Abstract
GerD of Bacillus subtilis is a protein essential for normal spore germination with either l-alanine or a mixture of l-asparagine, d-glucose, d-fructose, and potassium ions. GerD's amino acid sequence suggests that it may be a lipoprotein, indicating a likely location in a membrane. Location in the spore's outer membrane seems unlikely, since removal of this membrane does not result in a gerD spore germination phenotype, suggesting that GerD is likely in the spore's inner membrane. In order to localize GerD within spores, FLAG-tagged GerD constructs were made, found to be functional in spore germination, and detected in immunoblots of spore extracts as not only monomers but also dimers and trimers. Upon fractionation of spore extracts, GerD-FLAG was found in the inner membrane fraction from dormant spores and was present at ∼2,000 molecules/spore. GerD-FLAG in the inner membrane fraction was solubilized by Triton X-100, suggesting that GerD is a lipoprotein, and the protein was also solubilized by 0.5 M NaCl. GerD-FLAG was not processed proteolytically in a B. subtilis strain lacking gerF (lgt), which encodes prelipoprotein diacylglycerol transferase (Lgt), indicating that when GerD does not have a diacylglycerol moiety, signal sequence processing does not occur. However, unprocessed GerD-FLAG still gave bands corresponding to monomers and dimers of slightly higher molecular weight than that of GerD-FLAG from a strain with Lgt, further suggesting that GerD is a lipoprotein. Upon spore germination, much GerD became soluble and then appeared to be degraded as the germinated spores outgrew and initiated vegetative growth. All of these results suggest that GerD is a lipoprotein associated with the dormant spore's inner membrane that may be released in some fashion from this membrane upon spore germination.
Spores of Bacillus subtilis are extremely resistant to heat, radiation, desiccation, pH extremes, and toxic chemicals (31). Like many endospore formers, B. subtilis is primarily a soil organism and may be subject to large fluctuations in environmental conditions, in particular nutrient availability. Consequently, spore formation in times of low nutrient availability can be advantageous, as the dormant spore can ride out bad times and, when conditions are favorable, return to growth through spore germination and outgrowth. Germination of B. subtilis spores normally begins with the binding of specific nutrient germinants, either l-alanine or a mixture of l-asparagine, d-glucose, d-fructose, and potassium ions (AGFK), to specific receptors located in the spore's inner membrane (7, 16, 19, 23). Three functional nutrient receptors are present in B. subtilis spores, each encoded by the homologous tricistronic gerA, gerB, and gerK operons. The GerA nutrient receptor responds to l-alanine, while the GerB and GerK nutrient receptors cooperate in some fashion to respond to AGFK. The GerD protein also plays a role in germination, as spores of strains with both point and deletion mutations in gerD germinate poorly with both l-alanine and AGFK and are blocked very early in the spore germination pathway (18, 19, 25).
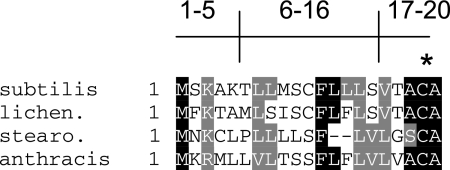
The gerD gene encodes an ∼20-kDa protein with a putative 11-amino-acid signal sequence as well as a likely recognition sequence for diacylglycerol addition to a specific cysteine residue near the protein's amino terminus (Fig. 1) (33, 38). GerD homologs are present throughout the bacilli (Fig. 1), although there is no obvious GerD homolog in Clostridium species. The GerD sequence also does not resemble that of any of the nutrient receptors. Transcription of gerD takes place only in the developing forespore compartment of the sporulating cell and is directed by the forespore-specific RNA polymerase sigma factor, σG, as is transcription of the gerA, gerB, and gerK operons (9, 10, 15, 30). Although components of the GerA and GerB receptors have been localized to the spore's inner membrane, the location of GerD is not yet known (7, 23). One report suggests that GerD is in the spore's integument fraction, i.e., the coat plus the cortex (17). However, the putative lipobox in GerD that appears to be essential for GerD function (25) suggests that this protein likely resides in a membrane.
FIG. 1.
Alignment of amino acid sequences of the amino-terminal regions of GerDs from various species. The sequences shown are from B. subtilis, B. licheniformis, Geobacillus stearothermophilis, and B. anthracis. Gray shading indicates similar residues, and black shading indicates identical residues. Residues 1 to 5 and 6 to 16 correspond to the n region and h region of the signal peptide, respectively, and residues 17 to 20 correspond to the lipobox. The asterisk denotes the invariant cysteine that is the site for diacylglycerol addition.
As noted above, GerD is most likely a lipoprotein that has a diacylglycerol linked to a specific cysteine residue (25, 33, 38) (Fig. 1). This type of lipoprotein has been characterized in the following four ways: (i) by sequence data indicating the presence of a conserved signal sequence and lipobox; (ii) by mutation of the invariant cysteine, the site for diacylglycerol addition, within the lipobox region, leading to abrogation of function; (iii) by globomycin treatment, which inhibits processing by signal peptidase II (Lsp); and (iv) by radiolabeling with fatty acids, usually palmitic acid (33). Typically, these lipoproteins contain a signal peptide sequence followed by a lipobox (28, 29, 32, 33). The signal peptide sequence is characterized by an n domain, consisting of the basic amino acids lysine and arginine, followed by a central hydrophobic h domain and, finally, a cleavage (c) domain. The cleavage domain contains a lipobox characterized by the consensus amino acid sequence Leu-Ala/Ser-Gly/Ala-Cys, with cleavage just prior to the cysteine residue (32, 33). GerD contains all three domains, as well as a similar lipobox in its cleavage domain (Fig. 1), and changing the putative diacylglycerylated cysteine to alanine results in a gerD spore germination phenotype (25). During the maturation of these types of lipoproteins, the diacylglycerol moiety from phosphatidylglycerol is initially transferred to the sulfydryl group of cysteine by a prelipoprotein diacylglycerol transferase (Lgt). The modified lipoprotein precursor is then cleaved by signal peptidase II, Lsp, and after cleavage of the signal peptide, the N-terminal cysteine may be N-acylated (28, 29, 33, 34, 35). Both lgt and lsp genes have been identified in B. subtilis; however, no gene encoding an N-acyltransferase (Lnt) has been found (11, 13, 27).
Since GerD appears to be a lipoprotein and is synthesized in the same compartment and at approximately the same time as the nutrient germinant receptors, it seems most likely that GerD is located in the spore's inner membrane. Indeed, in this work we demonstrate that GerD is a lipoprotein that is associated with the dormant spore's inner membrane. This localization of GerD in the same membrane as the spore's nutrient germinant receptors may assist in our understanding of the role of GerD in the nutrient receptor-mediated spore germination pathway.
MATERIALS AND METHODS
B. subtilis strain construction.
The B. subtilis strains used in this study are listed in Table 1; all are isogenic derivatives of strain PS832, a prototrophic derivative of strain 168. To add the FLAG epitope to the carboxy terminus of the gerD open reading frame (ORF), the gerD coding region, lacking only the translational stop codon and including the gerD promoter region (10, 38), encompassing ∼95 bp upstream of the translational start codon, was PCR amplified from chromosomal DNA of strain PS832 (all primer sequences are available on request). The forward primer, pFLAGF, introduced a HindIII site into the construct. The reverse primer, pFLAGR, introduced a FLAG tag followed by a translational stop codon and then an EcoRV site. After PCR amplification, the fragment was cloned into plasmid PCR2.1-TOPO (Invitrogen, Carlsbad, CA) in Escherichia coli, giving plasmid pPP01. Upon sequencing of the cloned gerD PCR product, we found that while the third codon in the annotated B. subtilis gerD sequence encodes a lysine, in our strain of B. subtilis this lysine had been replaced by an arginine. Arginine is also found in this position in GerDs from Bacillus anthracis, Bacillus licheniformis, and Geobacillus stearothermophilus (Fig. 1), as well as in GerDs from many other strains and species (data not shown). The gerD fragment in plasmid pPP01 was recovered as a HindIII-EcoRV fragment and cloned into plasmid pDG364 that had been linearized with EcoRI, its ends made blunt with T4 DNA polymerase, and then further digested with HindIII, giving plasmid pPP02. Plasmid pPP02 was then used to transform (1) B. subtilis strain FB62, in which the endogenous gerD gene has been replaced by an Spr cassette, as well as strain PS832, resulting in the insertion of the FLAG-tagged gerD gene at the amyE locus (14). Transformants in which gerD-flag was integrated at the amyE locus were identified by their amyE Cmr phenotype by patching them on plates containing starch and chloramphenicol (5).
TABLE 1.
Strains used in this study
| Strain | Genotype | Phenotypea | Source or reference |
|---|---|---|---|
| PS832 | Wild type | Laboratory stock | |
| FB62 | ΔgerD::spc | Spr | 6 |
| PP16 | amyE::gerD-flag | amyE Cmr | This work |
| PP17 | ΔgerD::spc amyE::gerD-flag | amyE Cmr Spr | This work |
| PP18 | amyE::PsspB-gerD-flag | amyE Cmr | This work |
| PP19 | ΔgerD::spc amyE::PsspB-gerD-flag | amyE Cmr Spr | This work |
| PP20 | Δlgt::erm amyE::gerD-flag | amyE Cmr Emr | This work |
| PP21 | ΔgerD::spc Δlgt::erm amyE::gerD-flag | amyE Cmr Emr Spr | This work |
| PP22 | Δlgt::erm amyE::PsspB-gerD-flag | amyE Cmr Emr | This work |
| PP23 | ΔgerD::spc Δlgt::erm amyE::PsspB-gerD-flag | amyE Cmr Emr Spr | This work |
Abbreviations: Cmr, chloramphenicol (5 μg/ml) resistant; Emr, erythromycin (1 μg/ml) resistant; Spr, spectinomycin (100 μg/ml) resistant.
Plasmid pPP01 was used to place gerD-flag under the control of the strong forespore-specific sspB promoter (4, 15), as follows. A fragment from 200 bp upstream of the translational start site of the sspB gene, encompassing the ribosome binding site and promoter to 12 bp downstream of the sspB translational start site, was PCR amplified from PS832 chromosomal DNA. The forward primer, pSspBF, included a HindIII site found within the sspB sequence and a PstI site incorporated into the primer. The reverse primer, pSspBR, had both XhoI and NdeI sites. This construct was digested with PstI and XhoI and cloned into similarly digested plasmid FE47 [pBluescript KS (−)], giving plasmid pPP05. Plasmid pPP01 containing the gerD-flag sequence was amplified with a forward primer, OFLAGF, that included an NdeI site, and a reverse primer, OFLAGR, that included an XhoI site. This PCR fragment was digested with NdeI and XhoI and cloned into similarly digested plasmid pPP05, giving plasmid pPP06. The insert in plasmid pPP06 was sequenced, recovered as a HindIII-EcoRV (an EcoRV site was previously incorporated into pPP01) fragment, and cloned into plasmid pDG364 that had been linearized with EcoRI, its ends made blunt with T4 DNA polymerase, and further digested with HindIII, giving plasmid pPP07. Plasmid pPP07 was used to transform B. subtilis strains FB62 and PS832 to an amyE Cmr phenotype as described above.
Spore preparation, cleaning, and decoating.
Spores were produced at 37°C on 2× SG medium agar plates without antibiotics, and spores were harvested and purified as described previously (20). All spore preparations were >98% free of vegetative or sporulating cells, as determined by phase-contrast microscopy. Spores were decoated by treatment for 30 min at 70°C with 0.15 M NaCl-0.1 N NaOH-0.5% sodium dodecyl sulfate (SDS)-0.1 M dithiothreitol and then washed five to seven times with 1 ml water (36). This decoating procedure removes not only the spore coat but also the spore's outer membrane proteins (3, 23, 37).
Spore germination.
Prior to nutrient germination, spores at an optical density at 600 nm (OD600) of 10 in water were heat activated at 70°C for 30 min and cooled on ice. Germination was done routinely at 37°C with spores at an initial OD600 of 1 in 25 mM Tris-HCl (pH 8.4) with either 10 mM l-alanine or AGFK, a mixture of l-asparagine (2.5 mM), d-glucose (5 mg/ml), d-fructose (5 mg/ml), and KCl (50 mM). Spore germination was measured by monitoring the OD600 of spore cultures, which falls ∼60% upon complete spore germination (2, 4). The extent of spore germination was also confirmed by phase-contrast microscopy. The rate of spore germination was determined from the maximum rate of fall in the OD600 of cultures, as determined from the maximum slope of a plot of OD600 versus time (2, 4).
Preparation and fractionation of dormant spore extracts.
Ten milligrams (dry weight) of spores was decoated in 1 ml, washed as described above, and lyophilized. The dry decoated spores were pulverized with 100 mg of glass beads in a dental amalgamator (Wig-L-Bug) at high speed for 20 pulses of 30 s each, with 30-s pauses between pulses (23). Once >80% of the spores were disrupted, as seen by phase-contrast microscopy, the dry powder was suspended in 0.25 to 0.5 ml of 4°C extraction buffer (containing the following [per ml]: 2 μg pancreatic RNase, 2 μg bovine DNase I in 10 mM MgCl2, 50 mM Tris-HCl [pH 7.4], and 1 mM phenylmethylsulfonyl fluoride) (23, 37). After allowing the glass beads to settle for 1 min on ice, the mix was centrifuged at 4°C for 10 min at 14,000 × g. The insoluble fraction was suspended in 0.25 to 0.5 ml extraction buffer and frozen, as was a small amount of supernatant fluid. The remaining supernatant fluid was further centrifuged at 4°C for 1 h at 100,000 × g, giving a pelleted inner membrane fraction (P100) and a soluble fraction (S100) (23, 37). The P100 fraction was suspended by brief sonication in 0.25 to 0.5 ml extraction buffer, and all fractions were mixed 1:1 with 2× SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (12) and boiled for 5 min prior to SDS-PAGE and Western blot analysis (see below).
In some experiments, spores were lyophilized, disrupted, and extracted as described above, and Triton X-100 was added to 1% or NaCl was added to 0.5 M to either the suspended P100 or initial insoluble fraction that had been suspended by brief sonication in 0.5 ml extraction buffer. Fractions were incubated at 4°C for 30 min and then centrifuged at 100,000 × g for 1 h as described above.
Preparation of germinated spore extracts.
For preparation of extracts from germinating spores, 1-ml aliquots of heat-activated spores (OD600 of 100) were incubated at 37°C in 60 ml of Luria-Bertani (LB) medium (14) plus 10 mM l-alanine. Spores were harvested at various times by centrifugation at 4°C for 15 min at 10,000 × g and washed five to seven times with 1 ml of water, and the pellet was suspended in 800 μl of 20% Nycodenz (Sigma-Aldrich, St. Louis, MO). Aliquots of ∼200 μl of the 20% Nycodenz suspension were layered over 2 ml of 50% Nycodenz in ultracentrifuge tubes, and the tubes were centrifuged at 20°C at 100,000 × g for 45 min. The germinated spores, which float because of their low wet density, were isolated and washed several times with water by centrifugation in a microcentrifuge to remove Nycodenz. The pelleted germinated spores were then lyophilized, disrupted, extracted, and fractionated as described above.
Western blot analysis.
Samples boiled in SDS-PAGE loading buffer were run by SDS-PAGE (12% acrylamide) (12), and proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA) (6). Western blots were probed with a 1:1,000 dilution of anti-FLAG M2 monoclonal antibody (Sigma-Aldrich, St. Louis, MO) overnight at room temperature and then with a 1:5,000 dilution of goat anti-mouse immunoglobulin G-alkaline phosphatase conjugate (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at room temperature, each in 1× Tris-buffered saline (100 mM Tris-HCl [pH 7.5], 150 mM NaCl) with 2% blocking reagent (Roche, Indianapolis, IN) and 0.1% Tween added as described previously (6). Alkaline phosphatase activity was detected on X-ray film by using the chemiluminescent substrate disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro) tricyclo [3.3.1.1.3,7] decan}-4-yl) phenyl phosphate (Roche, Indianapolis, IN). Each SDS-PAGE gel and Western blot also included a sample of a carboxy-terminal FLAG-BAP fusion protein (Sigma-Aldrich, St. Louis, MO) as a positive control and 5 μl of Precision Plus protein standards (Bio-Rad, Hercules, CA) as molecular weight markers.
RESULTS
GerD-FLAG fusion functionality.
To localize GerD within the B. subtilis spore, we generated FLAG-tagged versions of gerD that would allow us to detect GerD-FLAG by Western blot analysis. Two gerD-flag strains were generated, including strain PP17 (gerD amyE::gerD-flag), in which gerD-flag was expressed from the gerD promoter (10), and PP19 (gerD amyE::PsspB-gerD-flag), in which gerD-flag was under the control of the strong forespore-specific sspB promoter (4, 15); both strains also lacked wild-type gerD. To test the functionality of GerD-FLAG, the germination of spores of these two strains with l-alanine and AGFK was measured (Table 2). Since spores of both strains germinated as well as wild-type spores, and much better than spores lacking GerD, GerD-FLAG appeared to be functional.
TABLE 2.
Germination of spores with GerD-FLAGa
| Strain | Genotype | Rate of spore germination (%/h) with:
|
|
|---|---|---|---|
| l-Alanine | AGFK | ||
| PS832 | Wild type | 84 | 61 |
| FB62 | gerD | <5 | <5 |
| PP17 | gerD amyE::gerD-flag | 79 | 59 |
| PP19 | gerD amyE::PsspB-gerD-flag | 78 | 56 |
| PP21 | gerD lgt amyE::gerD-flag | <5 | 10 |
| PP23 | gerD lgt amyE::PsspB-gerD-flag | <5 | 13 |
Spore germination was initiated by addition of either l-alanine or AGFK, germination was monitored by measuring the OD600 of cultures, and rates of germination were calculated as described in Materials and Methods.
Detection of GerD-FLAG in spores.
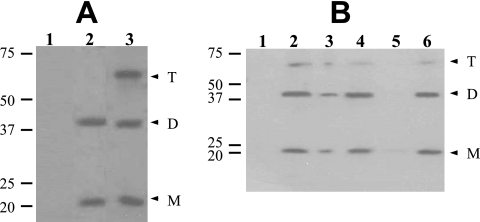
To demonstrate that GerD-FLAG could be detected in spores, spores of strains PP17 (gerD amyE::gerD-flag), PP19 (gerD amyE::PsspB-gerD-flag), and PS832 (wild type) were decoated, disrupted, and extracted, and samples were analyzed by Western blotting with anti-FLAG antibody (Fig. 2A). GerD-FLAG was readily detected in the total extracts of PP17 and PP19 spores, while there was no FLAG signal in extracts from PS832 spores. Thus, we could specifically detect GerD-FLAG in spores. Interestingly, not only were there immunoreactive bands at the expected molecular size of the GerD-FLAG monomer (∼21 kDa) from both PP17 and PP19 spores, but bands were also seen at the molecular sizes of GerD-FLAG dimers and, in PP19 spores, also trimers, although higher GerD-FLAG oligomers were not detected (Fig. 2A, lanes 2 and 3). Most subsequent experiments in this work used spores of strain PP19, in which GerD-FLAG was overexpressed. While GerD-FLAG expressed from the gerD promoter behaved in a similar manner (data not shown), the overexpressed protein was easier to detect.
FIG. 2.
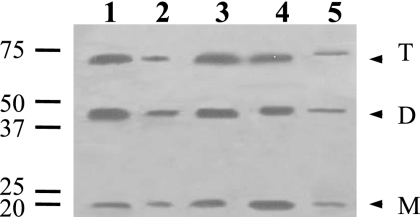
Detection and localization of GerD-FLAG in spores. (A) Dormant spores of strains PS832 (wild type), PP17 (gerD amyE::gerD-flag), and PP19 (gerD amyE::PsspB-gerD-flag) were disrupted and extracted, aliquots of the total extracts were run by SDS-PAGE, proteins were transferred to a PVDF membrane, and GerD-FLAG was detected with anti-FLAG antibody as described in Materials and Methods. Samples run in the various lanes were from spores of PS832 (1), PP17 (2), and PP19 (3). The numbered lines to the left of the figure denote the migration positions of molecular mass markers, in kDa, and the arrowheads on the right indicate the predicted migration positions of GerD-FLAG monomers (M), dimers (D), and trimers (T). Twice as much extract from PP17 spores was run as the amount of extract from PS832 and PP19 spores. Note that although aliquots with twice the amount of extract from spores of strain PP17 were run in this experiment, the bands from the strains appear to exhibit approximately the same intensity. However, in this experiment, the amount of extract analyzed was very high, probably saturating the assay. (B) Localization of GerD-FLAG in spores. Spores of strain PP19 (gerD amyE::PsspB-gerD-flag) were disrupted, extracted, and fractionated, aliquots of fractions from equal amounts of spores were run by SDS-PAGE, proteins were transferred to a PVDF membrane, and GerD-FLAG was detected as described in Materials and Methods. Samples run in the various lanes were from the following fractions: 1, coat/outer membrane; 2, total extract; 3, insoluble; 4, soluble; 5, S100; and 6, P100. The numbered lines to the left of the figure denote the migration positions of molecular mass markers, in kDa, and the arrowheads on the right indicate the predicted migration positions of GerD-FLAG monomers (M), dimers (D), and trimers (T).
GerD-FLAG localizes to the spore's inner membrane.
Since GerD-FLAG was functional and could readily be detected, this protein could be used to determine where GerD is located in the spore. Spores from strain PP19 (gerD amyE::PsspB-gerD-flag) were lyophilized, disrupted, extracted, and fractionated, fractions were run by SDS-PAGE, and GerD-FLAG was detected by Western blot analysis. No GerD-FLAG was detected in the coat/outer membrane fraction (Fig. 2B, lane 1). However, GerD-FLAG was again readily detected in the total extract, migrating at a position corresponding to the predicted monomer size as well as at sizes of GerD-FLAG dimers and trimers (Fig. 2B, lane 2). While some GerD-FLAG did pellet upon low-speed centrifugation (Fig. 2B, lane 3), the majority remained soluble (Fig. 2B, lane 4). The small amount of protein that remained insoluble could be due to a small percentage of spores that were not fully broken during disruption or to protein trapped in the large pellet fraction. However, almost all GerD-FLAG in the low-speed supernatant fluid pelleted during high-speed centrifugation (Fig. 2B, lanes 5 and 6). These results strongly suggest that GerD is located in the spore's inner membrane.
Solubilization of GerD-FLAG from the spore's inner membrane.
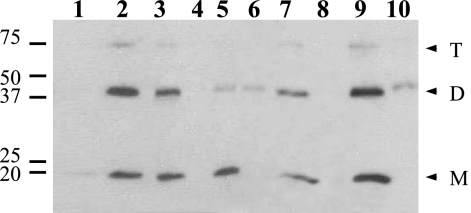
To further characterize the location of GerD-FLAG in the spore, the effects of both Triton X-100 and high salt on GerD-FLAG in the P100 fraction and the insoluble fraction from the total extract of spores of strain PP19 (gerD amyE::PsspB-gerD-flag) were assessed (Fig. 3). Treatment of the P100 fraction with Triton X-100 moved the majority of GerD-FLAG into the S100 fraction (Fig. 3, lanes 3 and 4). Treatment of the initial insoluble fraction with Triton X-100 also moved the great majority of the GerD-FLAG into the S100 fraction (Fig. 3, lanes 5 and 6). Treatment of the initial P100 fraction with 0.5 M NaCl also moved the GerD-FLAG into the S100 fraction (Fig. 3, lanes 7 and 8), as did treatment of the initial insoluble fraction with 0.5 M NaCl, although at least some of the GerD-FLAG dimer remained insoluble (Fig. 3, lanes 9 and 10).
FIG. 3.
Solubilization of GerD-FLAG from insoluble and P100 fractions. Spores of strain PP19 (gerD amyE::PsspB-gerD-flag) were disrupted, extracted, and fractionated, and fractions were treated with either 1% Triton X-100 or 0.5 M NaCl as described in Materials and Methods. Aliquots of fractions from equal amounts of spores were run by SDS-PAGE, proteins were transferred to a PVDF membrane, and GerD-FLAG was detected with anti-FLAG antibody. Samples run in the various lanes were as follows: 1, initial S100; 2, initial P100; 3, S100 from initial P100 plus Triton X-100; 4, P100 from initial P100 plus Triton X-100; 5, S100 from initial insoluble fraction plus Triton X-100; 6, P100 from initial insoluble fraction plus Triton X-100; 7, S100 from initial P100 plus NaCl; 8, P100 from initial P100 plus NaCl; 9, S100 from initial insoluble fraction plus NaCl; and 10, P100 from initial insoluble fraction plus NaCl. The numbered lines to the left of the figure denote the migration positions of molecular mass markers, in kDa, and the arrowheads to the right indicate the predicted migration positions of GerD-FLAG monomers (M), dimers (D), and trimers (T). Lanes 1 to 6 and 7 to 10 came from different gels.
The movement of GerD from the inner membrane to the soluble fraction upon the addition of Triton X-100 is certainly consistent with GerD being a lipoprotein. Although the primary sequence of GerD does not show any potential hydrophobic membrane-spanning regions, GerD likely contains a diacylglycerylated cysteine residue, as noted above, and this could anchor the protein to the inner membrane. The solubilization of GerD-FLAG from the inner membrane by 0.5 M NaCl further suggests that GerD-FLAG is not an integral membrane protein and is likely anchored to the inner membrane only by the diacylglycerol moiety. However, there are no data that we are aware of describing the behavior of membrane lipoproteins upon high-salt treatment.
GerD abundance in spores.
Previous work has shown that there are 24 to 40 molecules of the GerBA subunit of the GerB nutrient receptor per spore (23). If GerD does interact with the nutrient receptors, it would be helpful to also know the number of GerD molecules in spores. To estimate this number, the GerD-FLAG signals from different amounts of total extract from spores of strain PP17 (gerD amyE::gerD-flag) were compared to those from different amounts of a FLAG-BAP fusion protein (data not shown). This analysis indicated that there are ∼2,000 molecules of GerD per spore. We then compared the GerD-FLAG level in spores of strain PP19 (gerD amyE::PsspB-gerD-flag) to that in PP17 spores, and this comparison indicated that the level of GerD-FLAG overexpression from the sspB promoter is ∼20-fold, giving ∼40,000 molecules in each spore overexpressing GerD-FLAG.
Processing of GerD in lgt mutant B. subtilis spores.
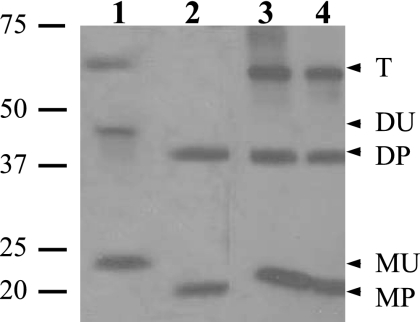
As noted above, B. subtilis GerD contains the signal sequence and lipobox characteristic of bacterial lipoproteins, and mutation of the invariant cysteine to an alanine inactivates GerD (25). One way of further characterizing a bacterial lipoprotein is by the addition of the antibiotic globomycin, which inhibits Lsp, thus preventing signal peptide processing. However, it is not clear that globomycin would enter the developing spore and inhibit Lsp during B. subtilis sporulation. Therefore, we chose to analyze GerD in spores of a strain with a deletion of the lgt (gerF) gene. Since lgt encodes the only prelipoprotein diacylglycerol transferase of B. subtilis, the diacylglycerylation of GerD will not take place in an lgt strain. Consequently, signal peptidase cleavage of the unmodified protein will not take place, and this should result in a GerD-FLAG protein that is ∼2 kDa larger than the fully processed protein. Indeed, GerD-FLAG in extracts from lgt spores was ∼2 kDa larger than GerD-FLAG from spores that contained Lgt (Fig. 4). However, with spores of strains that contained Lgt, even overexpressed GerD-FLAG ran at the lower molecular weights of the processed monomer, dimer, and trimer, as did GerD-FLAG from germinated spores with overexpressed GerD-FLAG (Fig. 4, lanes 3 and 4). This indicates that during sporulation, Lgt and Lsp are able to fully process even overexpressed GerD-FLAG. It was notable, however, that with lgt spores only overexpressed but unprocessed GerD-FLAG was detected, as the lower level of GerD-FLAG expressed from the gerD promoter was not detected in lgt spores (data not shown). However, whether the absence of GerD-FLAG from the latter spores was due to GerD-FLAG degradation or simply the removal of the FLAG tag is not clear. Analysis of the germination of spores that lacked Lgt with either wild-type or overexpressed levels of GerD-FLAG indicated that these spores did not germinate with l-alanine, although they did germinate slowly with AGFK (Table 2; see Discussion).
FIG. 4.
Processing of GerD-FLAG in spores that lack Lgt. Spores of strains PP23 (gerD lgt amyE::PsspB-gerD-flag), PP17 (gerD amyE::gerD-flag), and PP19 (gerD amyE::PsspB-gerD-flag) were disrupted, and total extracts were prepared as described in Materials and Methods. Spores of strain PP19 were also germinated for 45 min, harvested, purified, lyophilized, disrupted, and extracted as described in Materials and Methods. Aliquots of extracts from equal amounts of spores (except for PP17 samples, where twice as much was run) were run by SDS-PAGE, proteins were transferred to a PVDF membrane, and GerD-FLAG was detected with anti-FLAG antibody. Samples of total extract run in the various lanes were from spores of strains PP23 (dormant) (1), PP17 (dormant) (2), PP19 (germinated) (3), and PP19 (dormant) (4). The numbered lines to the left of the figure denote the migration positions of molecular mass markers, in kDa, and the arrowheads to the right indicate the predicted migration positions of both processed (P) and unprocessed (U) GerD-FLAG monomers (M), dimers (D), and trimers (T). Note that lanes 1 and 2 and lanes 3 and 4 are from different gels and that although extract from twice the amount of spores from strain PP17 was run in this experiment, the bands from the strains appear to exhibit approximately the same intensity. However, in this experiment, the amount of extract analyzed was very high, probably saturating the assay. Also note the gel artifact in lane 3 at the monomer weight (the band curves up slightly).
GerD is largely soluble in germinated/outgrowing spores.
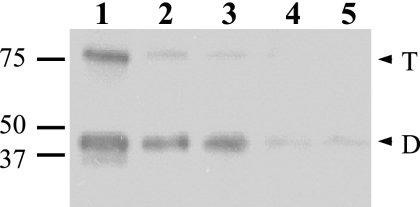
While GerD-FLAG in spore extracts prepared by mechanical disruption of spores was largely in the inner membrane fraction, in extracts prepared by lysozyme lysis of decoated spores most GerD-FLAG was present in the soluble (S100) fraction (data not shown). Since lysozyme lysis of spores also mimics and can even induce spore germination (30), we further examined the location of GerD-FLAG in germinated spores. Strikingly, after incubation of spores with l-alanine for 45 min, removal of dormant spores, and extraction of the purified spores, followed by fractionation and Western blot analysis, very little GerD-FLAG was in the P100 fraction from the germinated spores, as the majority of the protein was in the S100 fraction (Fig. 5). These results suggest that the location of GerD-FLAG, and thus by inference GerD itself, may change upon spore germination. However, the size of GerD-FLAG in the S100 fraction from germinated spores appeared to be identical to that of GerD-FLAG in the P100 fraction (Fig. 5).
FIG. 5.
Localization of GerD in germinated spores. Spores of strain PP19 (gerD amyE::PsspB-gerD-flag) were germinated in LB medium plus 10 mM l-alanine for 45 min, and germinated spores were purified as described in Materials and Methods. The germinated spores were lyophilized, disrupted, extracted, and fractionated as described in Materials and Methods. Aliquots of fractions from equal amounts of spores were run by SDS-PAGE, proteins were transferred to a PVDF membrane, and GerD-FLAG was detected with anti-FLAG antibody. Samples run in the various lanes were as follows: 1, total extract; 2, insoluble fraction; 3, soluble fraction; 4, S100; and 5, P100. The numbered lines to the left of the figure denote the migration positions of molecular mass markers, in kDa, and the arrowheads to the right indicate the predicted migration positions of GerD-FLAG monomers (M), dimers (D), and trimers (T).
Fate of GerD-FLAG following spore germination.
The previous experiment examined GerD-FLAG in extracts from spores incubated in germination medium for 45 min. Since GerD is likely not needed for spore outgrowth and vegetative growth, it seemed worthwhile to follow the fate of GerD-FLAG over an even longer period, in particular when the majority of the spores had gone through the first cell division. Strikingly, the majority of GerD-FLAG disappeared from spores after 3 h of incubation in germination/growth medium (Fig. 6). In this medium, the majority of cells were dividing by 2 h (data not shown). The disappearance of GerD-FLAG was not an artifact of culture dilution, as samples from the same culture volume were used for Western blot analysis of samples taken at each time point, suggesting that either GerD was being degraded or the FLAG tag was being removed. Again, there were no changes in the sizes of the GerD-FLAG oligomers as spore outgrowth and vegetative growth proceeded, and no immunoreactive bands smaller than that of GerD-FLAG dimers were seen in this experiment (Fig. 6 and data not shown).
FIG. 6.
Fate of GerD-FLAG following spore germination. Spores of strain PP19 (gerD amyE::PsspB-gerD-flag) were incubated in LB medium plus 10 mM l-alanine for 0 (lane 1), 0.75 (lane 2), 1 (lane 3), 2 (lane 4), 2.5 (lane 5), and 3 h (lane 6), and any dormant spores were removed as described in Materials and Methods. The purified germinated spores and/or cells were lyophilized, disrupted, and extracted, aliquots of total extracts from equal amounts of starting dormant spores were run by SDS-PAGE, proteins were transferred to a PVDF membrane, and GerD-FLAG was detected with anti-FLAG antibody. The numbered lines to the left of the figure present the migration positions of molecular mass markers, in kDa, and the arrowheads to the right indicate the predicted migration positions of GerD-FLAG dimers (D) and trimers (T).
DISCUSSION
The gerD gene encodes an ∼20-kDa protein containing a likely signal sequence, a lipobox, and an invariant cysteine to which lipid is probably added. These features, together with the knowledge that gerD transcription (10) is under the control of σG, a forespore-specific sigma factor, suggest that GerD is located in the spore's inner membrane. Indeed, GerD-FLAG was associated with the dormant spore's inner membrane. The solubilization of GerD-FLAG from the inner membrane by Triton X-100 suggests that GerD is indeed a lipoprotein. The solubilization by high salt further suggests that GerD is not an integral membrane protein, consistent with the relatively hydrophilic character of GerD, except for its signal peptide region (38).
The analysis of GerD processing in wild-type and lgt spores further strengthens the argument that GerD is a lipoprotein. B. subtilis contains only one prelipoprotein diacylglycerol transferase, Lgt, and thus when Lgt is not present, diacylglycerol cannot be added to GerD's Cys20 and Lsp will not cleave the protein just prior to this modified cysteine. The unprocessed lipoprotein will be ∼2 kDa larger than the processed form, as observed for overexpressed GerD-FLAG in lgt spores. Interestingly, when GerD-FLAG was expressed at wild-type levels in lgt spores, the protein was not detected and spores exhibited a germination phenotype similar to that of spores of a gerD deletion mutant, suggesting that this lgt phenotype may be due to the absence of GerD for some reason, as opposed to the lack of Lgt. However, spores of strain PP23 (gerD lgt amyE::PsspB::gerD-flag), in which GerD is overexpressed and readily detected, had a germination phenotype with both l-alanine and AGFK that was similar to that of lgt spores in which l-alanine germination is almost eliminated and the rate of AGFK germination is decreased 5- to 10-fold (8). It is difficult, however, to determine definitively whether the germination phenotype of lgt spores containing high levels of GerD-FLAG is due to the lack of diacylglycerylation of this protein or of the C protein components of the nutrient receptors, since the latter are also almost certainly lipoproteins (8). Indeed, replacement of the likely diacylglycerylated cysteine residue in the C proteins of the nutrient receptors with alanine reduces the rate of spore germination with l-alanine ≥98%, although it reduces the rate of AGFK germination only 5- to 10-fold (8). Consequently, the low but significant rate of AGFK germination of lgt spores with GerD-FLAG suggests that unprocessed GerD-FLAG may retain some function, since the rate of AGFK germination is reduced >95% by a gerD deletion mutation (25). However, the differences seen in rates of germination with AGFK in these experiments are relatively small.
While GerD does appear to be a lipoprotein located in the spore's inner membrane, this location appears to change upon spore germination, as most GerD appeared to be released from the inner membrane and then probably degraded. It is unclear whether release from the inner membrane is due to lipid cleavage or cleavage of the protein after the diacylglycerylated cysteine moiety. The migration positions in SDS-PAGE gels appeared to be essentially identical for GerD-FLAG in both the S100 and P100 fractions of dormant and germinated spores. However, removal of the diacylglycerol moiety alone or even with the modified cysteine residue would have only a minimal effect on the protein's molecular weight, and there might also be anomalous migration of these protein forms due to the diacylglycerol moiety. Lipoprotein release from membranes is not unheard of for gram-positive organisms (21, 22, 24, 26). Indeed, β-lactamase III of Bacillus cereus 569 exists in both membrane-bound and secreted forms (22). The membrane-bound form is a glyceride-cysteine lipoprotein, whereas the secreted form arises from the cleavage of the lipoprotein on the carboxyl side of the cysteine (22). Several other lipoproteins are also released from membranes, some in forms that retain the lipid modification, and these may well be functional (24, 26). It is, however, formally possible that the GerD-FLAG is actually not released from the inner membrane upon spore germination but that the germinated spore's inner membrane itself is more readily fragmented into such small pieces that they pellet poorly in the high-speed centrifugation step used to isolate the inner membrane. However, this was not the case when the location of the GerBA protein, a likely integral inner membrane protein, was determined in B. subtilis spores, as this protein remained in the inner membrane fraction in extracts from germinated spores (23). It is also possible that the reason for GerD-FLAG release from the inner membrane upon spore germination is that GerD is tightly associated with some other inner membrane protein, and it is this other protein that is released upon spore germination and brings GerD with it. However, neither the identity nor even the existence of any protein interacting with GerD has been established.
Spores lacking GerD are blocked prior to Ca2+-dipicolinic acid release during germination, a very early event in this process, suggesting that GerD is needed only early in germination. The release of GerD from the spore's inner membrane early in germination, coupled with the degradation of GerD during spore outgrowth and subsequent vegetative growth, suggests that GerD release from the inner membrane could be a mechanism to either shut off GerD function or effect removal of a protein that is no longer needed. However, we do not know if the degradation of GerD that may be occurring following spore germination is specific. It is also interesting that during spore germination no smaller processed forms of GerD were observed. This may be because any initial GerD cleavage is rate limiting in GerD degradation or because the FLAG sequence itself is being removed.
The fact that we found higher-molecular-weight forms of GerD on Western blots from SDS-PAGE gels run under reducing conditions suggests that GerD may function as a multimer, although the relevance of multimerization to GerD function is unknown, since we do not yet know precisely what GerD does. Interestingly, throughout our work we did not always see GerD-FLAG monomers, dimers, and trimers. Sometimes we saw monomers and dimers, and sometimes we saw dimers and trimers. Possible reasons for those differences, such as various amounts of GerD-FLAG present in extracts or slight differences in preparation of samples for SDS-PAGE, have not been investigated in detail but may be worth further study.
Our work has shown that there are ∼2,000 molecules of GerD per spore, compared to only 24 to 40 molecules of the GerBA subunit of the GerB receptor (23). However, another group of molecules involved in germination, the SpoVA proteins that may be involved in Ca-dipicolinic acid release, are present at ∼10,000 molecules per spore (37). One possible model for flow of information in spore germination is that GerD amplifies a signal received from the germinant receptors and then sends this amplified signal to downstream components of the germination pathways, such as SpoVA proteins. If this is indeed the case, then it would be appropriate for these different components to be present in increasing amounts to facilitate signal amplification. The major goal now is to determine how communication occurs between these germination components. Is communication direct or indirect, and how might GerD function, whatever that is, be regulated? These are certainly major challenges for future work.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM-19698) (to P.S.).
Footnotes
Published ahead of print on 13 June 2008.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atluri, S., K. Ragkousi, D. E. Cortezzo, and P. Setlow. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 18828-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan, C. E., and S. L. Neyman. 1986. Correlation of penicillin-binding protein composition with different functions of two membranes in Bacillus subtilis forespores. J. Bacteriol. 165498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabrera-Martinez, R.-M., F. Tovar-Rojo, V. R. Vepachedu, and P. Setlow. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 1852457-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 6.Harlow, E., and D. Lane. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 7.Hudson, K. D., B. M. Corfe, E. H. Kemp, I. M. Feavers, P. J. Coote, and A. Moir. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol. 1834317-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Igarashi, T., B. Setlow, M. Paidhungat, and P. Setlow. 2004. Effects of a gerF mutation on the germination of spores of Bacillus subtilis. J. Bacteriol. 1862984-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igarashi, T., and P. Setlow. 2006. Transcription of the Bacillus subtilis gerK operon encoding a spore germinant receptor and comparison with that of operons encoding other germinant receptors. J. Bacteriol. 1884131-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemp, E. H., R. L. Sammons, A. Moir, D. Sun, and P. Setlow. 1991. Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J. Bacteriol. 1734646-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390249-256. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 13.Leskelä, S., E. Wahlström, V. P. Kontinen, and M. Sarvas. 1999. Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the lgt gene. Mol. Microbiol. 311075-1085. [DOI] [PubMed] [Google Scholar]
- 14.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 15.Mason, J. M., R. H. Hackett, and P. Setlow. 1988. Studies on the regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores using lacZ gene fusions. J. Bacteriol. 170239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moir, A., B. M. Corfe, and J. Behravan. 2002. Spore germination. Cell. Mol. Life Sci. 59403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moir, A., E. H. Kemp, C. Robinson, and B. M. Corfe. 1994. The genetic analysis of bacterial spore germination. J. Appl. Bacteriol. 769S-16S. [PubMed] [Google Scholar]
- 18.Moir, A., E. Lafferty, and D. A. Smith. 1979. Genetic analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype with map location. J. Gen. Microbiol. 111165-180. [DOI] [PubMed] [Google Scholar]
- 19.Moir, A., and D. A. Smith. 1990. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44531-553. [DOI] [PubMed] [Google Scholar]
- 20.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 21.Nielsen, J. B., and J. O. Lampen. 1982. Glyceride-cysteine lipoproteins and secretion by gram-positive bacteria. J. Bacteriol. 152315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen, J. B., and J. O. Lampen. 1983. Beta-lactamase III of Bacillus cereus 569: membrane lipoprotein and secreted protein. Biochemistry 224652-4656. [DOI] [PubMed] [Google Scholar]
- 23.Paidhungat, M., and P. Setlow. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 1833982-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearce, B. J., A. M. Naughton, and H. R. Masure. 1994. Peptide permeases modulate transformation in Streptococcus pneumoniae. Mol. Microbiol. 12881-892. [DOI] [PubMed] [Google Scholar]
- 25.Pelczar, P. L., T. Igarashi, B. Setlow, and P. Setlow. 2007. Role of GerD in germination of Bacillus subtilis spores. J. Bacteriol. 1891090-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perego, M., C. F. Higgins, S. R. Pearce, M. P. Gallagher, and J. A. Hoch. 1991. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol. Microbiol. 5173-185. [DOI] [PubMed] [Google Scholar]
- 27.Pragai, Z., H. Tjalsma, A. Bolhuis, J. M. van Dijl, G. Venema, and S. Bron. 1997. The signal peptidase II (lsp) gene of Bacillus subtilis. Microbiology 1431327-1333. [DOI] [PubMed] [Google Scholar]
- 28.Sankaran, K., S. D. Gupta, and H. C. Wu. 1995. Modification of bacterial lipoproteins. Methods Enzymol. 250683-697. [DOI] [PubMed] [Google Scholar]
- 29.Sankaran, K., and H. C. Wu. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 26919701-19706. [PubMed] [Google Scholar]
- 30.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6550-556. [DOI] [PubMed] [Google Scholar]
- 31.Setlow, P. 2006. Spores of Bacillus subtilis: their resistance to radiation, heat and chemicals. J. Appl. Microbiol. 101514-525. [DOI] [PubMed] [Google Scholar]
- 32.Sutcliffe, I. C., and D. J. Harrington. 2002. Pattern searches for the identification of putative lipoprotein genes in gram-positive bacterial genomes. Microbiology 1482065-2077. [DOI] [PubMed] [Google Scholar]
- 33.Sutcliffe, I. C., and R. R. B. Russell. 1995. Lipoproteins of the gram-positive bacteria. J. Bacteriol. 1771123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tjalsma, H., V. S. Kontinen, Z. Pragai, H. Wu, R. Meima, G. Venema, S. Bron, M. Sarvas, and J. M. van Dijl. 1999. The role of lipoprotein processing by the signal peptidase II in the gram-positive eubacterium Bacillus subtilis. J. Biol. Chem. 2741698-1707. [DOI] [PubMed] [Google Scholar]
- 35.Tokunaga, M., H. Tokunaga, and H. C. Wu. 1982. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc. Natl. Acad. Sci. USA 792255-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vary, J. C. 1973. Germination of Bacillus megaterium spores after various extraction procedures. J. Bacteriol. 116797-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vepachedu, V. R., and P. Setlow. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J. Bacteriol. 1875677-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yon, J. R., R. L. Sammons, and D. A. Smith. 1989. Cloning and sequencing of the gerD gene of Bacillus subtilis. J. Gen. Microbiol. 1353434-3445. [DOI] [PubMed] [Google Scholar]