Abstract
The filaments of spirochete periplasmic flagella (PFs) have a unique structure and protein composition. In most spirochetes, the PFs consist of a core of at least three related proteins (FlaB1, FlaB2, and FlaB3) and a sheath of FlaA protein. The functions of these filament proteins remain unknown. In this study, we used a multidisciplinary approach to examine the role of these proteins in determining the composition, shape, and stiffness of the PFs and how these proteins impact motility by using the spirochete Brachyspira (formerly Treponema, Serpulina) hyodysenteriae as a genetic model. A series of double mutants lacking combinations of these PF proteins was constructed and analyzed. The results show the following. First, the diameters of PFs are primarily determined by the sheath protein FlaA, and that FlaA can form a sheath in the absence of an intact PF core. Although the sheath is important to the PF structure and motility, it is not essential. Second, the three core proteins play unequal roles in determining PF structure and swimming speed. The functions of the core proteins FlaB1 and FlaB2 overlap such that either one of these proteins is essential for the spirochete to maintain the intact PF structure and for cell motility. Finally, linear elasticity theory indicates that flagellar stiffness directly affects the spirochete's swimming speed.
The spirochetes constitute a very diverse but monophlytic group of bacteria that have common morphological attributes (37). All species of spirochetes have periplasmic flagella (PFs), and these organelles have been shown to be essential for motility (8, 25). One to several hundred PFs are subterminally attached near each end and extend toward the center of the cell, where they may or may not overlap. The number of PFs, and whether they overlap at the center of the cell, is dependent on the species. Surrounding the entire cell is a lipid bilayer membrane. This outer membrane is often referred to as an outer membrane sheath, since it surrounds the entire spirochete including the PFs. Thus, spirochetes differ from other bacteria in that the spirochete organelles for motility—the PFs—reside within the periplasmic space. There is a real advantage to being a spirochete, since these organisms can efficiently swim through highly viscous gel-like media which inhibit the motility of most other motile bacterial species (3, 8, 34).
PFs consist of three parts: basal body, hook, and filament. Consequently, the PFs are similar in structure to the flagella of other bacteria (8, 26, 32). Flagella in other species of bacteria often consist of a polymer of a protein referred to as flagellin; within the flagella is a narrow channel 20 Å in diameter whereby new flagelllin monomers pass through and assemble at the growing tip (53). However, the PF filament is unique and is among the most complex of bacterial flagellar filaments thus far analyzed (8, 25, 26). Specifically, in most spirochete species, the PFs contain one, and sometimes two, flagellar sheath proteins referred to as FlaA, and most often two to three core proteins designated FlaB1, FlaB2, and FlaB3 (6, 8, 25, 36, 41, 50). The spirochete Borrelia burgdorferi is unique, since it has only one FlaA protein and one FlaB protein (16). FlaA proteins are 37- to 44-kDa and are similar between species based on their amino acid sequences and antigenic cross-reactivity. These proteins are likely exported to the periplasmic space by the type II secretion pathway, as their N-terminal amino sequences are cleaved, and a typical peptidase I cleavage site is present near the N terminus (7, 8, 24, 36). In contrast, FlaB proteins are exported to the periplasmic space through the basal bodies via a type III secretion pathway (8, 36). These core proteins are generally 33 to 41 kDa and immunologically cross-react between one another in a given species and also between species (8, 25, 36). There is no sequence similarity or antigenic cross-reactivity between FlaA and FlaB proteins (8, 25, 36).
FlaB proteins comprise a family of related proteins. For example, FlaB proteins of Treponema pallidum, Treponema denticola, and Brachyspira hyodysenteriae have between 57 and 84% deduced amino acid sequence identity. In some cases, the sequence identity of FlaB proteins is higher between species than within a given species. In any given species, each FlaB protein is encoded by an individual gene (15, 24, 35, 39, 44). Because FlaB proteins, but not FlaA proteins, have sequence similarities to the flagellin of other bacteria, especially at the N- and C-terminal regions, FlaB proteins are considered to have an identical function in forming helical flagellar filaments that rotate (36, 52) and are assumed to have a narrow channel for new monomers to pass through. Along these lines, PFs have indeed been shown to rotate (9, 18), and FlaB proteins devoid of the FlaA sheath have been shown to be left-handed helices (9, 18, 24). The arrangement of the FlaB proteins within the filament is unknown: thus, it is unclear whether FlaB1, FlaB2, and FlaB3 reside on separate domains within a PF, or whether they are dispersed along the filament.
The roles of the individual filament proteins with respect to both PF structure and motility in spirochetes are not understood. We and others have begun to analyze the function of these filament proteins using B. hyodysenteriae as a model system (20, 24, 41). B. hyodysenteriae is the cause of swine dysentery, and evidence suggests that its motility is necessary for colonization of the intestine to bring about disease (20, 42). Furthermore, it is readily manipulated by both allelic exchange mutagenesis and by generalized transduction via a defective transducing phage (20, 41, 42, 46). Finally, the genes encoding these filament proteins are monocistronic; thus, downstream affects of insertion mutations are minimized (24). Because the noncultivable syphilis spirochete T. pallidum and the oral spirochete T. denticola have PFs similar to that of B. hyodysenteriae with respect to having one FlaA protein and three FlaB proteins and because these proteins show significant sequence similarity between species as noted above, the information obtained from B. hyodysenteriae should be relevant to these spirochetes as well (4, 36, 43).
In previous studies with B. hyodysenteriae, the effects of single deletion mutations in each of the filament genes (flaA, flaB1, flaB2, and flaB3) were analyzed in detail (24, 41). Rosey et al. partially analyzed the double flaA flaB1 mutant (42). We found that the loss of one of the FlaB proteins did not influence the assembly and the helical shape of the PFs. In contrast, the PF morphology of the flaA mutant was dramatically different (24). These results indicated that FlaA did not just passively form a sheath around a FlaB core but rather contributed to PF helicity. Furthermore, because all of the single mutants had a slight decrease in motility, each filament protein is evidently essential for full motility. To further examine the role of these proteins in PF structure and cell motility, we analyzed a series of double mutants in the genes that encode these proteins. Here we show that FlaB1 and FlaB2 overlap with respect to function, i.e., in the absence of either one of these two proteins, the other is able to carry out that function. The analysis also indicates that FlaB1, FlaB2, and FlaB3 are most likely dispersed along the entire PF. Furthermore, and most surprisingly, in the absence of both FlaB1 and FlaB2, a hollow tubular structure is formed that is primarily composed of a polymer of FlaA. Finally, we used linear elasticity theory to calculate the stiffness of the mutant PFs relative to the wild type. Our results suggest that flagellar stiffness directly affects the swimming speed of these spirochetes.
MATERIALS AND METHODS
Bacterial strains, generalized transduction, and culture conditions.
The B. hyodysenteriae B204 (wild type), flaA (flaA::kan or flaA::cat), flaB1 (flaB1::kan), flaB2 (flaB2::cat), and flaB3 (flaB3::cat) single-mutant, and flaA flaB1 double-mutant (flaA::cat-flaB1::kan) strains have been previously described. Each insertion of the resistance cassette had the following accompanying deletion (the genes varied from 0.85 to 1.2 kb in size) (24, 41, 42): flaA (169 bp), flaB1 (589 bp), flaB2 (345 bp), and flaB3 (278 bp). The bacteriophage VSH-1 was used for generalized transduction and construction of the flaA flaB2 (flaA::kan-flaB2::cat), flaA flaB3 (flaA::kan-flaB3::cat), flaB1 flaB2 (flaB1::kan-flaB2::cat), and flaB1 flaB3 (flaB1::kan-flaB3::cat) double mutants (46). Cells were grown in an atmosphere of 1 to 2% oxygen in a Coy anaerobic chamber and in media previously described, and all manipulations were done to minimize normal atmospheric exposure (24, 41, 42).
DNA manipulation and PCR conditions.
B. hyodysenteriae chromosomal DNA isolation, PCR, DNA sequencing, and Southern blot analysis were used to characterize the respective double mutants as previously described (24, 41). The primer sequences, target loci, and conditions for DNA amplification have been described in previous publications (24, 41).
Purification of PFs, SDS-PAGE, and Western blotting.
The PFs from the wild type and the mutants were purified and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), densitometry, and Western blotting, as previously described (24). Rabbit antisera directed to B. hyodysenteriae FlaA and FlaB proteins were kindly provided by M. Jacques (University of Montreal, Montreal, Quebec, Canada), and a monoclonal antibody that reacts with B. hyodysenteriae FlaB1, FlaB2, and FlaB3 was generously provided by G. Duhamel (University of Nebraska). The procedures for PF purification using shearing and differential centrifugation (without CsCl banding) have been previously described (24).
Measurement of the helicity of the PFs.
The PF morphology was analyzed by using dark-field microscopy. The helix handedness, helix pitch, and helix diameter of the PFs were determined as previously described (24). At least 25 PFs were measured from each strain, and the results are expressed as mean ± the standard error of the mean (SEM). Significant differences were established by using analysis of variance (ANOVA) and post-hoc Tukey-Kramer tests.
Motion analysis and swarm plate assay.
Two methods were used to measure spirochete motility. First, the motility of spirochete cells was measured by swarm agar blood plates. Swarm diameters were determined after 36 h of incubation (24). Second, the velocity of the wild type and mutants was measured by using a software package marketed as Volocity (Improvision, Inc., Coventry, United Kingdom). This software package has recently been used to successfully track B. burgdorferi cells (2, 31). Briefly, 1 μl of late-logarithmic-phase B. hyodysenteriae cultures was added into 20 μl of prereduced saline buffer with 1% methylcellulose (4,000 mesh). The spirochete cells were visualized by dark-field microscopy with a Zeiss Axioskop 2 microscope (Carl Zeiss, Inc., Jena, Germany) at ×200 magnification at 35°C equipped with a heated stage (Physitemp, Inc., Clifton, NJ). Approximately 100 cells of the wild type and all of the mutants were tracked and analyzed. For all of the strains, the velocity of the fastest 25 swimming cells was measured, and the results are expressed as means ± the standard deviations (SD) of the mean; this group of cells was chosen since, during the time interval for tracking (between 1 and 2 min), the swim paths of these cells appeared to be unhindered by the glass surface compared to slower-swimming cells.
Standard TEM and cryo-TEM.
To view the PFs, purified PFs using negative staining were examined by electron microscopy as previously described (24, 41). Cryo-transmission electron microscopy (cryo-TEM) was used to analyze the ultrastructure of the purified PFs from the wild type and the mutant strains. As an internal control to determine PF diameter, 1 μl of tobacco mosaic virus (TMV), freshly resuspended in distilled water, was added to 32 μl of each sample. The freshly prepared PFs containing TMV were applied to lacy carbon grids covered with fresh 10-nm-thick carbon films and then plunge-frozen in liquid ethane cooled by liquid nitrogen (13, 51). A 5-μl volume was applied to one side of the grid, and the grid was double blotted for 2 s before plunging. The plunging was performed at room temperature. Grids were stored in liquid nitrogen after plunging. Images were recorded with a JEOL JEM-4000FX at 400 kV (wild type) and an FEI Tecnai F20 at 200 kV (mutants). The underfocus setting on both microscopes was such that the contrast-transfer function first minimum corresponded to a spacing of about 4 nm. However, the effect of the higher coherence of the field emission gun in the case of the F20 was quite noticeable in the images. In both cases, images were recorded at −179°C, with an incident electron dose of 12 e-/Å2. The images were saved as digital format. The diameters of the PFs and derived structure were directly measured from the digital images by using the software Openlab (Improvision). No more than two measurements were made per filament, and at least 30 filaments were measured per sample. The measurements were calibrated by the diameters of TMV and were expressed as means ± the standard error of the mean. Significant differences were evaluated by using ANOVA and post-hoc Tukey-Kramer tests.
RESULTS
Construction of double mutants mediated by B. hyodysenteriae VSH-1 bacteriophage.
We and Rosey et al. had previously constructed the following single PF insertion-deletion mutants using kanamycin (kan) or chloramphenicol (cat) cassettes: flaA::kan, flaA::cat, flaB1::kan, flaB2::cat, and flaB3::cat (24, 41). To construct double mutations using the generalized defective transducing phage VSH-1, combinations of single mutants with different antibiotic resistance determinants were cocultured overnight and spread onto plates containing both antibiotics (46). The following double mutants were constructed: flaA flaB2, flaA flaB3, flaB1 flaB2, and flaB1 flaB3. Rosey et al. previously constructed a flaA flaB1 mutant (42); thus, all of the combinations of flaA with the three flaB mutants and two of the three possible combinations of the flaB mutants were obtained (we were unsuccessful in obtaining a flaB2 flaB3 mutant). PCR analysis using primers identical to those used to analyze single mutants (24) indicated that allelic exchange occurred at the targeted genes (data not shown).
Motility analysis of mutants.
Previous results using swarm plate assays indicated that single mutants were still motile but decreased in motility (24). To further investigate the role of these proteins in motility, we extended our analysis to velocity determinations of the wild type and single and double mutants. The velocities of the wild type and mutant strains were measured by the Volocity computer based tracking system, and the results are listed in the Table 1. Strains carrying single mutations in flaA, flaB1, or flaB2 had an approximate decrease in velocity of 28 to 38% compared to the wild type. The flaB3 mutant only decreased ca. 13%. These results suggest that FlaB3 is less important than other filament proteins in terms of motility. In the analysis of the double mutants, the combination of a mutation in flaA with any of the three flaB mutations revealed a pattern (Table 1). These three mutants (flaA flaB1, flaA flaB2, and flaA flaB3) were markedly slower than the single mutants; they had an approximate decrease in velocity of 50 to 60% compared to the wild type. The flaA flaB3 mutant was least affected (50%). Similarly, the flaB1 flaB3 double mutant also showed a decrease in velocity of 48%. Only the flaB1 flaB2 double mutant was completely nonmotile.
TABLE 1.
Correlation between cell speed and PF stiffness in wild type and the mutants
| Strain | Swarm diam (mm)a | Mean speed (μm/s) ± SD | Relative speedb | Helix (mean μm ± SEM)
|
Modulusc
|
||
|---|---|---|---|---|---|---|---|
| Pitch | Diam | RBM | RTM | ||||
| Wild type | 14.0 | 40.0 ± 4.0 | 1.00 | 2.84 ± 0.1 | 0.86 ± 0.06 | 1.00 | 1.00 |
| flaB3 mutant | 12.0 | 35.0 ± 2.5 | 0.87 | 2.78 ± 0.1 | 0.74 ± 0.08 | 0.93 | 1.24 |
| flaA mutant | 9.0 | 29.0 ± 3.0 | 0.72 | 2.40 ± 0.2 | 0.59 ± 0.06 | 0.76 | 0.62 |
| flaB1 mutant | 9.0 | 29.0 ± 3.5 | 0.72 | 2.70 ± 0.1 | 0.75 ± 0.04 | 0.81 | 1.02 |
| flaB2 mutant | 6.0 | 25.0 ± 4.5 | 0.62 | 2.70 ± 0.2 | 0.69 ± 0.06 | 0.87 | 1.16 |
| flaA flaB3 mutant | 9.0 | 20.0 ± 2.5 | 0.50 | 1.92 ± 0.1 | 0.44 ± 0.06 | 0.69 | 0.86 |
| flaA flaB1 mutant | 2.5 | 17.0 ± 2.0 | 0.42 | 1.77 ± 0.2 | 0.35 ± 0.05 | 0.57 | 0.64 |
| flaA flaB2 mutant | 2.5 | 16.0 ± 2.5 | 0.40 | 1.80 ± 0.1 | 0.38 ± 0.06 | 0.63 | 0.78 |
| flaB1 flaB3 mutant | 7.5 | 21.0 ± 2.5 | 0.52 | 2.70 ± 0.2 | 0.84 ± 0.07 | NA | NA |
| flaB1 flaB2 mutant | 0 | 0 | 0 | 0 | 0 | NA | NA |
The data are expressed as the swarm diameter of a given mutant after subtracting that of the nonmotile flaB1 flaB2 mutant.
Except for the flaB1 flaB3 mutant, the strains were ordered based on their decreasing relative speed. The relative speed is the mean swimming speed of the mutant bacteria divided by the mean swimming speed of the wild-type cells.
The relative bending modulus (RBM) and the relative twist modulus (RTM) are the stiffness of the flagellum to bending and twisting, respectively, divided by the bending or twisting modulus of the wild-type flagellum. NA, not applicable.
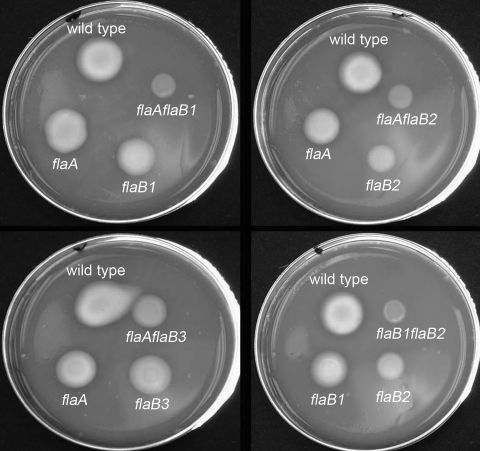
The results obtained by motion analysis were further tested by swarm plate assays. After 36 h of incubation, the swarm diameters of all of the double mutants were less than those of the wild type and single mutants (Fig. 1 and Table 1). Furthermore, only the flaB1 flaB2 double mutant failed to swarm. These results are in agreement with the velocity determinations noted above. Taken together, because single flaB1 and flaB2 mutants were still motile and the double flaB1 flaB2 mutant was nonmotile, there is functional overlap with respect to FlaB1 and FlaB2, i.e., if one of these proteins is missing, the other can supply in part the function necessary for motility. Furthermore, the results indicate that all double mutants were more severely inhibited in motility than the single mutants. Finally, because the single flaB3 mutant was least affected compared to the other single mutants, and in combination with mutations in flaA or flaB1, FlaB3 may not be as essential for motility as the other FlaB proteins.
FIG. 1.
Swarm plate assay of the wild type and the PF mutants. Approximately 4 μl of 4 × 105 washed cells were inoculated onto the surface of 0.3% agar prereduced plates by using a micropipette, followed by incubation for 36 h. This inoculum formed a drop size on the agar ∼4 mm in diameter. Due to space limitations, the swarm formed by the flaB1 flaB3 double mutant is not presented.
PF assembly in the mutants.
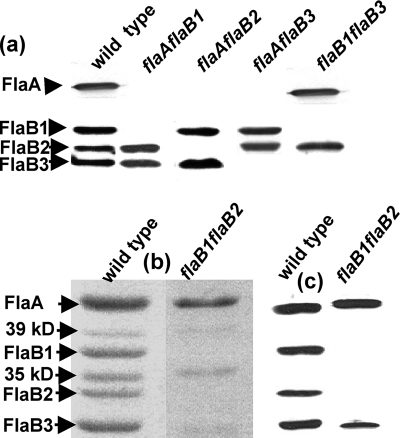
B. hyodysenteriae PFs are composed of four filament proteins: one FlaA sheath protein, and three FlaB core proteins (24). Each of these proteins is encoded by a separate gene that is monocistronic (24). In addition, other proteins of 35 and 39 kDa, which do not react with either FlaA or FlaB antisera, are associated with the PFs (Fig. 2b and c) (21, 24). The 39-kDa protein is considered to be weakly associated with the PFs, since it is lost on CsCl gradient centrifugation purification. In contrast, the 35-kDa protein is thought to be strongly associated with the PFs, since it is retained on gradient purification among all of the mutants (24). Because the 35-kDa protein is retained in PF preparations from mutants lacking FlaA, it evidently does not closely associate with FlaA. Preliminary results from peptide analysis indicate that the 35-kDa protein is a lipoprotein. We previously found that the single mutants still assembled the PFs (24). To analyze the composition and structure of the PFs from the double mutants, the PFs were purified and compared to the wild type and the single mutants. Western blot analysis indicated that all of the double mutants were specifically deficient in the proteins encoded by the targeted genes (Fig. 2a). For example, in all of the flaA flaB mutants, there was a complete loss of the FlaA and the corresponding FlaB protein. A similar pattern was found in whole-cell lysate analysis of these mutants probed with the FlaA and FlaB antisera from B. hyodysenteriae (data not shown). These results indicate that not only are all mutants deficient in the targeted filament proteins but also the remaining filament proteins are still expressed and assembled in a given PF from the respective mutants.
FIG. 2.
Analysis of the PF proteins in the mutants. (a) Western blot analysis of the purified PFs from wild type and four double mutants. (b and c) SDS-PAGE (b) and Western blot (c) analysis of the purified PFs from the wild type and the flaB1 flaB2 mutant. Rabbit antisera to FlaA, and monoclonal antibodies to FlaB were used as probes.
Because of the unique nonmotile phenotype of the flaB1 flaB2 mutant, its PF analysis is presented in more detail. SDS-PAGE analysis of the flaB1 flaB2 mutant was compared to that of the wild type. The PF fraction from the mutant was found to consist primarily of FlaA, a decreased amount of FlaB3, and small amounts of the 39- and 35-kDa proteins (Fig. 2b). Densitometry analysis indicated the ratio of FlaA to 35-kDa protein to FlaB3 to be 1:0.35:0.15 (for a reference, the wild type has a ratio of FlaA to FlaB1 to FlaB2 to FlaB3 of 1:0.42:0.22:0.45 [24] and to the 35-kDa protein of 0.4, but this latter ratio varied considerably from preparation to preparation). This conclusion was further supported by Western blot analysis of the PF fraction, which indicated strong reactivity to FlaA, and a decreased amount of FlaB3 (Fig. 2c).
PFs structure of mutants.
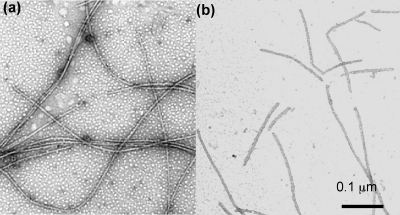
Spirochete PFs have been shown to be left-handed with a distinct helical pitch and helix diameter for a given species (9, 24). In B. hyodysenteriae, only the mutants deficient in flaA had a markedly altered PF helical shape; the PFs from single mutations in flaB1, flaB2, and flaB3 were similar or slightly less helical than that of the wild type (Table 1) (24). We analyzed the PFs of the double mutants as previously described for the single mutants. The helicity of the PFs isolated from the double mutants with flaA and flaB mutations were dramatically altered (Table 1). Specifically, the helix pitch of the PFs from the flaA flaB1, flaA flaB2, and flaA flaB3 mutants were 20 to 26% less than those of the single flaA mutant and 32 to 38% less than that of the wild type. Similarly, the helix diameters of PFs of these mutants were also dramatically affected: 25 to 41% less than that of the single flaA mutant and 48 to 59% less than that of the wild type. All of these mutants were left-handed (data not shown). These results indicate that the combinations of mutations of flaA with those of flaB cause a striking alteration in helicity of the PFs. The flaB1 mutation in combination with either the flaB2 or the flaB3 mutation yielded markedly different results. The flaB1 flaB3 mutant had a helix pitch and a diameter almost identical to that of the wild type. Thus, FlaA and FlaB2 together form PFs quite similar in shape to those of the wild type. In contrast to all other mutants, the filaments from the PF fraction of the flaB1 flaB2 mutant were completely nonhelical. TEM revealed that these filaments were short and appeared straighter than the wild type and other PF mutants (Fig. 3). Because the filaments of this mutant primarily consist of FlaA with only a small amount of FlaB3 (Fig. 2b and c), the results suggest that FlaA by itself is unable to confer a helical shape.
FIG. 3.
PF fraction stained with 1% phosphotungstate. (a) Wild type; (b) flaB1 flaB2 mutant.
Thin-section and negative-staining analysis of double mutants.
Previous studies indicate that the single mutations in these filament genes do not change the number of PFs. To test whether double mutations have altered the number of PFs in the given mutants, the number of PFs present in these mutants was determined by analysis of thin sections and by negative staining of lysed cells. Except for the flaB1 flaB2 mutant, all others had typical PF structures located in their periplasmic space, as can be seen in cross-sections and oblique sections (Fig. 4a and c). We found that these double mutants had 8 to 11 PFs per cell cross-section, which is similar to those found for the wild type (Fig. 4a). Thus, the double mutations in the given mutants did not influence the number of PFs per cell. Furthermore, the results also suggest that there was not a major variation from one mutant to another with respect to PF length; otherwise, we would have observed fewer PFs in the cross-section in the mutant compared to the wild type.
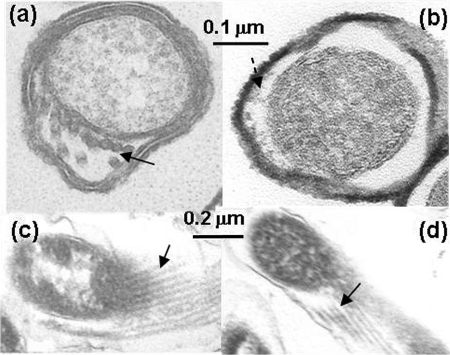
FIG. 4.
Electron microscopic analysis of the wild type and the flaB1 flaB2 mutant. (a and b) Thin section of the wild type and the flaB1 flaB2 mutant. The solid arrow points to PFs, and the dashed arrow points to amorphous staining material seen in the flaB1 flaB2 periplasmic space. (c and d) Oblique sections of the wild type and the flaB1 flaB2 mutant. Arrows point to PFs in panel c and to tubular material in panel d.
The flaB1 flaB2 mutant differed from the wild type and other double mutants, as seen in thin sections. Structures resembling PFs were not evident in cross-sections (Fig. 4b). In some cells, nondiscrete densely staining material was observed in the periplasmic space (Fig. 4b, dashed arrow). This periplasmic material lacked the definition of obvious filamentous structures as observed in the other mutants and the wild type (Fig. 4a). Interestingly, organized filamentous structures were observed in oblique sections similar to those found in the wild type (Fig. 4d) but were much more rarely observed than in the wild type and other mutants. However, when a gentle procedure to remove the outer membrane sheath with 0.5% Triton X-100 instead of the previously used 1% Triton X-100 to isolate PFs (24) was applied, no filamentous structures were seen attached to the cell cylinders (data not shown).
The flaB1 flaB2 double mutant forms hollow tubes.
To further characterize the filamentous structures found in the flaB1 flaB2 mutant, its PF fraction was isolated and analyzed in detail. Whereas the wild-type filaments stained primarily on the periphery using negative staining, the internal region of the filaments from the flaB1 flaB2 mutant stained dark (Fig. 5a and b). These results suggest that the filaments from the flaB1 flaB2 mutant were hollow tubular structures. The PF fractions were further analyzed by cryo-TEM. Whereas the wild-type PFs appeared dense throughout the filament image, only along the periphery of the filaments of the flaB1flaB2 was there an obvious increase in density (Fig. 5c and d). These results further substantiate that the flaB1 flaB2 mutant formed hollow tubular structures instead of intact PFs. Because SDS-PAGE and Western blot analysis indicated that this fraction contained primarily FlaA (Fig. 2b and c), FlaA evidently formed a PF sheath structure in the absence of FlaB1 and FlaB2. These results also suggest that in the absence of both FlaB1 and FlaB2, the core structure of the filaments is missing.
FIG. 5.
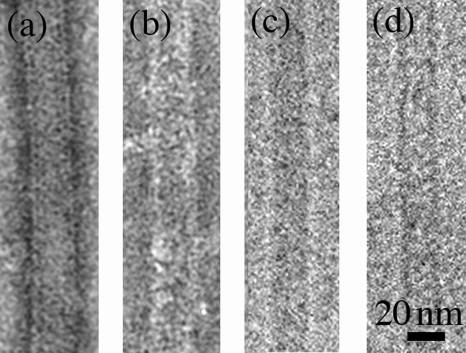
Ultrastructure of the PF fraction from the wild type and flaB1 flaB2 mutant. (a and b) Negative stain of PF fractions from the wild type (a) and the flaB1 flaB2 mutant (b) stained with uranyl acetate. (c and d) High-magnification cryo-TEM images of filaments from the PF fractions from wild type (c) and flaB1flaB2 mutant (d). The diameter flaB1 flaB2 filament was 20 nm.
The PF diameter was less in the mutants without FlaA.
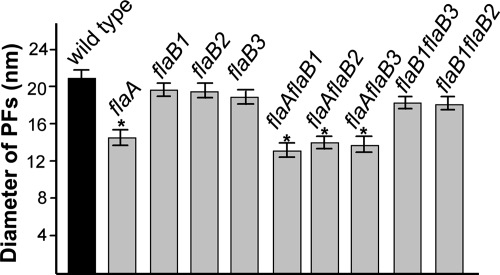
We previously measured the diameter of the PFs from the wild type and single mutants by negative staining. The results indicated that the diameters of the PFs from the flaA mutants but not the flaB mutants were thinner than that of the wild type (24). However, negative staining can yield inaccurate measurements, primarily due to flattening of the PFs on the carbon support film. Accordingly, cryo-TEM, in combination with TMV used as an internal control, allowed precise measurement of the PF diameter. Cryo-TEM measurements were found to be slightly less than previously found with negative staining (24). For example, the diameter of the wild-type PF measured by negative staining was approximate 25 nm, whereas the diameter measured by cryo-TEM was 21 nm. The single flaB mutants had approximately the same diameters as the wild type, which ranged from 18.0 to 20.0 nm (Fig. 6). In contrast, the diameters of the PFs from the flaA mutant of 14.8 nm were significantly thinner than that of the wild type (Fig. 6). Among the double mutants, the diameters of the PFs with the flaA mutation in combination with the flaB mutation were the same as for the single flaA mutant (Fig. 6). These results are consistent with the previous results and further support the conclusion that FlaA forms a sheath around the FlaB core (24). The diameters of the filaments from mutants with mutations in flaB1 in combination with mutations in flaB2 or flaB3 were also measured. The flaB1 flaB3 mutant had PFs with the same diameter as the wild-type PFs. This result was expected, since this mutant forms PFs with a helix pitch and diameter quite similar to that of the wild type (Table 1). We also measured the diameter of hollow tubes found in the flaB1 flaB2 PF fraction. These structures had the same diameter as the PFs of the wild type. The results indicate that FlaA can polymerize into hollow tubes independent of FlaB1 and FlaB2 and that these tubes have the same diameters as the wild-type PFs.
FIG. 6.
Measurement of PF diameters from the wild type and the PF mutants. For each strain, at least 30 filaments were measured, and the measurements were calibrated by the diameters of TMV.
PF stiffness correlates with speed.
The presence of a FlaA sheath about the PFs suggests that this sheath may provide a functional advantage for the function of the PFs. One possibility would be that the FlaA sheath makes the flagella stiffer, which would allow the spirochete to swim faster. To test this idea, we constructed a simple model for the helical shape of the PFs, which allowed us to estimate the stiffness of the mutant flagella relative to the wild-type flagella. The stiffness of a filamentary object can be defined by the bending modulus and the twist modulus. The bending modulus describes how much torque is required to impart a given amount of curvature to the filament. The twist modulus describes how much torque is required to twist the filament a given amount about its centerline. Given that the FlaA sheath is relatively straight (Fig. 3b) and by comparing the helical parameters of the wild-type and flaA mutant PFs, we calculated the stiffness (both for bending and twisting) of the FlaB core relative to the wild-type PF (Table 1). The difference between the two yields the stiffness of the FlaA sheath relative to the wild-type PF. In addition, by comparing the single flaB to the double flaA flaB mutants, we also calculated the relative stiffness of the PF core when one of the FlaB monomers is missing (Table 1; see the Appendix for a complete description of the mathematical model). The wild-type PFs were the stiffest structures with respect to bending, whereas the PFs from the flaA flaB1 mutant were the most pliable. With respect to twisting, the PFs of flaB3 mutant were the stiffest, and those from the flaA mutant were the most pliable. There was a strong agreement between the speed and the bending stiffness of the PFs such that stiffer PFs produced larger swimming speeds (Table 1). However, if this correlation were exactly true, then the flaB2 mutant should be faster than the flaA or flaB1 mutant. Likewise, the flaA flaB2 mutant should also be faster than the flaA flaB1 mutant. Note that for both of these cases, the relative twist modulus was larger in the slower mutant. These results suggest that larger bending moduli favor faster swimming speeds, but larger twist moduli favor slower swimming speeds.
DISCUSSION
Spirochetes are a medically and ecologically significant but poorly understood group of bacteria. Because their cell structure is unique, spirochete motility is different from that of other bacteria (8, 33). Cell movement is driven by two bundles of rotating PFs, which are complex structures consisting of FlaA sheath proteins, and multiple FlaB core proteins (8, 24, 25, 26). The functions of these filament proteins have not been characterized for several reasons. First, in general, spirochetes are a fastidious group of bacteria and are thus difficult to manipulate in the laboratory. For example, T. pallidum, several spirochete species found in the oral cavity, and others found in many habitants in nature have yet to be continuously grown in the laboratory. In addition, those that can be cultured have relatively long generation times. Second, the genetic tools are quite limited for spirochetes. For example, only recently have allelic exchange and transposon mutagenesis been developed for some spirochete species (5, 47). Finally, because the PFs normally reside in the periplasmic space, directly tracking their rotation, as done with externally located flagella such as those of Escherichia coli and Salmonella enterica serovar Typhimurium, is not currently possible.
The distribution of the FlaB proteins within the PF sheath is still unknown. Our previous results indicated that a given PF contains FlaA and all three FlaB proteins (24). Two possible structural models have been postulated based on the previous analysis of single mutants (24). One model states that each FlaB protein forms a specific domain within the sheath, e.g., FlaB1 forms one domain, FlaB2 forms a second, and FlaB3 forms a third. Alternatively, perhaps all three FlaB proteins interact with one another and are distributed along the entire PF core. We favor the latter hypothesis. If the FlaB subunits are arranged with each one in a specified domain, the double mutants with mutations in flaA and one of the flaB genes should have PFs that are not markedly different than those from the single flaA mutants. This was not the case, since the helical morphology of the PFs of the all the double flaA flaB mutants were markedly different from the wild-type and single FlaA mutants (Table 1). The results point to a given FlaA protein interacting with more than one FlaB protein to determine PF shape. Our results indicate that a given FlaB protein is distributed along the entire length of the PF. How do we explain the overlap in function with respect to FlaB1 and FlaB2? Perhaps FlaB1 and FlaB2 are both able to perform an essential function for PF morphogenesis and FlaB3 is unable to carry out this function. One likely region may be at the distal end of the hook region. Perhaps FlaB1 and FlaB2 can associate with the hook-associated proteins to permit growth of the growing flagellar filament and FlaB3 is unable to carry out this interaction. A similar suggestion has been made to account for flagellar deficient mutants that comprise the complex flagella of Sinorhizobium meliloti (22). Along these lines, the deduced C-terminal end of FlaB3 is markedly different from FlaB1 and FlaB2. Alternatively, either FlaB2 or FlaB1, but not FlaB3, could be capable of forming the lattice that gives rise to the normal (helical) filament.
Flagella containing multiple filament proteins are also found in other bacteria, such as Sinorhizobium spp., Caulobacter crescentus, Helicobacter spp., Campylobacter spp., Vibrio spp., and Bdellovibrio bacteriovorus (1, 12, 19, 22, 29, 40). Although inactivation of some of the genes encoding the filament proteins results in a complete loss in motility, others show a decrease in motility or have little or no effect. We find a similar result with B. hyodysenteriae flaA and flaB1, flaB2, and flaB3 genes—all single mutants are still motile but are less motile than the wild type. Our results indicate that FlaB1 and FlaB2 have at least a partial overlap in function with respect to flagellar morphogenesis and motility. Sequence analysis indicates that FlaB1 and FlaB2 have high identity with one another (76%) and a considerably lower identity with FlaB3 (57 to 60%). Because motility is strongly implicated as a virulence factor for B. hyodysenteriae (20, 42), redundancy of some of the filament genes is likely to have survival value.
A flagellar sheath has also been observed in several other bacterial genera, such as Vibrio, Helicobacter, and Caulobacter species. However, several lines of evidence indicate that the spirochete PF sheath differs from the flagellar sheath of these bacteria. Spirochete FlaA does not share significant homology to proteins found in these bacteria, and thus it is unique to the spirochetes. Second, in Vibrio and Helicobacter species, the flagellar sheath is fragile membrane material covering the organelle (27, 29). In most spirochete species, such as T. pallidum, FlaA is tightly associated with purified PFs and cannot be removed even after trypsin and detergent treatments (6, 10, 36). However, B. burgdorferi is different, since FlaA is readily dissociated from the core after treatment with Sarkosyl (16). C. crescentus is known to have a protein sheath around its flagellum, but only a small fraction appears to be sheathed, and it is likely that this sheath is composed of one of the flagellins that constitute the filament core (48). S. meliloti also has a sheath around the flagella, but this sheath is also likely to be derived from the core flagellar proteins (22, 38). Furthermore, our results indicate that formation of the sheath is not strictly dependent on the existence of the core. FlaB3 is still present in the flaB1 flaB2 mutant and could conceivably act as a start site for subsequent polymerization of FlaA. However, recent results with a fliG mutant of B. hyodysenteriae, which completely lacks detectable FlaB proteins, indicate that a core structure is not necessary for hollow tube formation (C. Li, N. W. Charon, and M. Marko, unpublished data).
The function of the flagellar sheath is not clear. The flagella with sheaths are relatively more rigid in Rhizobium lupini than in other bacteria, and the suggestion has been made that this increased rigidity may permit more efficient swimming in viscous environments (28, 49). What are the possible functions of the FlaA sheath in spirochetes? From our previous study with single mutants and the results presented here with double mutants, FlaA was shown to influence PF helicity (24). Our results suggest that the PF sheath increases the stiffness of the PFs and that this increased stiffness produces a larger swimming velocity. Since the PFs influence cell shape in several spirochete species (8, 30), and preliminary evidence suggests this is occurring in B. hyodysenteriae (C. Li and N. W. Charon, unpublished data), stiffer PFs can deform the cell cylinder more than weaker PFs. Larger deformations of the cell cylinder will produce more thrust. In addition, our analysis shows that the FlaA sheath has a larger effect on the twisting stiffness than it does on the bending stiffness. The PFs of B. burgdorferi have more than one polymorphic configuration (S. Shabata, S. Aizawa, S. Goldstein, and N. Charon, unpublished data), so it is likely that spirochete PF filaments are similar to other prokaryotic flagella in being polymorphic. A moderate twisting stiffness of the FlaA sheath could help stabilize a single conformation, since models for flagellar polymorphism suggest that change in handedness is driven by twisting rather than bending energy (11, 17, 45). Since the FlaA sheath is preferentially straight, a moderate stiffness with respect to twisting could even stabilize the PFs to one specific configuration. The increase of rigidity may also allow the organisms to efficiently swim in viscous gel-like media. Consistent with this conclusion, the motility of all the flaA mutants is significantly decreased in methylcellulose and on swarm plates (Fig. 1 and Table 1). Alternatively, because flagella are quasirigid and may undergo helical transformation, the major function of the sheath could be impacting the packing conformation of the FlaB subunits and thus stabilize its helical shape. However, a model for flagellar morphology based on monomer configuration suggests that changes in packing will also alter flagellar filament stiffness (45). We therefore expect that our estimates for both the bending and twisting moduli provide an accurate ordering of the mutant flagella from stiffest to most pliable. To our knowledge, the relationship between flagellar stiffness and motility has only been previously shown for E. coli mutants that encode the hook-associated HAP3 protein (14). Even though the flaB1 flaB3 mutant forms a filament, we were unable to evaluate the bending and twisting stiffness since this calculation would require knowing the conformation of the FlaB2 core in the absence of FlaA; i.e., a triple mutant (flaA flaB1 flaB3) would be required for this analysis. Because the changes in speed that we observe with respect to the helical parameters of the flagella are significantly larger than would be predicted for these same flagellar geometries rotating in a viscous fluid, we postulate that PF stiffness rather than flagellum shape per se directly impacts cell speed. Directly determining the stiffness of sheathed and unsheathed PFs using a methodology incorporating laser tweezers should provide the experimental test of this model.
APPENDIX
A helix can be defined by its radius (R) and pitch (P) or by its curvature (κ) and torsion (τ). Although the helix radius and helix pitch are a more natural way to visualize the configuration of the helix, the curvature and torsion provide a better description of the deformation that is required to produce a given shape. For example, to deform an initially straight filament into a helix requires that one bend it to have the curvature and twist it enough to have the torsion. The curvature and torsion of a filament are related to the helix radius and pitch by the following relations:
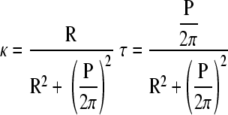 |
(1) |
Linear elasticity theory suggests that to change the curvature of a filament requires a moment that is proportional to the change in curvature, Δκ (for example, see reference 23):
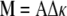 |
(2) |
where A is the bending modulus that describes the material stiffness; i.e., a steel rod has a larger value for A than a similar rod made out of rubber. Since a straight filament has zero curvature, the moment required to bend it to have curvature κ is Aκ. To deform a helical filament that has initial curvature κ0 requires a moment M = A(κ − κ0). Twisting a filament, and changing its torsion, requires a moment that is proportional to the change in torsion, M = CΔτ, where C is the twist modulus.
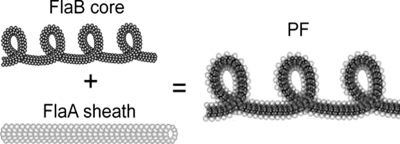
We describe the flagellum as a composite of two concentric filaments and assume that the FlaA sheath is preferentially straight, whereas the FlaB core prefers to be a helix with curvature κB and torsion τB (Fig. 7). When the FlaB core is inside the FlaA sheath, the FlaB core bends the FlaA sheath away from being straight; likewise, the FlaA sheath exerts a moment to the FlaB core that tends to straighten it. The sum of these two moments must equal zero:
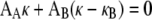 |
(3) |
where AA and AB are the bending moduli of the FlaA sheath and FlaB core, respectively. Therefore,
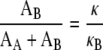 |
(4) |
Using the curvature of the wild-type flagellum for κ, we find that:
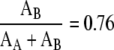 |
(5) |
So, the FlaB core provides 76% of the bending stiffness of the flagellum, and the FlaA sheath therefore provides 24% of the bending stiffness. A similar calculation can be done to calculate the torsional stiffness where we find that:
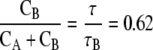 |
(6) |
Using this same procedure, the double mutant bending and twist stiffnesses can be calculated relative to the wild-type PF.
FIG. 7.
Schematic of the construction of the PF. The core of the flagellum is a helical left-handed filament composed of FlaB1, FlaB2, and FlaB3. FlaA forms a straight, hollow sheath. In a PF from the wild type, the FlaA sheath encloses the FlaB core which tends to straighten the core, leading to a flagellum with a larger helix pitch and helix diameter than the core by itself.
Acknowledgments
We thank L. Corum for technical assistance, E. Rosey and T. Stanton for providing strains and helpful discussions, and G. Hobbs for help with statistics. We appreciate the suggestions of D. Yelton and T. Elliott, and we thank G. Duhamel and M. Jacques for sharing antibodies. We also thank S. Goldstein, M. Motaleb, and M. Miller for comments on the manuscript.
This research was supported by Public Health Service (PHS) grants AI-29743 and DE12046 and USDA grant 95-37204-2132 awarded to N.W.C., PHS grant AR050656 awarded to C.L., PHS grant GM 0072004 awarded to C.W.W., and PHS grant RR01219 to J. Frank, P.I., supporting the Resource for Visualization of Biological Complexity.
Footnotes
Published ahead of print on 13 June 2008.
REFERENCES
- 1.Alm, R. A., P. Guerry, and T. J. Trust. 1993. Significance of duplicated flagellin genes in Campylobacter. J. Mol. Biol. 230359-363. [DOI] [PubMed] [Google Scholar]
- 2.Bakker, R. G., C. Li, M. R. Miller, C. Cunningham, and N. W. Charon. 2007. Identification of specific chemoattractants and genetic complementation of a Borrelia burgdorferi chemotaxis mutant: flow cytometry-based capillary tube chemotaxis assay. Appl. Environ. Microbiol. 731180-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, H. C., and L. Turner. 1979. Movement of microorganisms in viscous environments. Nature 278349-351. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, D. R., J. D. Radolf, M. A. Lovett, and J. N. Miller. 1986. The antigenic interrelationship between the endoflagella of Treponema phagedenis biotype Reiter and Treponema pallidum Nichols strain. I. Treponemicidal activity of cross-reactive endoflagellar antibodies against T. pallidum. J. Immunol. 1372973-2979. [PubMed] [Google Scholar]
- 5.Bourhy, P., H. Louvel, I. Saint-Girons, and M. Picardeau. 2005. Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J. Bacteriol. 1873255-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahamsha, B., and E. P. Greenberg. 1988. Biochemical and cytological analysis of the complex periplasmic flagella from Spirochaeta aurantia. J. Bacteriol. 1704023-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahamsha, B., and E. P. Greenberg. 1989. Cloning and sequence analysis of flaA, a gene encoding a Spirochaeta aurantia flagellar filament surface antigen. J. Bacteriol. 1711692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charon, N. W., and S. F. Goldstein. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 3647-73. [DOI] [PubMed] [Google Scholar]
- 9.Charon, N. W., S. F. Goldstein, S. M. Block, K. Curci, J. D. Ruby, J. A. Kreiling, and R. J. Limberger. 1992. Morphology and dynamics of protruding spirochete periplasmic flagella. J. Bacteriol. 174832-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cockayne, A., M. J. Bailey, and C. W. Penn. 1987. Analysis of sheath and core structures of the axial filament of Treponema pallidum. J. Gen. Microbiol. 1331397-1407. [DOI] [PubMed] [Google Scholar]
- 11.Coombs, D., G. Huber, J. O. Kessler, and R. E. Goldstein. 2002. Periodic chirality transformations propagating on bacterial flagella. Phys. Rev. Lett. 89118102. [DOI] [PubMed] [Google Scholar]
- 12.Driks, A., R. Bryan, L. Shapiro, and D. J. DeRosier. 1989. The organization of the Caulobacter crescentus flagellar filament. J. Mol. Biol. 206627-636. [DOI] [PubMed] [Google Scholar]
- 13.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21129-228. [DOI] [PubMed] [Google Scholar]
- 14.Fahrner, K. A., S. M. Block, S. Krishnaswamy, J. S. Parkinson, and H. C. Berg. 1994. A mutant hook-associated protein (HAP3) facilitates torsionally induced transformations of the flagellar filament of Escherichia coli. J. Mol. Biol. 238173-186. [DOI] [PubMed] [Google Scholar]
- 15.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, and J. C. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281375-388. [DOI] [PubMed] [Google Scholar]
- 16.Ge, Y., C. Li, L. Corum, C. A. Slaughter, and N. W. Charon. 1998. Structure and expression of the FlaA periplasmic flagellar protein of Borrelia burgdorferi. J. Bacteriol. 1802418-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein, R. E., A. Goriely, G. Huber, and C. W. Wolgemuth. 2000. Bistable helices. Phys. Rev. Lett. 841631-1634. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein, S. F., N. W. Charon, and J. A. Kreiling. 1994. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc. Natl. Acad. Sci. USA 913433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Josenhans, C., A. Labigne, and S. Suerbaum. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 1773010-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy, M. J., E. L. Rosey, and R. J. Yancey, Jr. 1997. Characterization of flaA− and flaB− mutants of Serpulina hyodysenteriae: both flagellin subunits, FlaA and FlaB, are necessary for full motility and intestinal colonization. FEMS Microbiol. Lett. 153119-128. [DOI] [PubMed] [Google Scholar]
- 21.Koopman, M. B., E. Baats, C. J. van Vorstenbosch, B. A. van der Zeijst, and J. G. Kusters. 1992. The periplasmic flagella of Serpulina (Treponema) hyodysenteriae are composed of two sheath proteins and three core proteins. J. Gen. Microbiol. 1382697-2706. [DOI] [PubMed] [Google Scholar]
- 22.Krupski, G., R. Gotz, K. Ober, E. Pleier, and R. Schmitt. 1985. Structure of complex flagellar filaments in Rhizobium meliloti. J. Bacteriol. 162361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landau, L. D., and E. M. Lifshitz. 1986. Theory of elasticity, 3rd ed. Butterworth-Heinenann, Ltd., Oxford, United Kingdom.
- 24.Li, C., L. Corum, D. Morgan, E. L. Rosey, T. B. Stanton, and N. W. Charon. 2000. The spirochete FlaA periplasmic flagellar sheath protein impacts flagellar helicity. J. Bacteriol. 1826698-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, C., A. Motaleb, M. Sal, S. F. Goldstein, and N. W. Charon. 2000. Spirochete periplasmic flagella and motility. J. Mol. Microbiol. Biotechnol. 2345-354. [PubMed] [Google Scholar]
- 26.Limberger, R. J. 2004. The periplasmic flagellum of spirochetes. J. Mol. Microbiol. Biotechnol. 730-40. [DOI] [PubMed] [Google Scholar]
- 27.Luke, C. J., and C. W. Penn. 1995. Identification of a 29-kDa flagellar sheath protein in Helicobacter pylori using a murine monoclonal antibody. Microbiology 141(Pt. 3)597-604. [DOI] [PubMed] [Google Scholar]
- 28.Maruyama, M., G. Lodderstaedt, and R. Schmitt. 1978. Purification and biochemical properties of complex flagella isolated from Rhizobium lupini H13-3. Biochim. Biophys. Acta 535110-124. [DOI] [PubMed] [Google Scholar]
- 29.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motaleb, M. A., L. Corum, J. L. Bono, A. F. Elias, P. Rosa, D. S. Samuels, and N. W. Charon. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. USA 9710899-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motaleb, M. A., M. R. Miller, R. G. Bakker, C. Li, and N. W. Charon. 2007. Isolation and characterization of chemotaxis mutants of the Lyme disease spirochete Borrelia burgdorferi using allelic exchange mutagenesis, flow cytometry, and cell tracking. Methods Enzymol. 422419-437. [DOI] [PubMed] [Google Scholar]
- 32.Murphy, G. E., J. R. Leadbetter, and G. J. Jensen. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 4421062-1064. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, G. E., E. G. Matson, J. R. Leadbetter, H. C. Berg, and G. J. Jensen. 2008. Novel ultrastructures of Treponema primitia and their implications for motility. Mol. Microbiol. 671184-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura, S., Y. Adachi, T. Goto, and Y. Magariyama. 2006. Improvement in motion efficiency of the spirochete Brachyspira pilosicoli in viscous environments. Biophys. J. 903019-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-de-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. de Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 1862164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norris, S. J., N. W. Charon, R. G. Cook, M. D. Fuentes, and R. J. Limberger. 1988. Antigenic relatedness and N-terminal sequence homology define two classes of periplasmic flagellar proteins of Treponema pallidum subsp. pallidum and Treponema phagedenis. J. Bacteriol. 1704072-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paster, B. J., and F. E. Dewhirst. 2000. Phylogenetic foundation of spirochetes. J. Mol. Microbiol. Biotechnol. 2341-344. [PubMed] [Google Scholar]
- 38.Pleier, E., and R. Schmitt. 1991. Expression of two Rhizobium meliloti flagellin genes and their contribution to the complex filament structure. J. Bacteriol. 1732077-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, G. Saint, I., R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422888-893. [DOI] [PubMed] [Google Scholar]
- 40.Rendulic, S., P. Jagtap, A. Rosinus, M. Eppinger, C. Baar, C. Lanz, H. Keller, C. Lambert, K. J. Evans, A. Goesmann, F. Meyer, R. E. Sockett, and S. C. Schuster. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303689-692. [DOI] [PubMed] [Google Scholar]
- 41.Rosey, E. L., M. J. Kennedy, D. K. Petrella, R. G. Ulrich, and R. J. Yancey, Jr. 1995. Inactivation of Serpulina hyodysenteriae flaA1 and flaB1 periplasmic flagellar genes by electroporation-mediated allelic exchange. J. Bacteriol. 1775959-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosey, E. L., M. J. Kennedy, and R. J. Yancey, Jr. 1996. Dual flaA1 flaB1 mutant of Serpulina hyodysenteriae expressing periplasmic flagella is severely attenuated in a murine model of swine dysentery. Infect. Immun. 644154-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruby, J. D., H. Li, H. Kuramitsu, S. J. Norris, S. F. Goldstein, K. F. Buttle, and N. W. Charon. 1997. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J. Bacteriol. 1791628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seshadri, R., G. S. Myers, H. Tettelin, J. A. Eisen, J. F. Heidelberg, R. J. Dodson, T. M. Davidsen, R. T. DeBoy, D. E. Fouts, D. H. Haft, J. Selengut, Q. Ren, L. M. Brinkac, R. Madupu, J. Kolonay, S. A. Durkin, S. C. Daugherty, J. Shetty, A. Shvartsbeyn, E. Gebregeorgis, K. Geer, G. Tsegaye, J. Malek, B. Ayodeji, S. Shatsman, M. P. McLeod, D. Smajs, J. K. Howell, S. Pal, A. Amin, P. Vashisth, T. Z. McNeill, Q. Xiang, E. Sodergren, E. Baca, G. M. Weinstock, S. J. Norris, C. M. Fraser, and I. T. Paulsen. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. USA 1015646-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srigiriraju, S. V., and T. R. Powers. 2005. Continuum model for polymorphism of bacterial flagella. Phys. Rev. Lett. 94248101. [DOI] [PubMed] [Google Scholar]
- 46.Stanton, T. B., E. G. Matson, and S. B. Humphrey. 2001. Brachyspira (Serpulina) hyodysenteriae gyrB mutants and interstrain transfer of coumermycin A1 resistance. Appl. Environ. Microbiol. 672037-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart, P. E., J. Hoff, E. Fischer, J. G. Krum, and P. A. Rosa. 2004. Genome-wide transposon mutagenesis of Borrelia burgdorferi for identification of phenotypic mutants. Appl. Environ. Microbiol. 705973-5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trachtenberg, S., and D. J. DeRosier. 1992. A three-start helical sheath on the flagellar filament of Caulobacter crescentus. J. Bacteriol. 1746198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trachtenberg, S., D. J. DeRosier, and R. M. Macnab. 1987. Three-dimensional structure of the complex flagellar filament of Rhizobium lupini and its relation to the structure of the plain filament. J. Mol. Biol. 195603-620. [DOI] [PubMed] [Google Scholar]
- 50.Trueba, G. A., C. A. Bolin, and R. L. Zuerner. 1992. Characterization of the periplasmic flagellum proteins of Leptospira interrogans. J. Bacteriol. 1744761-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagenknecht, T., R. Grassucci, and J. Frank. 1988. Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. J. Mol. Biol. 199137-147. [DOI] [PubMed] [Google Scholar]
- 52.Wilson, D. R., and T. J. Beveridge. 1993. Bacterial flagellar filaments and their component flagellins. Can. J. Microbiol. 39451-472. [DOI] [PubMed] [Google Scholar]
- 53.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424643-650. [DOI] [PubMed] [Google Scholar]