Abstract
The denitrifying “Aromatoleum aromaticum” strain EbN1 was demonstrated to utilize p-ethylphenol under anoxic conditions and was suggested to employ a degradation pathway which is reminiscent of known anaerobic ethylbenzene degradation in the same bacterium: initial hydroxylation of p-ethylphenol to 1-(4-hydroxyphenyl)-ethanol followed by dehydrogenation to p-hydroxyacetophenone. Possibly, subsequent carboxylation and thiolytic cleavage yield p-hydroxybenzoyl-coenzyme A (CoA), which is channeled into the central benzoyl-CoA pathway. Substrate-specific formation of three of the four proposed intermediates was confirmed by gas chromatographic-mass spectrometric analysis and also by applying deuterated p-ethylphenol. Proteins suggested to be involved in this degradation pathway are encoded in a single large operon-like structure (∼15 kb). Among them are a p-cresol methylhydroxylase-like protein (PchCF), two predicted alcohol dehydrogenases (ChnA and EbA309), a biotin-dependent carboxylase (XccABC), and a thiolase (TioL). Proteomic analysis (two-dimensional difference gel electrophoresis) revealed their specific and coordinated upregulation in cells adapted to anaerobic growth with p-ethylphenol and p-hydroxyacetophenone (e.g., PchF up to 29-fold). Coregulated proteins of currently unknown function (e.g., EbA329) are possibly involved in p-ethylphenol- and p-hydroxyacetophenone-specific solvent stress responses and related to other aromatic solvent-induced proteins of strain EbN1.
Alkylphenols, such as p-ethylphenol, are present in coal tars and crude oils (2, 45). Besides the accidental release of fuel-derived alkylphenols to the environment, phenolic compounds are also prominent constituents of petrochemical wastewaters arising from spent caustic and coal gasification. Alkylphenol concentrations in these effluents (mainly phenol, cresols, and ethylphenols) range from 100 to 68,000 mg liter−1, depending on the source (2). Moreover, p-ethylphenol can also be plant derived; e.g., native olive oils contain up to 52 mg p-ethylphenol per kg, and the content can reach 470 mg kg−1 during storage (5). p-Ethylphenol can also be formed from p-coumaric acid, a major component of cereal cell walls, by several yeast and Lactobacillus species (11, 50). Due to their cytotoxicity and relatively high water solubility (www.epa.gov/safewater/mcl.html), alkylphenols are of environmental concern.
Anaerobic degradation of aromatic compounds requires reactions independent of molecular oxygen. They are fundamentally different from the oxygenase-catalyzed reactions employed under oxic conditions (for an overview, see references 14, 17, and 18). A variety of anaerobic aromatic compound-degrading bacteria were newly isolated during the last decades (for an overview, see references 46 and 51). Most of them are denitrifiers belonging to the “Aromatoleum”/Azoarcus/Thauera cluster within Betaproteobacteria (R. Rabus et al., unpublished data). “Aromatoleum aromaticum” strain EbN1, a metabolically versatile representative of this group, was originally isolated with ethylbenzene under anoxic conditions and demonstrated to utilize a large variety of aromatic compounds (39). The complete genome sequence of strain EbN1 (38) and several proteomic studies (26, 49, 52) revealed a fine-tuned, substrate-specific regulation of its known catabolic pathways.
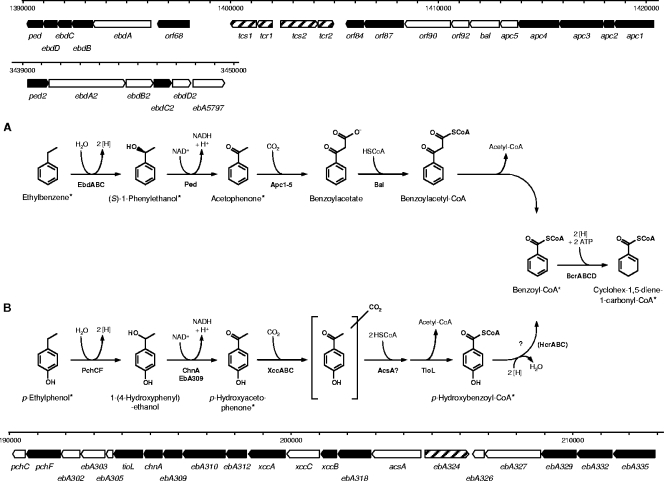
Anaerobic ethylbenzene degradation (Fig. 1) in strain EbN1 proceeds via initial oxidation of ethylbenzene to (S)-1-phenylethanol by ethylbenzene dehydrogenase (EbdABC) (23). A second oxidation step, catalyzed by (S)-1-phenylethanol dehydrogenase (Ped) (20, 24) then yields acetophenone, which is subsequently converted to benzoyl-coenzyme A (CoA) via carboxylation (acetophenone carboxylase [Apc1-5]), CoA activation (benzoylacetate CoA-ligase [Bal]), and thiolytic removal (benzoylacetyl-CoA thiolase) of acetyl-CoA (6). All proteins involved in anaerobic degradation of ethylbenzene are encoded in two close-by gene clusters at approximately 1.4 Mb of the chromosome (37, 38). One includes the ebd and ped genes for the “upper” part of the degradation pathway from ethylbenzene to acetophenone. The other contains apc and bal genes for the “lower” part from acetophenone to benzoyl-CoA. The expression of these two gene clusters is sequentially regulated in response to the presence of their specific substrates, i.e., ethylbenzene and acetophenone (26).
FIG. 1.
Anaerobic degradation pathways of ethylbenzene (A) and p-ethylphenol (B) in “A. aromaticum” strain EbN1. The two initial reactions are chemically analogous but involve different enzymatic catalysis. Both pathways converge at the common intermediate benzoyl-CoA (the ethylbenzene pathway is modified from reference 37). Enzyme names of the indicated gene products (shown in bold type) are as follows: Apc1-5, acetophenone carboxylase; AcsA, predicted acetoacetyl-CoA synthetase; Bal, benzoylacetate CoA-ligase; BcrABCD, benzoyl-CoA reductase; ChnA, predicted cyclohexanol dehydrogenase; EbA309, putative alcohol dehydrogenase; EbdABCD, ethylbenzene dehydrogenase; HcrABC, p-hydroxybenzoyl-CoA reductase; Ped, (S)-1-phenylethanol dehydrogenase; PchCF, predicted p-ethylphenol methylhydroxylase; TioL, predicted thiolase; XccABC, predicted p-hydroxyacetophenone carboxylase. A scale model for the organization of the involved genes is displayed for both pathways. In addition, the paralogous gene cluster for the “upper part” of the alkylbenzene degradation pathway is shown. The scale bar indicates the locations of the genes on the chromosome by the nucleotide positions. Products of genes marked in black were identified on the 2D DIGE gels. *, compounds identified by GC-MS analysis.
p-Ethylphenol has not been tested as a substrate for anaerobic growth of strain EbN1 before, but it was reported to support anaerobic growth of phylogenetically related strain EB1 (3). A pathway for anaerobic degradation of p-ethylphenol is presently unknown. In Pseudomonas putida JD1, the aerobic p-ethylphenol degradation pathway is initiated by an oxygen-independent hydroxylation (9). However, subsequent reaction steps include monooxygenase activity (48), necessitating a different pathway under anoxic conditions.
In the present study we investigated anaerobic degradation of p-ethylphenol in strain EbN1 to elucidate the degradation pathway, study substrate-specific regulation, and analyze involved proteins.
MATERIALS AND METHODS
Media and cultivation.
“A. aromaticum” strain EbN1 was cultivated under nitrate-reducing conditions as previously described (39). The soluble substrates were added from sterile stock solutions. The poorly water-soluble substrates ethylbenzene and p-ethylphenol were provided as dilutions in 2,2,4,4,6,8,8-heptamethylnonane (HMN) as an inert carrier phase. The chemicals used were of analytical grade.
Cultivation was carried out in 500-ml flat bottles containing 400 ml of medium and 10 ml carrier phase when required. Bottles were anoxically sealed with rubber stoppers under a N2/CO2 (9:1, vol/vol) atmosphere. Cells were adapted to anaerobic growth with acetophenone (2 mM), benzoate (4 mM), p-hydroxyacetophenone (2 mM), ethylbenzene (2% in HMN [vol/vol]) and p-ethylphenol (0.5% in HMN [wt/vol]) for at least five passages.
Mass cultivation was performed to supply sufficient cell material for RNA and proteomic analysis. Cells were harvested at midlinear growth phase as described previously (6). Cell pellets were immediately frozen in liquid nitrogen and stored at −80°C until further analysis. For each growth condition, a set of twelve parallel cultures generated independent biological samples.
Analysis of metabolites.
Formation of metabolites was studied in substrate-adapted cultures (see above), with benzoate-adapted cultures serving as a reference state. Unambiguous evidence for identified metabolites to be derived directly from p-ethylphenol was obtained from experiments using deuterated p-ethylphenol (2,3,5,6-d4-p-ethylphenol; Campro Scientific, Berlin, Germany) as a growth substrate. Metabolites of at least two independent cultures were extracted and analyzed per adaptation condition.
Respective cultures were incubated until nitrate and nitrite were nearly depleted, as determined by means of Merckoquant test stripes (Merck, Darmstadt, Germany). Subsequently, cultures were heat inactivated and chilled on ice, and the carrier phase was removed via a separatory funnel after heating, as described previously (40). The inactivated cultures were extracted repeatedly at pH 1.5 with diethyl ether. Extracts were derivatized prior to gas chromatographic-mass spectrometric (GC-MS) analysis using a solution of diazomethane in diethyl ether. GC-MS measurements were performed using a Trace GC-MS (Thermoelectron, Dreieich, Germany). The GC was equipped with a temperature-programmable injection system and a BPX5-fused silica capillary column (length, 50 m; inner diameter, 0.22 mm; film thickness, 0.25 μm). Helium was used as a carrier gas. The GC oven temperature was programmed from 50°C (1 min isothermal) to 310°C (20 min isothermal) at a rate of 3°C min−1. The MS was operated in electron impact mode at an ion source temperature of 230°C. Full-scan mass spectra were recorded over the mass range of 50 to 600 Da at a rate of 2.5 scans s−1. Identification of metabolites was based on comparisons of GC retention times and mass spectra with those of authentic standards.
Physiological adaptation experiments.
Substrate adaptation experiments were performed with cells of strain EbN1 adapted to anaerobic growth with ethylbenzene, acetophenone, p-ethylphenol, or p-hydroxyacetophenone shifted to any of these four substrates. Controls lacked either organic substrate or inoculum. During incubation, samples were withdrawn from the aqueous phase (1 ml) using N2-flushed sterile syringes at intervals of 2 to 3 h. Samples were directly used to monitor growth by measuring the optical density at 660 nm (UV-1202; Shimadzu, Duisburg, Germany). The end of incubation was indicated by the depletion of the electron acceptor nitrate and intermediary formed nitrite as well as lack of increase in optical density. Nitrate and nitrite concentrations were monitored with Merckoquant test strips (Merck). For each of the four test conditions, four replicate shift experiments were conducted (two replicates for each of two independently inoculated cultures). In all cases, the four replicates yielded highly similar time courses of growth.
Two-dimensional difference gel electrophoresis (2D DIGE).
Cell disruption with a PlusOne sample grinding kit (GE Healthcare, Munich, Germany) and preparation of protein extracts were performed as previously described (15). Protein concentration was determined according to the method described by Bradford (4).
Isoelectric focusing was carried out as described previously (15) using an IPG-phor system (GE Healthcare) and 24-cm IPG strips with a nonlinear pH gradient of 3 to 11 (GE Healthcare). DeStreak rehydration solution (GE Healthcare) was used to enhance resolution and reproducibility in the alkaline pH range. An EttanDalt II system (GE Healthcare) was used to carry out separation according to molecular mass in 12.5% acrylamide gels as described recently (52).
2D DIGE was essentially carried out as described by Gade et al. (15). A total of 200 pmol of CyDye Fluors was used to label 50 μg of protein sample. Preelectrophoretic labeling with different cyanine dyes allows coseparation of up to three samples in a single gel. An individual experiment in the present study contained (per gel) equal amounts of reference state, test state, and internal standard.
To account for biological variation, protein extracts from three independently grown cultures were prepared for each substrate condition. Three parallel gels for each protein extract were analyzed to also account for technical variation. Protein extracts from cells anaerobically grown with benzoate served as a reference state since the activated form, benzoyl-CoA, is the central intermediate of anaerobic aromatic compound degradation in strain EbN1. The reference state was labeled with Cy5. Protein extracts from cells anaerobically grown with ethylbenzene, acetophenone, p-ethylphenol, or p-hydroxyacetophenone served as test states and were each labeled with Cy3. All experiments contained the same internal standard, which was composed of equal amounts of all protein preparations of the reference and all test states and labeled with Cy2.
2D DIGE gels were scanned immediately after electrophoresis with a Typhoon 9400 scanner (GE Healthcare). Analysis of cropped images was performed with the DeCyder software (version 5.0; GE Healthcare). Parameters for codetection of protein spots were as described by Wöhlbrand et al. (52). Protein spots that were defined as significantly regulated fulfilled the following criteria: an average ratio (n-fold change) of <−2.5 or >2.5, an analysis of variance P value of <0.05, a t test value of <10−4, and matched in at least 27 gels. Overall, the DeCyder analysis included 36 gels (representing 108 gel images), which contained on average 1,063 detected protein spots.
In order to identify differentially regulated spots by MS, separate preparative gels (300-μg protein load) were run for all test states. These gels were stained with colloidal Coomassie brilliant blue according to the method described by Doherty et al. (12). The high reproducibility of spot patterns allowed MS-based identification of differentially regulated proteins as determined by 2D DIGE/DeCyder. Only unambiguously matching spots were included in this analysis.
Protein identification by MS.
Tryptic digest of excised proteins was performed as described previously (21). Peptide masses were determined by matrix-assisted laser desorption ionization-time of flight MS. Protein identification and genome analysis were based on the published list of annotated genes from the genome of strain EbN1 (38). Peptide mass fingerprints were mapped to the in silico digests of the predicted proteins by using the MS digest program (7).
Preparation of mRNA and real-time reverse transcription (RT)-PCR.
Total RNA was prepared from substrate-adapted cells as described by Oelmüller et al. (34). RNA was prepared from two independent cultures for each growth condition to account for biological variation. Total removal of DNA by DNase digestion was confirmed by PCR as described previously (26). The quality of RNA preparation was controlled with a RNA 6000 Nano assay using a 2100 Bioanalyzer (Agilent Technologies, Böblingen, Germany). Antisense primers of gene-specific primer pairs (Table 1) were used for RT of 2.5 μg total RNA using H minus M-MLuV reverse transcriptase (MBI Fermentas, St. Leon-Roth, Germany) according to the manufacturer's instructions. Real-time PCR was performed as described previously (26) using an iQ5 real-time PCR detection system (Bio-Rad, München, Germany) and qPCR Mastermix Plus for SYBR green I (Eurogentec, Köln, Germany). PCR efficiencies were determined as described by Ramakers et al. (41), and relative expression levels were calculated as described by Pfaffl (35). For each individual RNA preparation, at least two independent RTs were performed. For each generated cDNA, at least two real-time RT-PCR experiments with three parallels each were conducted.
TABLE 1.
Primer sequences of genes involved in anaerobic ethylbenzene and p-ethylphenol degradation of “A. aromaticum” strain EbN1
| Primera | Sequence (5′ → 3′) | Target gene | Product length (bp) |
|---|---|---|---|
| bcrC 108F | CAAGTGGTGGCAACGATGTGT | bcrC | 191 |
| bcrC 299R | GAAGGTCTGGCGATACTGGATGC | ||
| hcrB 405F | GCCATCGTCAACATAGAATT | hcrB | 202 |
| hcrB 607R | TGCCAGTACTTCAACCAGAGC | ||
| ebdA 300F | CTCCGCGGCGTCCTTGCT | ebdA | 186 |
| ebdA 486R | CGCGCCGTGCCCAGTTCTACC | ||
| ebdA2 2666F | GGCGGAGCTCGGTATCAA | ebdA2 | 190 |
| ebdA2 2856R | GGGCTTCCATTCAGGTAGTA | ||
| apc1 1441F | AGCGGCGCCATCTCACTG | apc1 | 153 |
| apc1 1594R | ACCGCCGTCGACATTCTCT | ||
| pchF 1099F | CCATCCGGGAGCACCACT | pchF | 237 |
| pchF 1336R | GGCCGGCAACGTCATCATC | ||
| xccA 1215F | GAATTCCGGGTGCTGGCTCTC | xccA | 198 |
| xccA 1413R | GCGCGACGGGGCTCAATA |
Antisense primers (“R”) were used for RT.
Expression of genes encoding the catalytic subunits of the initial enzymes of both pathways were studied, i.e., ebdA (encoding the α-subunit of ethylbenzene dehydrogenase) and pchF (encoding the α-subunit of the p-cresol methylhydroxylase-like protein) as well as subunits of the enzymes involved in carboxylation of acetophenone (apc1, encoding the β-subunit of acetophenone carboxylase) and p-hydroxyacetophenone (xccA, encoding the carboxyltransferase subunit of the biotin carboxylase). Also, transcription of hcrB (encoding the β-subunit of the predicted p-hydroxybenzoyl-CoA reductase) was analyzed, since reductive dehydroxylation of p-hydroxybenzoyl-CoA is assumed to feed into the central benzoyl-CoA pathway during p-ethylphenol degradation (Fig. 1). Benzoate-adapted cells served as a reference state, and bcrC (encoding the γ-subunit of benzoyl-CoA reductase) was selected as a reference gene, since benzoyl-CoA reductase represents the first common enzyme of the ethylbenzene and p-ethylphenol degradation pathways.
RESULTS AND DISCUSSION
Considering the chemical structures of p-ethylphenol and p-hydroxyacetophenone, a reaction sequence similar to that of anaerobic ethylbenzene degradation could be envisioned (Fig. 1). Thus, p-ethylphenol may be converted to benzoyl-CoA via 1-(4-hydroxyphenyl)-ethanol, p-hydroxyacetophenone, and p-hydroxybenzoyl-CoA (Fig. 1). To test this hypothesis, substrate-specific metabolites were analyzed (Fig. 2). Moreover, cultures of strain EbN1 adapted to anaerobic growth with p-ethylphenol, p-hydroxyacetophenone, ethylbenzene, and acetophenone were used to (i) determine adaptation times to growth with either of these substrates (Fig. 3), (ii) investigate substrate-dependent regulation of the p-ethylphenol pathway on the molecular level (Table 2 and Fig. 4 to 6), and (iii) elucidate possible functions of the specifically regulated proteins.
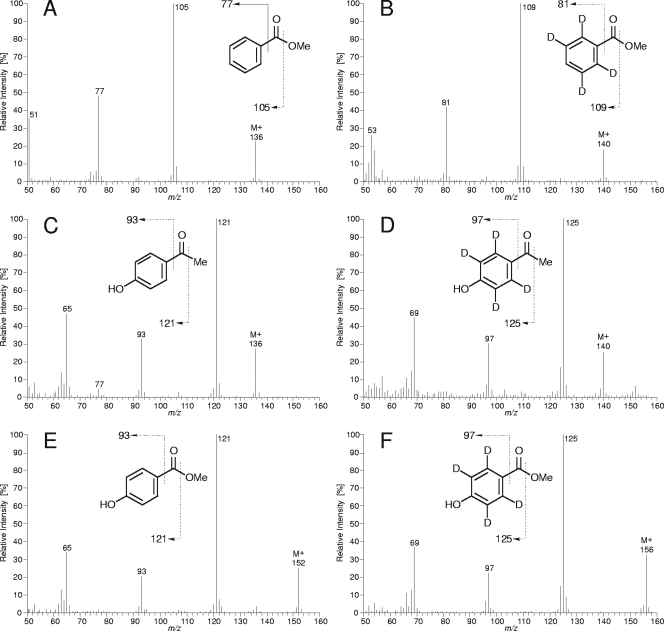
FIG. 2.
Mass spectra of identified metabolites of anaerobic p-ethylphenol degradation and their 2,3,5,6-d4-isotopomers derived from 2,3,5,6-d4-p-ethylphenol utilization. (A) Benzoic acid methyl ester. (B) 2,3,5,6-d4-Benzoic acid methyl ester (please note that the MS evidence demonstrates four d atoms on the aromatic ring but does not necessarily prove the depicted labeling pattern). (C) p-Hydroxyacetophenone. (D) 2,3,5,6-d4-p-Hydroxyacetophenone. (E) p-Hydroxybenzoic acid methyl ester. (F) 2,3,5,6,-d4-p-Hydroxybenzoic acid methyl ester.
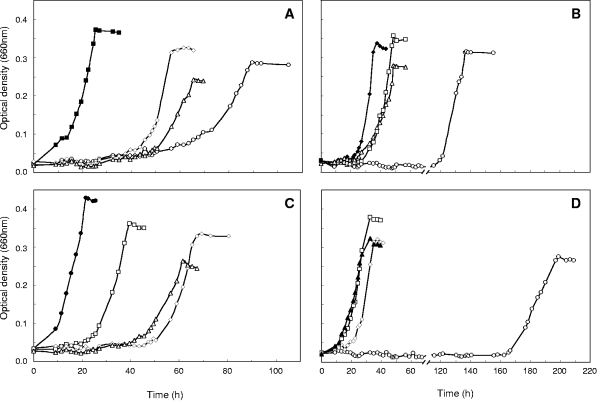
FIG. 3.
Anaerobic growth of “A. aromaticum” strain EbN1 with aromatic substrates under nitrate-reducing conditions. Cultures were inoculated with cells adapted to anaerobic growth with ethylbenzene (A), p-ethylphenol (B), acetophenone (C), or p-hydroxyacetophenone (D). Inoculated fresh anoxic media contained as a single aromatic substrate ethylbenzene (□), acetophenone (○), p-ethylphenol (⋄), or p-hydroxyacetophenone (▵). Filled symbols represent the adaptation substrate of the respective inoculum culture.
TABLE 2.
Changes (n-fold) in abundances of proteins involved in anaerobic ethylbenzene and p-ethylphenol degradation in substrate-adapted cells of “A. aromaticum” strain EbN1 as determined by 2D DIGE
| Assumed function of identified proteinsa | Fold change with adaptation substrateb:
|
|||
|---|---|---|---|---|
| Etb | Acp | pEp | pAc | |
| Ethylbenzene degradation | ||||
| EbdB (c1A63) | 60.3 | 1.3 | 2.4 | 1.6 |
| EbdC (c1A62)c | 154.0 | 1.3 | 1.7 | 1.4 |
| EbdD (c1A60) | 17.8 | 1.4 | 1.4 | 1.4 |
| Ped (c1A58) | 17.5 | 1.7 | 1.5 | 1.8 |
| Orf68 (c1A68)c | 311.2 | 3.0 | 21.0 | 39.1 |
| Acetophenone degradation | ||||
| Apc1 (c1A105)c | 32.2 | 21.6 | 1.3 | −1.4 |
| Apc2 (c1A200)c | 73.4 | 29.9 | −1.0 | 1.1 |
| Apc3 (c1A102)c | 35.8 | 33.9 | −2.4 | −2.6 |
| Apc4 (c1A100)c | 95.9 | 45.7 | 1.4 | −1.4 |
| Orf84 (c1A84) | 53.8 | 1.3 | 2.1 | 1.4 |
| Orf87 (c1A87) | 101.3 | 1.7 | −1.1 | 1.2 |
| p-Ethylphenol degradation | ||||
| PchF (EbA300) | 1.9 | 1.4 | 18.9 | 23.1 |
| ChnA (EbA307) | 1.1 | −1.0 | 7.2 | 6.9 |
| EbA309 | 1.5 | 1.6 | 12.4 | 13.4 |
| EbA310 | −1.5 | −1.4 | 9.1 | 8.7 |
| EbA312 | 1.4 | 1.2 | 48.4 | 47.5 |
| p-Hydroxyacetophenone degradation | ||||
| XccA (EbA314) | 1.6 | 1.9 | 35.5 | 35.6 |
| XccB (EbB9) | 1.7 | 1.6 | 88.3 | 85.3 |
| TioL (EbA306) | 1.3 | 1.2 | 28.4 | 32.5 |
| EbA318 | 1.7 | 1.6 | 41.6 | 40.7 |
| pEp and pAc-related proteins | ||||
| EbA329 | 2.7 | 2.6 | 15.7 | 32.3 |
| EbA332 | 4.2 | 2.1 | 102.1 | 259.7 |
| EbA335 | 6.7 | 1.3 | 4.9 | 9.4 |
| Paralogous ethylbenzene gene cluster | ||||
| EbdC2 (EbA5793) | 2.0 | 1.4 | 3.5 | 3.4 |
| Ped2 (EbA5789) | 2.9 | 1.6 | 20.6 | −1.0 |
Proteins are listed according to the reaction sequence. Protein names and open reading frame numbers (e.g., EbA309) are as described previously (38).
Cells were adapted to anaerobic growth with the selected substrates for at least five passages. Substrate abbreviations: Acp, acetophenone; Etb, ethylbenzene; pAc, p-hydroxyacetophenone; pEp, p-ethylphenol. Changes marked in bold are above the set threshold of significance (>|2.5|).
For protein species separated as two spots with different pIs, the n-fold change of the most-abundant spot is indicated. Data not shown revealed a similar ratio of n-fold changes.
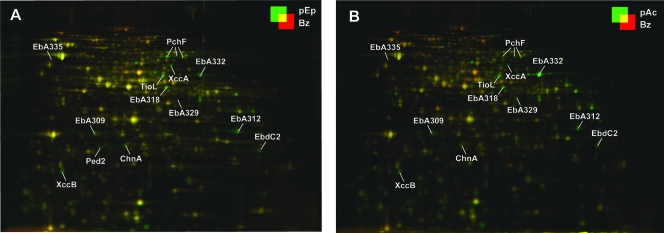
FIG. 4.
Differential protein profiles (2D DIGE) of substrate-adapted cells of “A. aromaticum” strain EbN1. Adaptation substrates for anaerobic growth were p-ethylphenol (A; pEp) and p-hydroxyacetophenone (B; pAc). Cultures adapted to anaerobic growth with benzoate (Bz) served as a reference state. Protein abbreviations are as described in the legend to Fig. 1. Changes (n-fold) in protein abundances of annotated proteins are indicated in Table 2.
FIG. 6.
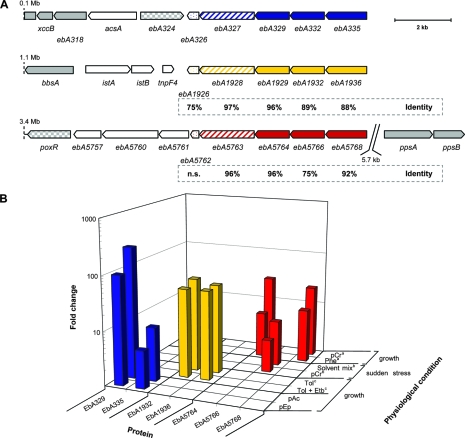
Gene products possibly involved in solvent stress tolerance of “A. aromaticum” strain EbN1. (A) Chromosomal gene locations are indicated by their nucleotide positions. Genes encoding stress-related proteins are highlighted in blue, yellow, and red. Hypothetical proteins, filled; predicted efflux pump, hatched; and predicted universal stress protein, dotted. Amino acid sequence identities of gene products (referring to EbA326-335) are indicated below the paralogous genes (n.s., no significant similarity). Genes encoding proteins of aromatic compound catabolism are highlighted in gray (filled), and respective regulatory proteins are checkered. (B) Changes (n-fold) in abundances of identified hypothetical proteins (EbA329-335, EbA1932-1936, and EbA5764-5768) during anaerobic growth shock with different aromatic substrates, as determined by 2D DIGE. Substrate abbreviations: pEp, p-ethylphenol; pAc, p-acetophenone; Tol, toluene; Etb, ethylbenzene; pCr, p-cresol; Phe, phenol. “a,” anaerobic growth with benzoate represents the reference state. No upregulation during anaerobic growth with phenylalanine, phenylacetate, benzyl alcohol, benzaldehyde, p-hydroxybenzoate, m-hydroxybenzoate, and o-aminobenzoate (52). “b,” anaerobic growth with succinate represents the reference state (49). “c,” anaerobic growth with benzoate represents the reference state. No induction during anaerobic growth with ethylbenzene (26).
Metabolite formation during anaerobic p-ethylphenol degradation.
The intermediates p-hydroxyacetophenone, p-hydroxybenzoate, and benzoate were identified in p-ethylphenol-utilizing cultures by GC-MS analysis. This finding was further verified by identification of corresponding 2,3,5,6-d4-isotopomers upon incubation with 2,3,5,6-d4-p-ethylphenol (Fig. 2). Accordingly, p-hydroxybenzoate and benzoate were identified when cells were grown with p-hydroxyacetophenone. Overall, the observed metabolites are in agreement with the proposed pathway (Fig. 1).
Adaptation to aromatic growth substrates.
Under all four studied adaptation conditions (i.e., p-ethylphenol, p-hydroxyacetophenone, ethylbenzene, and acetophenone), the primary adaptation substrate was readily utilized, while anaerobic growth with the three other aromatic substrates occurred in most cases only after prolonged incubation (Fig. 3), indicating that the respective degradation capacities had to be induced. Ethylbenzene- as well as acetophenone-adapted cultures required pronounced adaptation times (∼50 h) to start utilizing p-ethylphenol and p-hydroxyacetophenone for growth (Fig. 3A and C). Conversely, p-ethylphenol-adapted cultures displayed distinct adaptation times for the utilization of ethylbenzene and acetophenone (Fig. 3B). p-Hydroxyacetophenone-adapted cultures required an adaptation time of >160 h to start growing with acetophenone (Fig. 3D). Similarly long adaptation times (>100 h) to begin acetophenone utilization were observed for toluene-adapted cultures of strain EbN1 (data not shown), pointing to a special toxicity of acetophenone. Overall, these adaptation experiments already indicate the presence of two different degradation pathways which operate only in response to their respective substrates. The only exception to these congruent adaptation patterns was the simultaneous adaptation of p-hydroxyacetophenone-adapted cells to the utilization of ethylbenzene (Fig. 3D), which contrasts the absent expression of ethylbenzene-related genes under this adaptation condition (see Table 2 and Fig. 5).
FIG. 5.
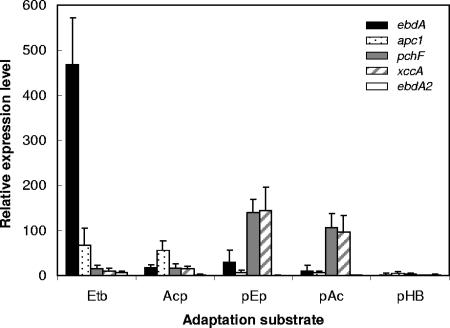
Relative expression levels of ebdA, apc1, pchF, and xccA in substrate-adapted cells of “A. aromaticum” strain EbN1. The selected genes encode catalytic subunits ethylbenzene dehydrogenase (ebdA), acetophenone carboxylase (apc1), predicted p-ethylphenol methylhydroxylase (pchF), predicted p-hydroxyacetophenone carboxylase (xccA), and paralogous catalytic subunit of ethylbenzene dehydrogenase (ebdA2). Benzoate-adapted cells were used as a reference state, and bcrC (encoding the γ-subunit of benzoyl-CoA reductase) served as a reference gene. Relative expression levels were determined by real-time RT-PCR. Abbreviations of adaptation substrates: Etb, ethylbenzene; Acp, acetophenone; pEp, p-ethylphenol; pAc, p-acetophenone; pHB, p-hydroxybenzoate.
Regulation of the degradation pathways.
The soluble proteomes of substrate-adapted cells of strain EbN1 were analyzed by 2D DIGE. In total, 87 protein spots were differentially regulated (n-fold change of >|2.5| compared to benzoate-adapted cells), of which 71 were identified by mapping of peptide masses. The identified proteins represented 51 different protein species, 25 of which could be assigned to the two studied degradation pathways (Table 2 and Fig. 4). In addition to the proteomic analysis, gene expression of the catalytic subunits of the initial enzymes of both pathways as well as the respective carboxylating enzymes was investigated on the RNA level (Fig. 5).
(i) Substrate-specific regulation of the ethylbenzene degradation pathway.
Subunits of both enzymes of the “upper” part of the ethylbenzene degradation pathway (EbdBCD, ethylbenzene dehydrogenase; Ped, (S)-1-phenylethanol dehydrogenase) were specifically increased in abundance (17- to 154-fold) during anaerobic growth with ethylbenzene but not with p-ethylphenol (Table 2). Accordingly, the relative expression level of ebdA was strongly increased in cells grown with ethylbenzene (>460-fold) and to a lower extent with acetophenone (17-fold) (Fig. 5), agreeing well with previously reported ethylbenzene-specific expression of ebdA and formation of EbdC and EbdD in strain EbN1 (26). Conversely, in p-ethylphenol- and p-hydroxyacetophenone-adapted cells, only a low-level induction of ebdA transcription was observed (30- and 9-fold, respectively), agreeing with the concurrent lack of significant EbdBCD and Ped formation.
Significantly elevated abundances (up to 95-fold) (Table 2) of identified subunits of acetophenone carboxylase (Apc1234) as well as an increased apc1 expression level (up to 66-fold) (Fig. 5) were only observed in cells grown with ethylbenzene and acetophenone, agreeing with previous observations (26). The lack of significant formation of the Apc proteins during anaerobic growth with p-ethylphenol and p-hydroxyacetophenone (ranging from −2.6 to 1.4-fold) and the marginally increased expression of apc1 (7- and 6-fold, respectively) point to a narrow effector spectrum of the predicted sensor/regulator system Tcs1/Tcr1 for acetophenone.
(ii) Paralogous ethylbenzene dehydrogenase (EbdABC2) is not involved in anaerobic p-ethylphenol degradation.
The chromosome of strain EbN1 contains a second gene cluster related to the “upper” part of the ethylbenzene degradation pathway encoded at approximately 3.4 Mb of the chromosome (Fig. 1) (38). The physiological functions of the encoded paralogs (EbdABCD2, Ped2, and EbA5797) are currently unknown. The previously observed induction of paralogous (S)-1-phenylethanol dehydrogenase (Ped2) during anaerobic growth with p-cresol, in analogy to induction of ebd genes by toluene, led to the assumption that the paralogous ethylbenzene dehydrogenase (EbdABCD2) could be involved in p-ethylphenol degradation (52). Agreeing with such an assumption, p-ethylphenol was oxidized at high rates by ethylbenzene dehydrogenase, though with a significantly higher Km than ethylbenzene (40.5 μM versus 0.45 μM) (47). However, the abundances of identified EbdC2 and Ped2 were only slightly increased during anaerobic growth with p-ethylphenol and p-hydroxyacetophenone (up to 3.5-fold for EbdC2 and 20.6-fold for Ped2, respectively) (Table 2). Also, expression of ebdA2 (encoding the α-subunit of paralogous ethylbenzene dehydrogenase) was only marginally increased during growth with ethylbenzene (7.0-fold) but not with any other substrate (Fig. 5). Considering the strong increase in EbdC abundance (154.0-fold) and the high expression level of ebdA (>460-fold) in ethylbenzene-grown cells, a metabolic function of the paralogous enzyme in anaerobic p-ethylphenol degradation seems unlikely.
(iii) Coordinated and substrate-specific regulation of the p-ethylphenol and p-hydroxyacetophenone degradation pathway.
Due to their specific upregulation (up to 259-fold) in p-ethylphenol- and p-hydroxyacetophenone-adapted cells (Table 2 and Fig. 4), 12 proteins could be related to a common degradation pathway for both compounds. The predicted functions of these proteins are as follows: subunit of a p-cresol methylhydroxylase-like protein (PchF), thiolase (TioL), cyclohexanol dehydrogenase (ChnA), putative alcohol dehydrogenase (EbA309), flavin adenine dinucleotide-linked oxidase (EbA310), sugar phosphatase (EbA312), biotin-dependent carboxylase (XccAB), and four hypothetical proteins (EbA318, EbA329, EbA332, and EbA335). Analogously, expression analysis of pchF and xccA revealed a specific upregulation as well as highly similar relative expression levels during anaerobic growth with p-ethylphenol (140- and >140-fold, respectively) and p-hydroxyacetophenone (107- and 97-fold, respectively) (Fig. 5). In contrast, their expression was only slightly increased in ethylbenzene- and acetophenone-adapted cells (<16-fold) (Fig. 5), agreeing well with the proteomic data.
Most of these proteins are encoded in a large operon-like structure (∼15 kb) on the chromosome of strain EbN1 (38). The only exceptions are the hypothetical proteins EbA329, EbA332, and EbA335, which are encoded in a second operon-like structure, at a distance of only 2.5 kb upstream of the first one (Fig. 1). Interestingly, in between these two operons lies the EbA324 gene, encoding a putative sigma 54-dependent transcriptional regulator, possibly involved in gene regulation of the p-ethylphenol degradation pathway.
The organization of all genes of the p-ethylphenol degradation pathway in a single operon-like structure is indicative of a coordinated regulation, which contrasts with the described sequential regulation of the two catabolic operons of the ethylbenzene degradation pathway (26). This assumption is supported by the highly similar pchF and xccA expression levels as well as the conspicuously similar increased abundances of all identified proteins encoded in the gene cluster (Table 2 and Fig. 5) during anaerobic growth with p-ethylphenol and p-hydroxyacetophenone. Moreover, RT analysis applying only the pchF antisense primer enables PCR amplification of both pchF and xccA, pointing to expression of the whole operon as a polycistronic transcript (see Fig. S1 in the supplemental material). Overall, the differential proteomic and gene expression data demonstrate a clear difference between the ethylbenzene and p-ethylphenol degradation pathways from a regulatory point of view.
(iv) Sigma 54-dependent regulation of the anaerobic p-ethylphenol degradation pathway.
Regulation of aromatic compound degradation pathways is known to occur on the transcriptional level, mediated by a variety of transcriptional regulators sensing the aromatic stimulus (16, 26). The two gene clusters, induced by p-ethylphenol and p-hydroxyacetophenone, are separated by the EbA324 gene, encoding a putative sigma 54-dependent transcriptional regulator in trans (Fig. 1) (38). Interestingly, both gene clusters harbor the −12/−24 consensus sequence of sigma 54-dependent promoters (5′-TGGC-N7-TTGCA-3′) (19) approximately 100 bp upstream of the translational start of acsA and the EbA335 gene, suggesting involvement of EbA324 in initiation of their transcription. Sequence analysis of EbA324 classified it as an NtrC-type transcriptional regulator, comprising a C-terminal helix-turn-helix, a sigma 54 interaction, and an N-terminal receptor domain (31). NtrC-type regulators are known to be involved in transcription of genes with diverse physiological functions (27). The sigma 54-dependent XylR regulator of aerobic toluene and xylene degradation in Pseudomonas putida is activated upon recognition of toluene and xylenes as well as their benzyl alcohol and benzaldehyde derivatives (1). Interestingly, the amino acid sequence of EbA324 shares 40% sequence identity with XylR, supporting its predicted function. Since p-ethylphenol and p-hydroxyacetophenone induce expression of the p-ethylphenol-related gene cluster to the same extent, one may speculate that both molecules are recognized by EbA324, agreeing with the known broad effector spectrum of related XylR (1). However, chemically related ethylbenzene and acetophenone apparently do not belong to the effector spectrum of EbA324.
Enzyme candidates of anaerobic p-ethylphenol and p-hydroxyacetophenone degradation.
Based on their predicted functions, most of the specifically upregulated proteins can be correlated to individual reactions within the proposed p-ethylphenol degradation pathway (Fig. 1).
(i) p-Ethylphenol methylhydroxylase.
The initial dehydrogenation of p-ethylphenol, forming 1-(4-hydroxyphenyl)-ethanol, could be catalyzed by the p-cresol methylhydroxylase-like protein (PchCF). Dioxygen-independent dehydrogenation of p-ethylphenol by an ethylphenol methylhydroxylase (EPMH) has been reported for Pseudomonas putida JD1 (43) and Aspergillus fumigatus (22). Interestingly, P. putida JD1 possesses two different, but highly similar, hydroxylases for p-cresol and p-ethylphenol degradation (43), for which gene sequences are presently not available. Indeed, PchF of strain EbN1 shares 31% sequence identity with the p-cresol methylhydroxylase (PchF; PCMH) from P. putida NCIMB 9866, which is also active on p-ethylphenol (30). Due to the hydroxyl group in para position, enabling intermediary formation of a quinone methide, the redox potential of the EPMH flavin cofactor (+280 mV) is sufficient for initial dehydrogenation of p-ethylphenol (43). In contrast, dehydrogenation of ethylbenzene necessitates the molybdenum cofactor and an electron acceptor with a higher redox potential (+380 mV) to achieve reasonable oxidation rates of the chemically stable substrate (23). Thus, oxidation of p-ethylphenol by the PCMH-like protein (PchCF) in strain EbN1 seems reasonable.
(ii) 1-(4-Hydroxyphenyl)-ethanol dehydrogenase.
The oxidation of 1-(4-hydroxyphenyl)-ethanol to p-hydroxyacetophenone might also be performed by PchCF, since purified EPMH from P. putida JD1 forms p-hydroxyacetophenone as a major oxidation product from p-ethylphenol (43). However, in the initial reaction, EPMH stereospecifically forms the R(+)-1-(4-hydroxyphenyl)-ethanol enantiomer in high excess (>90%), which is only slowly oxidized by EPMH (44). Rapid oxidation of the R(+)-enantiomer in crude extracts of P. putida JD1 was suggested to result from the involvement of a second, so-far-unknown enzyme (44). In strain EbN1, the two predicted alcohol dehydrogenases (ChnA and EbA309) may catalyze this oxidation, since both display high sequence similarity to (S)-1-phenylethanol dehydrogenase of strain EbN1 (Ped; 33% and 36% identity, respectively), catalyzing the analogous reaction in the pathway for anaerobic ethylbenzene degradation (24).
(iii) Carboxylase.
Carboxylation of p-hydroxyacetophenone is most probably catalyzed by the biotin-dependent carboxylase XccABC. This enzyme does not share any homology with acetophenone carboxylase (Apc1-5), acetone carboxylase (AcxABC), or biotin-independent phenylphosphate carboxylase (PpcABCD) of strain EbN1 (38). Thus, it represents the fourth type of carboxylase in strain EbN1 and can be expected to employ a distinct catalytic mechanism. Biotin-dependent carboxylases are a diverse group of enzymes found in various biosynthetic pathways in pro- and eukaryotes (8). The two-step catalysis involves (i) carboxylation of biotin and (ii) transcarboxylation from biotin to the respective acceptor molecules.
(iv) Further enzymes.
Activation of the carboxylation product to its respective CoA-ester could be performed by the acetoacetyl-CoA synthetase-like protein AcsA, which is not homologous to the benzoylacetate CoA-ligase (Bal) of the acetophenone degradation pathway. Subsequently, the predicted thiolase (TioL) may thiolytically cleave the CoA-activated carboxylation product to p-hydroxybenzoyl-CoA and acetyl-CoA. At present, however, a hydrolytic cleavage of the carboxylation product without prior CoA activation as known from oxaloacetate hydrolase (29) cannot be excluded.
The final reductive dehydroxylation, yielding benzoyl-CoA, is supposed to be catalyzed by the predicted hydroxybenzoyl-CoA reductase (HcrABC), as suggested for the anaerobic phenol and p-cresol degradation pathways (38, 52). However, subunits of the enzyme from strain EbN1 could not be detected on 2D DIGE gels until now, and a transcript of hcrB could not be detected in significant amounts in p-hydroxybenzoate-grown cells (data not shown; also see the supplemental material). Thus, the last step in the transformation of p-ethylphenol (also phenol and p-cresol) to benzoyl-CoA in strain EbN1 is presently not well understood.
It should be noted, however, that aerobic utilization of m-hydroxybenzoate also induced formation of the proteins of the p-ethylphenol pathway (PchF, TioL, ChnA, EbA310, XccAB, EbA318, and EbA332) (52), though to a markedly lower extent (maximum 30-fold). The physiological meaning of this regulatory pattern remains elusive at present.
Possible involvement of a specific solvent stress system in p-ethylphenol and p-hydroxyacetophenone metabolism.
Besides the genes predicted to be involved in the anaerobic degradation of p-ethylphenol and p-hydroxyacetophenone, three further gene products, EbA329, EbA332, and EbA335, displayed strongly increased abundances during anaerobic growth of strain EbN1 with both aromatic substrates (Table 2 and Fig. 4). All of them represent proteins of unknown function, encoded together with another hypothetical protein (EbA326) and a putative transport protein (EbA327) in an operon-like structure shortly upstream of the p-ethylphenol gene cluster (Fig. 1) (38).
Further analysis of EbA326 revealed domains related to a cytoplasmic universal stress protein (UpsA) from Escherichia coli (31), the expression of which is enhanced during prolonged exposure to stress agents (33). Furthermore, analysis of EbA327 revealed two transmembrane domains and a sensory domain related to an RND-type efflux pump (AcrB) from E. coli (31). AcrB is assumed to capture its substrate either from within the inner membrane or from the cytoplasm (53), followed by excretion through an outer membrane channel (TolC). The AcrB-TolC interaction is mediated by a periplasmic accessory protein (AcrA), and the transport process is driven by proton motive force (36). Energy-dependent efflux pumps of the RND family are known to be involved in solvent tolerance (e.g., n-hexane or toluene) in several gram-negative bacteria, in particular E. coli and Pseudomonas sp. (42).
Interestingly, the hypothetical proteins EbA332 and EbA335 possess signal sequences for periplasmic localization (13). Though sequence analysis of EbA332 and EbA335 revealed no significant similarity to AcrA and TolC, respectively, the proteins are relatively similar in size (400 and 500 amino acids [aa] for AcrA and TolC, respectively, and 450 and 550 aa for EbA332 and EbA335, respectively). In addition, prediction of EbA335 secondary structure elements indicated the formation of numerous beta-sheets and turn regions, possibly forming a beta-barrel as seen in outer membrane porins (10). Considering the specific formation of these proteins in the presence of aromatic solvents, one may speculate that the EbA327, EbA332, and EbA335 genes encode a so-far-uncharacterized, solvent-specific efflux system, analogous to the AcrAB-TolC system of E. coli and Pseudomonas sp. (see Fig. S2 in the supplemental material).
Remarkably, the chromosome of strain EbN1 harbors two additional gene clusters which display high sequence similarity (76 to 97% aa sequence identities) (Fig. 6A) to the EbA326-335 cluster and exhibit the same gene order (Fig. 6) (38). The first cluster (EbA1926-1936) is located adjacent to the catabolic genes of anaerobic toluene degradation and the second (EbA5762-5768) is near ppsB (encoding a subunit of the predicted phenylphosphate synthetase) (Fig. 6) (52). In addition, the three paralogous gene clusters are specifically expressed in the presence of different aromatic compounds, which correlate well with the genomic context in each case (Fig. 6B). The products of genes EbA326-335, EbA1926-1936, and EbA5762-5768 were specifically formed during anaerobic growth with, e.g., p-ethylphenol, toluene (26), and phenol/p-cresol (52), respectively. Formation of EbA5762-5768 was also recently observed in succinate-utilizing cells of strain EbN1 suddenly stressed with phenolic compounds (49). These analogous regulatory patterns support the predicted solvent-related function of the encoded proteins and point to a fine-tuned system of solvent stress tolerance in strain EbN1.
Notably, highly similar gene clusters (42 to 88% aa sequence identity; see Fig. S3 in the supplemental material) were detected in the genomes of plant-associated Azoarcus sp. strain BH72 (42 to 80% aa identity) (25), anaerobic aromatic compound-degrading Thauera aromatica K172 (80 to 88% aa) (28), and aerobic aromatic compound-degrading and solvent-tolerant P. putida KT2440 (32). In the case of Azoarcus sp. strain BH72 and T. aromatica K172, these genes are also located adjacent to genes of aerobic phenol and anaerobic toluene degradation, respectively (see Fig. S3 in the supplemental material), further supporting a general solvent stress-related function of these gene clusters.
Supplementary Material
Acknowledgments
We are grateful to D. Lange and internship student Heiko Nacke (both from Bremen) and C. Karger and A. Sobotta (both from Potsdam) for technical assistance. We are indebted to F. Widdel for general support of proteomic studies in our group.
This study was supported by the Deutsche Forschungsgemeinschaft and the Max-Planck-Gesellschaft.
Footnotes
Published ahead of print on 6 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abril, M. A., C. Michan, K. N. Timmis, and J. L. Ramos. 1989. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 1716782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acuna-Arguelles, M. E., P. Olguin-Lora, and E. Razo-Flores. 2003. Toxicity and kinetic parameters of the aerobic biodegradation of the phenol and alkylphenols by a mixed culture. Biotechnol. Lett. 25559-564. [DOI] [PubMed] [Google Scholar]
- 3.Ball, H. A., H. A. Johnson, M. Reinhard, and A. M. Spormann. 1996. Initial reactions in anaerobic ethylbenzene oxidation by a denitrifying bacterium, strain EB1. J. Bacteriol. 1785755-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 5.Brenes, M., C. Romero, A. García, F. J. Hidalgo, and M. V. Ruiz-Méndez. 2004. Phenolic compounds in olive oils intended for refining: formation of 4-ethylphenol during olive paste storage. J. Agric. Food Chem. 528177-8181. [DOI] [PubMed] [Google Scholar]
- 6.Champion, K. M., K. Zengler, and R. Rabus. 1999. Anaerobic degradation of ethylbenzene and toluene in denitrifying strain EbN1 proceeds via independent substrate-induced pathways. J. Mol. Microbiol. Biotechnol. 1157-164. [PubMed] [Google Scholar]
- 7.Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 712871-2882. [DOI] [PubMed] [Google Scholar]
- 8.Cronan, J. E., and G. L. Waldrop. 2002. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 41407-435. [DOI] [PubMed] [Google Scholar]
- 9.Darby, J. M., D. G. Taylor, and D. J. Hopper. 1987. Hydroquinone as the ring-fission substrate in the catabolism of 4-ethylphenol and 4-hydroxyacetophenone by Pseudomonas putida JD1. J. Gen. Microbiol. 1332137-2146. [Google Scholar]
- 10.Delcour, A. H. 2002. Structure and function of pore-forming beta-barrels from bacteria. J. Mol. Microbiol. Biotechnol. 41-10. [PubMed] [Google Scholar]
- 11.Dias, L., S. Dias, T. Sancho, H. Stender, A. Querol, M. Malfeito-Ferreira, and V. Loureiro. 2003. Identification of yeasts isolated from wine-related environments and capable of producing 4-ethylphenol. Food Microbiol. 20567-574. [Google Scholar]
- 12.Doherty, N. S., B. H. Littman, K. Reilly, A. C. Swindell, J. M. Buss, and N. L. Anderson. 1998. Analysis of changes in acute-phase plasma proteins in an acute inflammatory response and in rheumatoid arthritis using two-dimensional gel electrophoresis. Electrophoresis 19355-363. [DOI] [PubMed] [Google Scholar]
- 13.Emanuelsson, O., S. Brunak, G. von Heijne, and H. Nielsen. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protocols 2953-971. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, G. 2008. Anaerobic metabolism of aromatic compounds. Ann. N. Y. Acad. Sci. 112582-99. [DOI] [PubMed] [Google Scholar]
- 15.Gade, D., J. Thiermann, D. Markowsky, and R. Rabus. 2003. Evaluation of two-dimensional difference gel electrophoresis for protein profiling. Soluble proteins of the marine bacterium Pirellula sp. strain 1. J. Mol. Microbiol. Biotechnol. 5240-251. [DOI] [PubMed] [Google Scholar]
- 16.Gerischer, U. 2002. Specific and global regulation of genes associated with the degradation of aromatic compounds in bacteria. J. Mol. Microbiol. Biotechnol. 4111-121. [PubMed] [Google Scholar]
- 17.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56345-369. [DOI] [PubMed] [Google Scholar]
- 18.Heider, J. 2007. Adding handles to unhandy substrates: anaerobic hydrocarbon activation mechanisms. Curr. Opin. Chem. Biol. 11188-194. [DOI] [PubMed] [Google Scholar]
- 19.Helmann, J. D., and M. J. Chamberlin. 1988. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 57839-872. [DOI] [PubMed] [Google Scholar]
- 20.Höffken, H. W., M. Duong, T. Friedrich, M. Breuer, B. Hauer, R. Reinhardt, R. Rabus, and J. Heider. 2006. Crystal structure and enzyme kinetics of the (S)-specific 1-phenylethanol dehydrogenase of the denitrifying bacterium strain EbN1. Biochemistry 4582-93. [DOI] [PubMed] [Google Scholar]
- 21.Jeno, P., T. Mini, S. Moes, E. Hintermann, and M. Horst. 1995. Internal sequences from proteins digested in polyacrylamide gels. Anal. Biochem. 22475-82. [DOI] [PubMed] [Google Scholar]
- 22.Jones, K. H., P. W. Trudgill, and D. J. Hopper. 1994. 4-Ethylphenol metabolism by Aspergillus fumigatus. Appl. Environ. Microbiol. 601978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kniemeyer, O., and J. Heider. 2001. Ethylbenzene dehydrogenase, a novel hydrocarbon-oxidizing molybdenum/iron-sulfur/heme enzyme. J. Biol. Chem. 27621381-21386. [DOI] [PubMed] [Google Scholar]
- 24.Kniemeyer, O., and J. Heider. 2001. (S)-1-Phenylethanol dehydrogenase of Azoarcus sp. strain EbN1, an enzyme of anaerobic ethylbenzene catabolism. Arch. Microbiol. 176129-135. [DOI] [PubMed] [Google Scholar]
- 25.Krause, A., A. Ramakumar, D. Bartels, F. Battistoni, T. Bekel, J. Boch, M. Bohm, F. Friedrich, T. Hurek, L. Krause, B. Linke, A. C. McHardy, A. Sarkar, S. Schneiker, A. A. Syed, R. Thauer, F.-J. Vorholter, S. Weidner, A. Puhler, B. Reinhold-Hurek, O. Kaiser, and A. Goesmann. 2006. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 241385-1391. [DOI] [PubMed] [Google Scholar]
- 26.Kühner, S., L. Wöhlbrand, I. Fritz, W. Wruck, C. Hultschig, P. Hufnagel, M. Kube, R. Reinhardt, and R. Rabus. 2005. Substrate-dependent regulation of anaerobic degradation pathways for toluene and ethylbenzene in a denitrifying bacterium, strain EbN1. J. Bacteriol. 1871493-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kustu, S., E. Santero, J. Keener, D. Popham, and D. Weiss. 1989. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 53367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leuthner, B., C. Leutwein, H. Schulz, P. Horth, W. Haehnel, E. Schiltz, H. Schagger, and J. Heider. 1998. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica—a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol. Microbiol. 28615-628. [DOI] [PubMed] [Google Scholar]
- 29.Lu, Z., X. Feng, L. Song, Y. Han, A. Kim, O. Herzberg, W. R. Woodson, B. M. Martin, P. S. Mariano, and D. Dunaway-Mariano. 2005. Diversity of function in the isocitrate lyase enzyme superfamily: the Dianthus caryophyllus petal death protein cleaves α-keto and α-hydroxycarboxylic acids. Biochemistry 4416365-16376. [DOI] [PubMed] [Google Scholar]
- 30.McIntire, W., D. J. Hopper, and T. P. Singer. 1985. p-Cresol methylhydroxylase. Assay and general properties. Biochem. J. 228325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bork, V. Buillard, L. Cerutti, R. Copley, E. Courcelle, U. Das, L. Daugherty, M. Dibley, R. Finn, W. Fleischmann, J. Gough, D. Haft, N. Hulo, S. Hunter, D. Kahn, A. Kanapin, A. Kejariwal, A. Labarga, P. S. Langendijk-Genevaux, D. Lonsdale, R. Lopez, I. Letunic, M. Madera, J. Maslen, C. McAnulla, J. McDowall, J. Mistry, A. Mitchell, A. N. Nikolskaya, S. Orchard, C. Orengo, R. Petryszak, J. D. Selengut, C. J. A. Sigrist, P. D. Thomas, F. Valentin, D. Wilson, C. H. Wu, and C. Yeats. 2007. New developments in the InterPro database. Nucleic Acids Res. 35D224-D228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. P. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, C. M. Fraser, et al. 2003. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 5630. [DOI] [PubMed] [Google Scholar]
- 33.Nyström, T., and F. C. Neidhardt. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11537-544. [DOI] [PubMed] [Google Scholar]
- 34.Oelmüller, U., N. Krüger, A. Steinbüchel, and G. C. Friedrich. 1990. Isolation of procaryotic RNA and detection of specific mRNA with biotinylated probes. J. Microbiol. Methods 1173-81. [Google Scholar]
- 35.Pfaffl, M. W. 2001. A new mathematical model for relative quantification of realtime RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piddock, L. J. V. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4629-636. [DOI] [PubMed] [Google Scholar]
- 37.Rabus, R., M. Kube, A. Beck, F. Widdel, and R. Reinhardt. 2002. Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 178506-516. [DOI] [PubMed] [Google Scholar]
- 38.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 18327-36. [DOI] [PubMed] [Google Scholar]
- 39.Rabus, R., and F. Widdel. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 16396-103. [DOI] [PubMed] [Google Scholar]
- 40.Rabus, R., H. Wilkes, A. Behrends, A. Armstroff, T. Fischer, A. J. Pierik, and F. Widdel. 2001. Anaerobic initial reaction of n-alkanes in a denitrifying bacterium: evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical in n-hexane metabolism. J. Bacteriol. 1831707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramakers, C., J. M. Ruijter, R. H. L. Deprez, and A. F. M. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 33962-66. [DOI] [PubMed] [Google Scholar]
- 42.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-González, A. Rojas, W. Terán, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56743-768. [DOI] [PubMed] [Google Scholar]
- 43.Reeve, C. D., M. A. Carver, and D. J. Hopper. 1989. The purification and characterization of 4-ethylphenol methylenehydroxylase, a flavocytochrome from Pseudomonas putida JD1. Biochem. J. 263431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeve, C. D., M. A. Carver, and D. J. Hopper. 1990. Stereochemical aspects of the oxidation of 4-ethylphenol by the bacterial enzyme 4-ethylphenol methylenehydroxylase. Biochem. J. 269815-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seifert, W. K., and W. G. Howells. 1969. Interfacially active acids in a California crude oil. Anal. Chem. 41554-562. [Google Scholar]
- 46.Spormann, A. M., and F. Widdel. 2000. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation 1185-105. [DOI] [PubMed] [Google Scholar]
- 47.Szaleniec, M., C. Hagel, M. Menke, P. Nowak, M. Witko, and J. Heider. 2007. Kinetics and mechanism of oxygen-independent hydrocarbon hydroxylation by ethylbenzene dehydrogenase. Biochemistry 467637-7646. [DOI] [PubMed] [Google Scholar]
- 48.Tanner, A., and D. J. Hopper. 2000. Conversion of 4-hydroxyacetophenone into 4-phenyl acetate by a flavin adenine dinucleotide-containing Baeyer-Villiger-type monooxygenase. J. Bacteriol. 1826565-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trautwein, K., S. Kühner, L. Wöhlbrand, T. Halder, K. Kuchta, A. Steinbüchel, and R. Rabus. 2008. Solvent stress response of denitrifying “Aromatoleum aromaticum” strain EbN1. Appl. Environ. Microbiol. 742267-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Beek, S., and F. G. Priest. 2000. Decarboxylation of substituted cinnamic acids by lactic acid bacteria isolated during malt whisky fermentation. Appl. Environ. Microbiol. 665322-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12259-276. [DOI] [PubMed] [Google Scholar]
- 52.Wöhlbrand, L., B. Kallerhoff, D. Lange, P. Hufnagel, J. Thiermann, R. Reinhardt, and R. Rabus. 2007. Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1. Proteomics 72222-2239. [DOI] [PubMed] [Google Scholar]
- 53.Yu, E. W., J. R. Aires, and H. Nikaido. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 1855657-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.