, L. Van Simaey, C. De Ganck, M. Temmerman, and M. Vaneechoutte. 2004. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobiurn vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. Report of the ad-hoc-committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 33.Zhou, X., S. J. Bent, M. G. Schneider, C. C. Davis, M. R. Islam, and L. J. Forney. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150:2565-2573. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, X., C. J. Brown, Z. Abdo, C. C. Davis, M. A. Hansmann, P. Joyce, J. A. Foster, and L. J. Forney. 2007. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1:121-133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.