Abstract
Wall teichoic acids are anionic phosphate-rich polymers that are part of the complex meshwork of carbohydrates that make up the gram-positive cell wall. These polymers are essential to the proper rod-shaped morphology of Bacillus subtilis and have been shown to be an important virulence determinant in the nosocomial opportunistic pathogen Staphylococcus aureus. Together, sequence-based studies, in vitro experiments with biosynthetic proteins, and analyses of the chemical structure of wall teichoic acid have begun to shed considerable light on our understanding of the biogenesis of this polymer. Nevertheless, some paradoxes remain unresolved. One of these involves a putative duplication of genes linked to CDP-ribitol synthesis (tarI′J′ and tarIJ) as well as poly(ribitol phosphate) polymerization (tarK and tarL) in S. aureus. In the work reported here, we performed careful studies of the dispensability of each gene and discovered a functional redundancy in the duplicated gene clusters. We were able to create mutants in either of the putative ribitol phosphate polymerases (encoded by tarK and tarL) without affecting teichoic acid levels in the S. aureus cell wall. Although genes linked to CDP-ribitol synthesis are also duplicated, a null mutant in only one of these (tarI′J′) could be obtained, while tarIJ remained essential. Suppression analysis of the tarIJ null mutant indicated that the mechanism of dysfunction in tarI′J′ is due to poor translation of the TarJ′ enzyme, which catalyzes the rate-limiting step in CDP-ribitol formation. This work provides new insights into understanding the complex synthetic steps of the ribitol phosphate polymer in S. aureus and has implications on specifically targeting enzymes involved in polymer biosynthesis for antimicrobial design.
Carbohydrates are ubiquitously found on the exterior of living cells, from glycoproteins and glycolipids on eukaryotic cells to capsular polysaccharides, teichoic acid, lipoteichoic acid, and lipopolysaccharide on prokaryotic bacteria (3, 23-25). Cell wall teichoic acids are a chemically diverse group of anionic phosphate-rich polymers that are covalently linked to peptidoglycan and are found only in gram-positive organisms. Wall teichoic acids account for up to 60% of the gram-positive cell wall dry weight (13), but the function of these polymers has yet to be explained. Regardless of its function, teichoic acid synthesis has increasingly been implicated as a reasonable antibacterial target. Wall teichoic acid has been shown to be a critical shape determinant in Bacillus subtilis and a factor in the virulence of Staphylococcus aureus (10, 11, 31, 32).
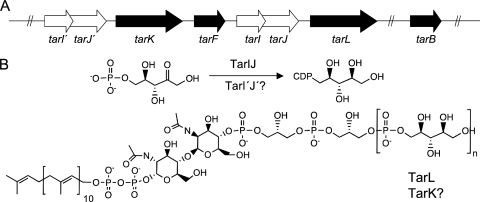
Cell wall teichoic acid polymers often consist of repeats of glycerol phosphate or ribitol phosphate linked through a phosphodiester bond from the 1 position carbon to the terminal phosphate (24). While the model gram-positive strain B. subtilis 168 has a poly(glycerol phosphate) polymer, both B. subtilis W23 and S. aureus have a poly(ribitol phosphate) teichoic acid that is attached via a linkage unit to the 6′ position of N-acetyl muramic acid on peptidoglycan. Biochemical and genetic studies suggest that the biosynthesis of cell wall teichoic acid in S. aureus commences with the creation of a disaccharide of N-acetylglucosamine-1-phosphate and N-acetylmannosamine on the membrane-bound isoprenoid undecaprenyl-phosphate by the concerted action of the enzymes TarO and TarA. Subsequently, the enzymes TarB and TarF catalyze the addition of the first and a subsequent residue of glycerol phosphate (6). In the model gram-positive organism B. subtilis, the TarB equivalent (TagB) has been shown to provide a critical “priming” activity, where the addition of the first glycerol phosphate residue is necessary for further oligomerization of glycerol phosphate (5, 27). The S. aureus enzymes TarK and TarL are believed to be involved in synthesis of the ribitol phosphate polymer of cell wall teichoic acid by using the activated precursor, CDP-ribitol. We have previously shown that TarIJ from S. aureus catalyzes a bifunctional reaction involving reduction of ribulose 5-phosphate to ribitol 5-phosphate and subsequent cytidylyl transfer to form CDP-ribitol (26). Thus, TarIJ (TarI′J′) and TarKL are thought to be critical to the polymerization of ribitol phosphate on an oligomer of glycerol phosphate (Fig. 1).
FIG. 1.
Poly(ribitol phosphate) synthesis in S. aureus. (A) The region of the S. aureus chromosome involved in ribitol phosphate polymer synthesis for cell wall teichoic acid contains a putatively duplicated gene cluster (tarI′J′K and tarIJL). The genes in this region encode the enzymes involved in CDP-ribitol synthase (TarI′J′ and TarIJ), denoted by the white arrows, and genes involved in glycerol phosphate (TarB and TarF) and ribitol phosphate (TarK and TarL) transferase activities, denoted by the black arrows. The gene tarF, encoding a glycerol phosphate transferase, separates the two gene clusters. (B) Cell wall teichoic acid is assembled on the membrane-bound prenyl-linked disaccharide. The activities of the ribitol phosphate transferases and the CDP-ribitol synthase enzymes are indicated. It is unknown if TarI′J′ and TarK are capable of efficiently catalyzing their predicted functions.
Although the gene products involved in poly(ribitol phosphate) teichoic acid biosynthesis in B. subtilis strain W23 have been assigned functions based on homology to the well-characterized enzymes of B. subtilis strain 168 (18), the biosynthetic pathway for poly(ribitol phosphate) teichoic acid in S. aureus remains unresolved. The major difficulty in translating our understanding from B. subtilis to the S. aureus system lies in inconsistencies in the organization of biosynthetic genes between the two organisms and in the apparent duplicated loci tarI′J′ and tarK. This issue was first addressed in a comparative bioinformatic study of poly(ribitol phosphate) teichoic acid biosynthetic enzymes from B. subtilis W23 and S. aureus by Qian et al. (28). These researchers suggested that two polycistronic gene clusters are involved in the synthesis of the ribitol phosphate polymer. Furthermore, they reported that the high sequence similarities between the gene products (79% identity between TarK and TarL, 76% identity between TarI′ and TarI, and 80% identity between TarJ′ and TarJ) were most readily explained by a duplication of the tarI, tarJ, and tarL genes, resulting in a highly similar locus carrying tarI′, tarJ′, and tarK (28). This putative duplication was present in all strains of S. aureus for which sequence data were available. While this study provided some important insight into the genetic organization of teichoic acid synthesis in S. aureus, functional assignments of these duplicated gene clusters remained unresolved. Recently, Suzanne Walker's group took steps to address this and other questions with genetic and biochemical studies of poly(ribitol phosphate) teichoic acid synthesis in S. aureus (6, 22). They tested the dispensability of both tarK and tarL by routine gene deletion methods and reported that the former could be deleted and the latter could not. Similarly, they reported that tarI′J′ but not tarIJ could be deleted. The results suggested that these were not simply redundant, duplicated loci. Interestingly, when expressed at a high copy number, tarK was able to suppress the lethal phenotype associated with the deletion of tarL (22). Also puzzling was a lack of ribitol phosphate transferase activity for pure recombinant TarK in vitro (6). From this work, a model was developed where TarK and TarL from B. subtilis W23 each catalyze a separate priming and polymerase step in ribitol-phosphate polymer formation, whereas TarK and TarL from S. aureus are each bifunctional enzymes that can catalyze both of the reactions. The model further proposes that although the S. aureus enzymes are bifunctional, they are not functionally redundant in the cell due to differences in expression (22). In the work reported here, we have revisited the questions of gene function and dispensability for the apparently duplicated loci (tarI′J′K and tarIJL). In contrast to the study by Meredith et al. (22), our work suggests that the two S. aureus polycistronic gene clusters (tarIJL and tarI′J′K) are functionally redundant and that each is sufficient for ribitol phosphate polymer formation in S. aureus.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Bacterial strains and plasmids used for this study are described in Table 1. Sequences of primers used in this study are provided in Table S1 in the supplemental material. S. aureus strains were grown at 37°C on Mueller-Hinton medium (BD, Sparks, MD) supplemented when necessary with the following compounds: 10 μg/ml erythromycin, 20 μg/ml kanamycin, 15 μg/ml chloramphenicol, 300 μg/ml spectinomycin, 5% (wt/vol) sucrose, and 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG), unless otherwise indicated. Cloning was done with Escherichia coli strain Novablue (Novagen, Madison, WI) grown on Luria-Burtani (LB) medium supplemented with 50 μg/ml ampicillin or 50 μg/ml kanamycin for selection.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus strains | ||
| RN4220 (EBII13) | Parent strain | 16 |
| SA178RI (EBII16) | RN4220 containing T7 polymerase | 11 |
| tar gene integrants | ||
| EBII43 | SA178RI tarIJ::pSAKO-ΔtarIJ (Ermr Kanr) | 11 |
| EBII48 | SA178RI tarI′J′::pSAKO-ΔtarI′J′ (Ermr Kanr) | This study |
| EBII93 | SA178RI tarK::pSAKO-ΔtarK (Ermr Kanr) | This study |
| EBII94 | SA178RI tarL::pSAKO-ΔtarL (Ermr Kanr) | This study |
| EBII105 | EBII101 tarK::pSAKO-ΔtarK (Ermr Specr Kanr) | This study |
| Noncomplemented deletion mutants | ||
| EBII100 | SA178RI tarK::Erm (Ermr Kans) | This study |
| EBII101 | SA178RI tarL::Erm (Ermr Kans) | This study |
| EBII49 | SA178RI tarI′J′::Erm (Ermr Kans) | This study |
| Complemented gene integrants | ||
| EBII50 | EBII43/pG164-tarIJ | 11 |
| EBII113 | EBII43/pG164-tarJ′ | This study |
| EBII114 | EBII43/pG164-tarI′J′ | This study |
| EBII162 | EBII43/pG164-168_tagDF | This study |
| EBII174 | EBII105/pLI50-tarK | This study |
| EBII175 | EBII105/pLI50-tarL | This study |
| EBII176 | EBII105/pG164-W23_tarKL | This study |
| EBII177 | EBII105/pG164-tarKL | This study |
| Complemented deletions | ||
| EBII56 | SA178RI tarIJ::Erm/pG164-tarIJ | 11 |
| EBII118 | SA178RI tarIJ::Erm/pG164-tarJ′ | This study |
| EBII178 | EBII101 tarK::Spec (Ermr Specr Kans)/pLI50-tarK | This study |
| EBII179 | EBII101 tarK::Spec (Ermr Specr Kans)/pLI50-tarL | This study |
| EBII180 | EBII101 tarK::Spec (Ermr Specr Kans)/pG164-tarKL | This study |
| EBII181 | EBII101 tarK::Spec (Ermr Specr Kans)/pG164-W23_tarKL | This study |
| E. coli strains | ||
| Novablue (EB1) | endA1 hsdR17(rK12− mK12+) supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proA+B+lacIqZΔM15::Tn10] Tcr | Novagen |
| MC1061 (EB30) | MC1061F′ lacIqlacZ::Tn5 (neoR) pro+ | 7 |
| Plasmids | ||
| pUS19 | Source for Specr cassette | 2 |
| pMUTIN4 | Source for Ermr cassette | 29 |
| pSAKO | E. coli replicating vector containing sacB [BamP] W29; Kanr | 11 |
| pG164-tarD | E. coli-S. aureus shuttle vector for T7-based expression (Ampr Cmr) | 11 |
| pLI50 | E. coli-S. aureus shuttle vector for native expression (Ampr Cmr) | 19 |
| pBluescript SKII (+) | Plasmid for cloning (Ampr) | Stratagene |
| pSAKO-ΔtarK | Plasmid for single integration into tarK flank (Ermr Kanr) | This study |
| pSAKO-ΔtarK | Plasmid for single integration into tarK flank (Specr Kanr) | This study |
| pSAKO-ΔtarL | Plasmid for single integration into tarL flank (Ermr Kanr) | This study |
| pG164-168_tagDF | pG164 containing tagD and tagF from B. subtilis 168 | This study |
| pG164-W23_tarK | pG164 containing tarK from B. subtilis W23 | This study |
| pG164-W23_tarL | pG164 containing tarL from B. subtilis W23 | This study |
| pG164-W23_tarKL | pG164 containing tarK from B. subtilis W23 | This study |
| pG164-tarKL | pG164 containing SACOL0238/0242 from S. aureus RN4220 | This study |
| PG164-tarIJ | pG164 containing tarIJ (SACOL0240/0241) from S. aureus RN4220 | 11 |
| PG164-tarI′J′ | pG164 containing tarI′J′ (SACOL0236/0237) from S. aureus RN4220 | This study |
| PG164-tarJ′ | pG164 containing tarI′J′ (SACOL0237) from S. aureus RN4220 | This study |
| pLI50-tarK | pLI50 containing SACOL0238 from S. aureus RN4220 | This study |
| pLI50-tarL | pLI50 containing SACOL0242 from S. aureus RN4220 | This study |
Construction of pSAKO gene-specific integration vectors.
pSAKO integration vectors were generated as previously described (11). Briefly, the 1-kb sequences flanking the gene of interest were amplified using gene-specific primer pairs A/B and C/D. A second PCR was performed to link the two flanks with an erythromycin resistance cassette amplified from the plasmid pMUTIN4. pSAKO and the PCR products were digested with XhoI and purified. The 3-kb fragments and pSAKO were ligated, transformed into E. coli strain MC1061 or Novablue, and selected on 100 μg/ml erythromycin or 50 μg/ml kanamycin, respectively. pSAKO-ΔtarK (Specr) was generated by replacement of the erythromycin resistance cassette with a spectinomycin resistance cassette amplified from plasmid pUS19, using the EcoRI site.
Construction of pG164 and pLI50 complementation vectors.
B. subtilis tarK and tarL genes were amplified from B. subtilis strain W23, tagD and tagF genes were amplified from strain 168, and tarK (SACOL0238), tarL (SACOL0242), and tarI′J′ genes were amplified from S. aureus, using the primers listed in Table S1 in the supplemental material, and subcloned into pBluescript. The resulting pBluescript clones were digested with BamHI and XhoI and subsequently cloned into the digested pG164 vector.
To create a bicistronic complementation vector, tarL (primer set 0242b) was cloned into pG164-tarK to make pG164-tarKL and tagD was cloned into pG164-168_tagF to make pG164-168_tagDF, each by use of the XhoI site. Plasmid pG164-W23_tarKL was created by amplifying tarK and tarL from B. subtilis W23 in tandem, using primers tarKFor and tarLRev. For native expression constructs, approximately 500 bp upstream of tarK and tarL was amplified from S. aureus along with each gene, using the primers listed in Table S1 in the supplemental material, and cloned into the SmaI-digested pLI50 plasmid.
Creation of S. aureus single-integrant and complemented strains.
SA178R1 was transformed by electroporation with gene-specific pSAKO integration plasmids, as described previously (11), and selected on Mueller-Hinton agar (MHA) supplemented with erythromycin and kanamycin. A tarK single integrant in a ΔtarL background (EBII105) was constructed by transformation of pSAKO-ΔtarK (Specr) directly into a ΔtarL background (EBII101) and selected on MHA supplemented with erythromycin, kanamycin, and spectinomycin. Successful integration of the plasmids was verified by PCR. Complementation plasmids were transformed into single-integrant strains and grown on MHA supplemented with erythromycin, kanamycin, and chloramphenicol. Resulting strains can be found in Table 1. Single-integrant strains were used to assay gene dispensability or to generate gene deletions.
Creation of deletion strains.
Complemented single-integrant strains (EBII50 and EBII113) were passed three times on MHA supplemented with sucrose, erythromycin, chloramphenicol, and IPTG to generate deletion strains (EBII56 and EBII118). Uncomplemented single integrants (EBII48, EBII93, and EBII94) were passed three times on MHA supplemented with sucrose and erythromycin to generate uncomplemented deletions (EBII49, EBII100, and EBII101).
To create complemented double deletion mutants of tarK and tarL, strains that were ΔtarL mutants and single integrants of pSAKO at tarK (EBII174, EBII175, EBII176, and EBII177) were grown overnight on MHA plates supplemented with erythromycin, spectinomycin, kanamycin, and chloramphenicol and were subsequently streaked onto MHA plates supplemented with sucrose for counterselection, as previously described (11), to generate double deletion strains (EBII178, EBII179, EBII181, and EBII180, respectively). Strains using the inducible complementation plasmid pG164 (EBII176 and EBII177) contained IPTG in the medium. All deletion strains were verified by PCR as previously described (11).
Purification and assay of CDP-ribitol synthase activity of TarI′J′ and TarIJ.
The tarI′J′ genes were amplified as a single PCR product and cloned into the pDEST14 expression vector by using primers gtarI′J′-F and gtarI′J′-R (see Table S1 in the supplemental material) and a Gateway cloning kit (Invitrogen, Carlsbad, CA). The TarI′J′ enzymes were purified and assayed as previously described for TarIJ (26).
Cell wall isolation and analysis.
Strains were grown in triplicate and harvested from 100 ml of overnight Mueller-Hinton broth culture. Cell wall isolation and determination of phosphate content were carried out as previously described (4). In summary, an S. aureus suspension was lysed by being sheared with glass beads (Sigma-Aldrich, St. Louis, MO), and cell walls were extracted by two rounds of boiling in sodium dodecyl sulfate (21), treated with RNase, DNase, and trypsin (17), and mineralized (1). Cell wall phosphate content was assayed by the protocol of Chen et al. (8), using KH2PO4 as a standard.
RESULTS
Mutants of tarK and tarL were obtained at equivalent frequencies.
We employed a previously described allelic replacement method that has been shown to effectively report on gene dispensability in S. aureus (11). This method relies on chromosomal integration of the plasmid pSAKO, carrying upstream and downstream flanks of the target gene surrounding a drug resistance cassette and a sucrose-inducible counterselectable marker. Integration results in a tandem duplication of the flanks surrounding the target gene, with the plasmid sequence between the two copies. Gene dispensability is assayed by the frequency of gene replacement or reversion to wild type after counterselection for the plasmid sequences. Because this method employs a screen rather than selection to isolate mutants, it has particular strength in the passive production of mutations without strong selection for suppressor mutations. Table 2 summarizes the frequency of excision events of viable clones upon counterselection for integrants in tarK and tarL. Using this screening methodology, we obtained 53% of clones and 38% of clones that displayed a phenotype consistent with a deletion of tarK and tarL, respectively, indicating that each gene is individually dispensable.
TABLE 2.
Dispensability analysis of S. aureus tarK and tarL genes
| S. aureus integrant strain genotyped | Complementing plasmidd | Phenotype (no. of colonies)
|
||
|---|---|---|---|---|
| Wild type | Nonexcisant | Mutant | ||
| tarK (SACOL0238) | 49 | 0 | 55 | |
| tarL (SACOL0242)a | 85 | 0 | 53 | |
| tarK ΔtarL* | 104 | 0 | 0 | |
| pLI50-tarK | 52 | 2 | 50 | |
| pLI50-tarL | 68 | 0 | 36 | |
| pG164-tarKLb | 66 | 1 | 37 | |
| pG164-W23_tarKc* | 104 | 0 | 0 | |
| pG164-W23_tarLc* | 104 | 0 | 0 | |
| pG164-W23_tarKLc | 76 | 0 | 28 | |
Total of 138 colonies selected.
pG164 induced with 0.4 mM IPTG.
pG164 induced with 1.0 mM IPTG.
*, statistically significant (P < 0.05).
A tarK and tarL double deletion cannot be created in a wild-type background.
To ensure that TarK and TarL perform the same cellular function, the allelic replacement method was used to excise the dispensable tarK gene cassette in a tarL null mutant. Of the 104 selected clones, all generated the wild-type locus upon excision of the plasmid, indicating that it is not possible to delete both tarK and tarL in the same genetic background. The inability to generate a double deletion indicates that the protein product of either one of these genes is necessary and sufficient for viability, suggesting functional redundancy between tarK and tarL.
To prevent suppression effects from high-copy expression, the low-copy shuttle vector pLI50 and native promoters were used for complementation. When either gene, along with the corresponding upstream 500-bp sequence, was placed individually on pLI50, we were able to generate a double deletion of tarK and tarL on the chromosome. In the presence of pLI50-tarK or pLI50-tarL, the double deletion phenotype was observed in 48% or 35% of clones, respectively, confirming that either open reading frame can complement the double deletion. Although it has been suggested that expression of tarI′J′K genes is controlled from a common promoter (22), complementation of the Δtark ΔtarL strain by use of the direct upstream regions of both tarK and tarL suggests that although the gene clusters appear to be operonic, the transcription of the tarK and tarL genes is likely regulated separately from that of tarI′J′ and tarIJ. While we cannot rule out the possibility of low levels of transcription being generated from cryptic promoter elements in the pLI50 vector DNA, given that the cloned sequence was able to generate sufficient levels to complement the double deletion it is likely that the tarK and tarL genes have their own promoter elements.
The putative B. subtilis W23 ribitol phosphate primase and polymerase were able to complement the double deletion when they were coexpressed from the pG164-W23_tarKL vector, but individual expression of either tarK or tarL from B. subtilis W23 was unable to rescue a lethal phenotype. Taken together, these results indicate that in S. aureus, tarK and tarL may be fully functionally redundant, capable of efficiently adding the first unit and subsequent polymer of ribitol phosphate.
Mutants of tarK (SACOL0238) and tarL (SACOL0242) are not compromised in growth or cell wall teichoic acid levels.
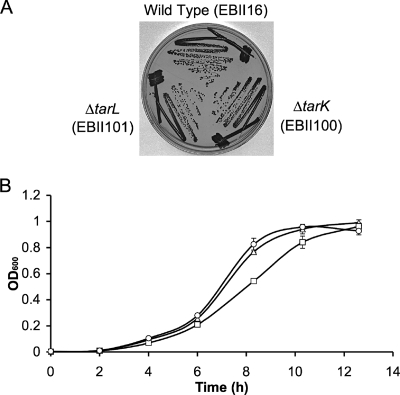
Both single mutant strains were characterized by monitoring growth rates in liquid and on solid medium (Fig. 2). Colony morphology and growth of the tarK and tarL mutants appeared identical to those of the wild-type strain on solid MHA medium; in liquid Mueller-Hinton broth, the tarL mutant was only slightly impaired compared to the wild type and the tarK mutant (doubling times of 1.46 h and 1.67 h for the tarK and tarL mutants, respectively, compared to 1.47 h for the parent strain). These results indicate that the viability of the mutant strains is not compromised despite their missing either gene product. To determine whether the mutant strains and the wild-type strain are also similar in their wall teichoic acid composition, cell wall phosphate content was assayed. The parent strain S. aureus SA178RI contained 0.66 ± 0.01 μmol phosphate/mg of cell wall, while tarK and tarL mutants contained 0.73 ± 0.03 and 0.65 ± 0.08 μmol phosphate/mg of cell wall, respectively. Thus, the deletion of either open reading frame did not affect cell wall teichoic acid levels, suggesting that gene dosage does not alter teichoic acid levels.
FIG. 2.
Growth analysis of tarK and tarL mutants. (A) S. aureus parent strain SA178R1 (EBII16) and ΔtarK (EBII100) and ΔtarL (EBII101) mutant strains were grown on MHA overnight at 37°C in the absence of complementation. Growth was assessed after 24 h. On solid medium, no discernible growth difference was observed for either mutant compared to the parent strain. (B) Growth of ΔtarK (▵) and ΔtarL (□) deletion strains and of parent strain SA178R1 (○) was also assessed in Mueller-Hinton broth. Each of these strains was subcultured to a starting optical density at 600 nm (OD600) of 0.002, and growth was monitored by measuring the optical density at 600 nm from a 96-well plate in triplicate at 37°C, with aeration, at 250 rpm. Both mutants showed similar lag times before exponential growth. The tarK mutant showed comparable exponential growth to the parent strain, while the tarL mutant showed a slightly lower rate of growth during this stage. Although there was a slight slow-growth phenotype, neither mutant appeared to be compromised on either liquid or solid medium.
TarI′J′ and TarIJ each synthesize CDP-ribitol, with comparable efficiencies.
The tarIJ locus has previously been shown to be essential (11), despite the duplicated locus tarI′J′. The inability of tarI′J′ to complement a tarIJ null mutant is unclear in light of the fact that a functional redundancy is observed between the TarK and TarL enzymes. We tested the in vitro activity of each gene product to ascertain whether a catalytically inactive TarI′J′ enzyme may impart the essentiality of tarIJ. TarIJ has been shown to form a hetero-oligomer of the TarI and TarJ enzymes to reduce d-ribulose-5-phosphate with NADPH to ribitol-5-phosphate and subsequently activate the intermediate with CTP to form CDP-ribitol. TarI′J′ may have been rendered an incompetent enzyme through either active-site mutation or prevention of hetero-oligomerization between the TarI′ and TarJ′ domains. Native purification of TarI′ and TarJ′, using published methods for TarIJ (26), indicated that the enzymes form a hetero-oligomer that is stable over three purification steps. In vitro assay of the reductase reactions catalyzed by TarIJ and TarI′J′ led to calculation of similar Michaelis constants for each NADPH and ribulose-5-phosphate, as well as to comparable rates of turnover. Similarly, the cytidylyltransferase reactions had comparable turnover rates and Kms for CTP, while the Michaelis constant for ribitol-5-phosphate was 10-fold lower for TarI′J′ than for TarIJ, giving the former a slightly more favorable specificity constant. In sum, the dispensable CDP-ribitol synthase encoded by tarI′ and tarJ′ is a catalytically active hetero-oligomer competent in the formation of CDP-ribitol to the same extent as TarIJ. Kinetic constants for the CDP-ribitol synthase reaction are listed in Table 3.
TABLE 3.
Comparison of CDP-ribitol synthase activities of different loci
| Activity, enzyme, and substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1/s−1) |
|---|---|---|---|
| Reductase activity | |||
| TarI′J′ | |||
| NADPH | 6.21 ± 1.07 | 0.41 ± 0.01 | 6.60 × 104 |
| Ribulose-5-P | 11.0 ± 0.9 | 3.73 × 104 | |
| TarIJa | |||
| NADPH | 6.74 ± 1.64 | 0.79 ± 0.06 | 1.17 × 105 |
| Ribulose-5-P | 28.5 ± 13.1 | 2.77 × 104 | |
| Cytidylyltransferase activity | |||
| TarI′J′ | |||
| CTP | 38.7 ± 6.20 | 66.7 ± 1.4 | 1.72 × 106 |
| Ribitol-5-P | 102.2 ± 9.6 | 6.53 × 105 | |
| TarIJa | |||
| CTP | 78.9 ± 33.2 | 76.8 ± 6.5 | 9.73 × 105 |
| Ribitol-5-P | 1,280 ± 390 | 5.98 × 104 |
Data for TarIJ were previously published by Pereira et al. (26).
Dispensability of CDP-ribitol synthase.
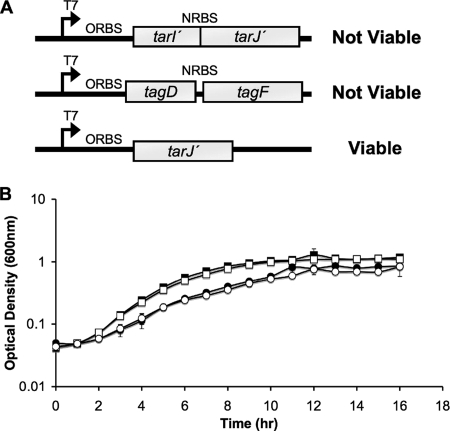
Since each TarI′J′ and TarIJ enzyme formed complexes able to efficiently synthesize CDP-ribitol in vitro, the necessity of tarIJ could conceivably be due to the lower level of expression from the promoter driving the expression of tarI′J′. Overexpression of TarI′J′ on the pG164 plasmid, however, was not successful in suppression of the lethal phenotype of a tarIJ mutant. Furthermore, the tarI′J′ null mutant did not display a growth phenotype (doubling time of 1.98 h, compared to 1.93 h for the parent strain), suggesting that the products of these genes do not affect ribitol phosphate biosynthesis (Fig. 3). Interestingly, overexpression of TarJ′ alone, using an optimized ribosome binding site (30) on pG164, was sufficient to suppress the lethal phenotype of the tarIJ null mutant. These results imply that TarI′ expressed from the chromosome was capable of associating with TarJ′ expressed from the plasmid to suppress the tarIJ null mutation. This suggests that the failure in tarI′J′ activity is due to a poor translation of the native tarJ′ product. Growth analysis of the mutants indicates that complementation with tarJ′ alone (doubling time of 3.15 h) returns the tarIJ mutant to a growth rate comparable to that for the strain complemented with tarIJ on a plasmid (doubling time of 2.88 h).
FIG. 3.
Suppression of tarIJ deletion with TarI′J′ overexpression constructs. (A) Summary of suppression of tarIJ null mutation through overexpression in trans on the plasmid pG164. Neither overexpression of TarI′J′ nor overexpression of B. subtilis TagD (CDP-glycerol synthase) and TagF (glycerol-phosphate polymerase) was sufficient to suppress a tarIJ deletion. Interestingly, providing only tarJ′ with an optimally engineered ribosome binding site was able to suppress the tarIJ mutant. (B) Growth rates of CDP-ribitol synthase mutants were compared in liquid medium. The tarI′J′ mutant (□) grew equivalently to the parent strain SA178R1 (▪). Complementation with pG164-TarJ′ (○) showed a similar growth rate to complementation with pG164-TarIJ (•), which gave slightly impaired growth compared to that of the parent strain. These results indicate that the low native levels of TarI′ and TarJ′ expressed in trans can functionally replace CDP-ribitol synthase activity from TarIJ.
DISCUSSION
The biosynthesis of ribitol phosphate teichoic acid in S. aureus has been studied for decades. Only recently, however, have we begun to appreciate the details of the complex nature of its biogenesis. Together, biochemical studies of the recombinant enzymes and phenotypic examinations of the genes involved are beginning to yield a picture of the biosynthetic steps. Nevertheless, recent work from our research group on wall teichoic acid biosynthetic genes in S. aureus and B. subtilis has revealed complex and contextual gene dispensability phenotypes. That work used careful gene dispensability testing to show that genes with a role in late steps in the biosynthetic pathway, i.e., tarB, -D, -F, -I, -J, and -H in S. aureus, were indispensable for cell viability in culture (11). Paradoxically, while these genes were essential for viability, the first step (tarO in S. aureus) proved to be dispensable in both B. subtilis and S. aureus (10, 11). This apparent contradiction was addressed when we demonstrated that the late-acting genes could be deleted in both B. subtilis and S. aureus, but only in the presence of an accompanying deletion in tarO, encoding the first enzyme in the teichoic acid biosynthetic pathway (10, 11). While many of the late-acting wall teichoic biosynthetic genes from S. aureus were tested in the previous work, those thought to be involved in ribitol phosphate priming and polymerization (tarK and tarL) were not. Indeed, these genes were recently proposed to exist along with the genes for CDP-ribitol synthesis (tarI and -J) in a duplicated region of the S. aureus chromosome to yield the tandem loci tarI′J′K and tarIJL (28). In the work reported here, we have leveraged the essential phenotype of late-acting tar genes to better understand the biosynthetic contributions of these genes and determined, in contrast to a recently published report (22), that these tandem loci are redundant. Our results reveal further complexity in wall teichoic acid genetics that is manifest in biosynthetic redundancy.
Proposals for the biosynthetic steps involved in ribitol phosphate teichoic acid biosynthesis in S. aureus have been based on what is understood from glycerol phosphate teichoic acid biosynthesis in B. subtilis 168 (18, 28). It has been shown that the TagB enzyme is required to add the first glycerol phosphate to the prenyl-linked disaccharide in a “priming” reaction, while TagF subsequently adds the polymer (5, 27). By analogy, these studies predicted a need for two separate ribitol phosphate transferase activities for poly(ribitol phosphate) synthesis. The prediction held that a primase (TarK) would add the first ribitol phosphate and a polymerase (TarL) would add subsequent residues. Recently, in vitro biochemical experiments with pure recombinant proteins failed to yield activity for TarK but suggested that TarL could catalyze both priming and polymerase activities (6). Genetic studies by the same group showed that the tarL gene was essential (22), suggesting that tarK and tarL were not functionally redundant.
In the work presented here, we employed an approach developed by our group for the study of gene dispensability in S. aureus (11) to show that mutants in the tarK and tarL genes can be obtained easily and with approximately equal frequencies. Since no significant differences were seen in growth rates or cell wall phosphate levels of the tarK and tarL mutants, we surmised that cell wall-associated teichoic acid was unperturbed by either deletion. These results suggest that the TarK and TarL enzymes are indeed functionally redundant. This finding agrees with the conclusion of Meredith et al. that only a single S. aureus enzyme is required for ribitol-phosphate polymer formation (22), which contradicts the two-enzyme model—with a primase and polymerase—for teichoic acid polymer biosynthesis that has been well developed for the model gram-positive bacterium B. subtilis 168 (4, 27).
We were very interested to see that complementation of the S. aureus ΔtarK ΔtarL deletion strain required both B. subtilis W23 tarK and tarL. That is, neither tarK nor tarL alone from B. subtilis could complement the defect. These data correspond with results and conclusions drawn from cross-species complementation experiments with B. subtilis W23 genes to rescue transduction-mediated mutations in S. aureus tarL (22) and are consistent with the primase and polymerase model for B. subtilis, where addition of the first polyol-phosphate unit (“priming”) is obligate to polymerization. This paradigm has held up to extensive genetic and biochemical experimentation with B. subtilis (4, 11, 22, 27). The work presented here, however, suggests that the S. aureus enzymes TarK and TarL have redundant functions in priming and polymerization, a feature not shared by other members of this enzyme class. These findings highlight some profound gaps in our understanding of the structural and functional characteristics of the enzymes that synthesize teichoic acid polymers.
Although tarI′J′K and tarIJL are chromosomally arranged in two adjoining gene clusters that likely resulted from a chromosome duplication event, the tarI′J′ locus cannot functionally complement the tarIJ locus, despite the observed redundancy of TarK and TarL. To probe the mechanism of the essentiality of tarIJ, we overexpressed and purified recombinant TarI′J′ and compared the steady-state kinetic parameters of the TarI′J′ enzyme complex to those of TarIJ. In vitro analysis of the two CDP-ribitol synthases showed approximately equivalent specificity constants for each substrate (Table 3), suggesting that the failure is not at the enzymatic level. We attempted to suppress the tarIJ lesion through overexpression in trans of TarI′J′, TarJ′, and TagDF enzymes. Only through separation of TarI′ and TarJ′ expression by providing exclusively tarJ′ in trans was it possible to suppress the tarIJ null mutation. These data suggest that despite the fact that the activity of the tarI′J′ promoter has been reported to be 30-fold lower than that of the tarIJ promoter (22), it is the low levels of TarJ′ that thwart activity from the tarI′J′ locus in vivo. The low cellular TarJ′ levels could be attributed to unstable mRNA, poor proteolytic stability of the protein, or possibly inadequate translational readthrough for the overlapping open reading frames of tarI′ and tarJ′ (overlap of seven nucleotides). The ability of the TarJ′ expression construct to suppress the tarIJ null mutation supports the latter notion.
Although polymer substitution has been observed in B. subtilis 168 by replacement of its glycerol phosphate teichoic acid biosynthetic genes with the ribitol phosphate teichoic acid biosynthetic genes from B. subtilis W23 (14, 33), we were unable to accomplish this with S. aureus. Since CDP-ribitol is essential for the poly(ribitol phosphate) polymerase action of both TarK and TarL, we attempted to suppress a deletion in tarIJ with the CDP-glycerol synthase (TagD) and poly(glycerol phosphate) polymerase (TagF) from B. subtilis 168 to form a “hybrid” strain. While a tagDF expression construct could suppress a tarF (glycerol phosphate polymerase) deletion (data not shown), suppression of a tarIJ null mutation was not possible. It is unknown whether the insufficiency lay in the ability of glycerol phosphate teichoic acid to functionally replace ribitol phosphate teichoic acid or in an inability to export the hybrid polymer. In Streptococcus pneumoniae, for example, the absence of choline as a decoration on the teichoic acid polymer prevents extracellular transport (9). It is possible that the formation of glycerol phosphate polymer by TagDF may not have met the requirements (e.g., length, decorations, or chemical composition) for the S. aureus teichoic acid transporter TarGH (11).
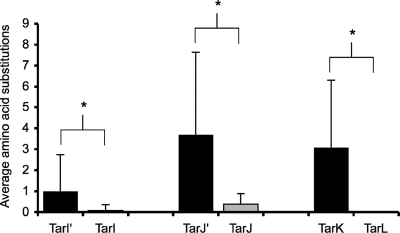
The functional redundancy between the tarK and tarL genes observed here raises questions on the selective pressure for maintaining two functional gene clusters capable of synthesizing a ribitol phosphate polymer. It has been postulated that the existence of the tarI′J′K cluster is likely due to a duplication event (28), a rare occurrence that is a major force in evolution. Duplicated genes initially benefit the organism due to gene dosage effects (15), but their prolonged existence can lead to either portioning of the ancestral gene's functions or silencing due to entopic mutation (12). In the case of the duplication observed here, we posit that both have played a role. Analysis of sequence drift of these genes showed a higher level of divergence leading to a larger number of substitution mutations among the TarI′, TarJ′, and TarK enzymes than among the TarI, TarJ, and TarL enzymes (Fig. 4), supporting the notion that the latter genes are stabilized by purifying selection while the former are mutating toward differentiation or silencing. The bifunctional nature of the TarK and TarL enzymes is a feature not shared by any other members of this enzyme class. It is conceivable that these enzymes are not as catalytically efficient as other members of the enzyme class due to a relaxed specificity toward substrates. Because the polymers formed by TarK are considerably shorter than those formed by TarL (22), it is possible that TarK is diverging toward the function of priming the ribitol phosphate polymer, allowing for efficient polymerization by TarL. In the case of TarIJ, it seems that the use of CDP-ribitol as a substrate for both TarK and TarL has made the second copy of TarI′J′ an unnecessary remnant of duplication, allowing for silencing by attenuation of translation of the gene encoding TarJ′, the enzyme catalyzing the rate-limiting step of CDP-ribitol formation, a common fate of duplicated genes (20).
FIG. 4.
Analysis of genetic drift for genes linked to ribitol phosphate polymer formation. The translated sequences of tarI′, tarI, tarJ′, tarJ, tarK, and tarL were analyzed for congruency among S. aureus strains. Protein sequences from 13 S. aureus strains were aligned pairwise, using BLAST (National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov]), to strain COL in order to assess primary sequence variance. Alignments were performed over the entire sequence length (TarI′, 238 amino acids [aa]; TarI, 238 aa; TarJ′, 341 aa; TarJ, 341 aa; TarK, 564 aa; and TarL, 562 aa). S. aureus strains COL, Newman, Mu50, N315, JH9, JH1, Mu3, USA300_TCH1516, USA 300, NCTC 8325, RF122, MRSA252, MW2, and MSSA476 were used in this study. The data plotted show the average numbers of amino acid substitutions observed for the translated products of the loci. From these data, it can be seen that the products of the tarI′J′K gene cluster have a higher level of divergence than the products of the tarIJL gene cluster. *, P < 0.05. For the products of the tarI′J′K genes, the various numbers of substitutions as well as differing types and locations of substitutions per strain are reflected in the large standard deviation, supporting the notion that these substitutions occurred randomly. The few small sequence variations observed for products of the tarIJL gene cluster, however, were all conservative sequence substitutions common to many strains in a group. These data suggest that only the tarIJL gene cluster is stabilized by purifying selection.
In the work reported here, we have demonstrated additional complexity in wall teichoic acid genetics that is evident in biosynthetic redundancy. While we have shown that TarK and TarL are functionally redundant, a careful kinetic analysis of TarK and TarL is needed to dissect the roles that each of these enzymes have in teichoic acid synthesis. This and other work aimed at a complete understanding of wall teichoic acid biogenesis in gram-positive bacteria will provide critical details for strategies aimed at subverting and exploiting this cellular machinery in antibacterial drug discovery.
Supplementary Material
Acknowledgments
This work was supported by Canadian Institutes of Health research grant MOP-15496, a Canada Research Chair held by E. D. Brown, and Canadian Institutes of Health Research Canada graduate scholarships held by M. P. Pereira and M. A. D'Elia.
Footnotes
Published ahead of print on 13 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ames, B. N. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8115-118. [Google Scholar]
- 2.Benson, A. K., and W. G. Haldenwang. 1993. Regulation of sigma B levels and activity in Bacillus subtilis. J. Bacteriol. 1752347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhavsar, A. P., and E. D. Brown. 2006. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol. Microbiol. 601077-1090. [DOI] [PubMed] [Google Scholar]
- 4.Bhavsar, A. P., L. K. Erdman, J. W. Schertzer, and E. D. Brown. 2004. Teichoic acid is an essential polymer in Bacillus subtilis that is functionally distinct from teichuronic acid. J. Bacteriol. 1867865-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhavsar, A. P., R. Truant, and E. D. Brown. 2005. The TagB protein in Bacillus subtilis 168 is an intracellular peripheral membrane protein that can incorporate glycerol phosphate onto a membrane-bound acceptor in vitro. J. Biol. Chem. 28036691-36700. [DOI] [PubMed] [Google Scholar]
- 6.Brown, S., Y. H. Zhang, and S. Walker. 2008. A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem. Biol. 1512-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138179-207. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P., T. Toribara, and H. Warner. 1956. Microdetermination of phosphorus. Anal. Chem. 181756-1758. [Google Scholar]
- 9.Damjanovic, M., A. S. Kharat, A. Eberhardt, A. Tomasz, and W. Vollmer. 2007. The essential tacF gene is responsible for the choline-dependent growth phenotype of Streptococcus pneumoniae. J. Bacteriol. 1897105-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Elia, M. A., K. E. Millar, T. J. Bevridge, and E. D. Brown. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J. Bacteriol. 1888313-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Elia, M. A., M. P. Pereira, Y. S. Chung, W. Zhao, A. Chau, T. J. Kenney, M. C. Sulavik, T. A. Black, and E. D. Brown. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 1884183-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan, and J. Postlethwait. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1511531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, I. C. 1997. Bacterial cell surface carbohydrates: structure and assembly. Biochem. Soc. Trans. 25183-187. [DOI] [PubMed] [Google Scholar]
- 14.Karamata, D., H. M. Pooley, and M. Monod. 1987. Expression of heterologous genes for wall teichoic acid in Bacillus subtilis 168. Mol. Gen. Genet. 20773-81. [DOI] [PubMed] [Google Scholar]
- 15.Kondrashov, F. A., I. B. Rogozin, Y. I. Wolf, and E. V. Koonin. 2002. Selection in the evolution of gene duplications. Genome Biol. 3RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 17.Kruyssen, F. J., W. R. de Boer, and J. T. Wouters. 1980. Effects of carbon source and growth rate on cell wall composition of Bacillus subtilis subsp. niger. J. Bacteriol. 144238-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarevic, V., F.-X. Abellan, S. B. Moller, D. Karamata, and C. Mauel. 2002. Comparison of ribitol and glycerol teichoic acid genes in Bacillus subtilis W23 and 168: identical function, similar divergent organization, but different regulation. Microbiology 148815-824. [DOI] [PubMed] [Google Scholar]
- 19.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103101-105. [DOI] [PubMed] [Google Scholar]
- 20.Lynch, M., and J. S. Conery. 2000. The evolutionary fate and consequences of duplicate genes. Science 2901151-1155. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald, K. L., and T. J. Beveridge. 2002. Bactericidal effect of gentamicin-induced membrane vesicles derived from Pseudomonas aeruginosa PAO1 on gram-positive bacteria. Can. J. Microbiol. 48810-820. [DOI] [PubMed] [Google Scholar]
- 22.Meredith, T. C., J. G. Swoboda, and S. Walker. 2008. Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus. J. Bacteriol. 1903046-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moxon, E. R., and J. S. Kroll. 1990. The role of bacterial polysaccharide capsules as virulence factors. Curr. Top. Microbiol. Immunol. 15065-85. [DOI] [PubMed] [Google Scholar]
- 24.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira, M. P., and E. D. Brown. 2004. Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase activities in Haemophilus influenzae and Staphylococcus aureus. Biochemistry 4311802-11812. [DOI] [PubMed] [Google Scholar]
- 27.Pereira, M. P., J. W. Schertzer, M. A. D'Elia, K. P. Koteva, D. W. Hughes, G. D. Wright, and E. D. Brown. 2008. The wall teichoic acid polymerase TagF efficiently synthesizes poly(glycerol phosphate) on the TagB product lipid III. Chembiochem 91385-1390. [DOI] [PubMed] [Google Scholar]
- 28.Qian, Z., Y. Yin, Y. Zhang, L. Lu, Y. Li, and Y. Jiang. 2006. Genomic characterization of ribitol teichoic acid synthesis in Staphylococcus aureus: genes, genomic organization and gene duplication. BMC Genomics 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 1443097-3104. [DOI] [PubMed] [Google Scholar]
- 30.Vellanoweth, R. L., and J. C. Rabinowitz. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 61105-1114. [DOI] [PubMed] [Google Scholar]
- 31.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturiya, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10243-245. [DOI] [PubMed] [Google Scholar]
- 32.Weidenmaier, C., A. Peschel, Y. Q. Xiong, S. A. Kristian, K. Dietz, M. R. Yeaman, and A. S. Bayer. 2005. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J. Infect. Dis. 1911771-1777. [DOI] [PubMed] [Google Scholar]
- 33.Young, M., C. Mauel, P. Margot, and D. Karamata. 1989. Pseudo-allelic relationship between nonhomologous genes concerned with biosynthesis of polyglycerol phosphate and polyglycerol phosphate teichoic acids in Bacillus subtilis strains 168 and W23. Mol. Microbiol. 31805-1812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.