Abstract
Cytochrome cbb3-type oxidases are members of the heme copper oxidase superfamily and are composed of four subunits. CcoN contains the heme b-CuB binuclear center where oxygen is reduced, while CcoP and CcoO are membrane-bound c-type cytochromes thought to channel electrons from the donor cytochrome into the binuclear center. Like many other bacterial members of this superfamily, the cytochrome cbb3-type oxidase contains a fourth, non-cofactor-containing subunit, which is termed CcoQ. In the present study, we analyzed the role of CcoQ on the stability and activity of Rhodobacter capsulatus cbb3-type oxidase. Our data showed that CcoQ is a single-spanning membrane protein with a Nout-Cin topology. In the absence of CcoQ, cbb3-type oxidase activity is significantly reduced, irrespective of the growth conditions. Blue native polyacrylamide gel electrophoresis analyses revealed that the lack of CcoQ specifically impaired the stable recruitment of CcoP into the cbb3-type oxidase complex. This suggested a specific CcoQ-CcoP interaction, which was confirmed by chemical cross-linking. Collectively, our data demonstrated that in R. capsulatus CcoQ was required for optimal cbb3-type oxidase activity because it stabilized the interaction of CcoP with the CcoNO core complex, leading subsequently to the formation of the active 230-kDa cbb3-type oxidase complex.
Aerobic and facultative aerobic bacteria usually contain multiple respiratory oxidases that facilitate their adaptation to different environmental O2 concentrations. Most of these terminal oxidases belong to the heme copper oxidase superfamily (12), which is characterized by the presence of a six-coordinated low-spin heme and a unique binuclear center composed of a high-spin heme and a copper atom (CuB) within the subunit I (37). Besides this common feature, the heme copper oxidase superfamily is quite diverse in terms of electron donor, oxygen affinity, subunit composition, and heme types. Based on structural analyses, the heme copper oxidases were originally classified into three subfamilies: the aa3/bo3-like oxidases (subfamily A), the ba3-like oxidases (subfamily B), and the cbb3-type oxidases (subfamily C) (32), but the accumulating genome sequence data indicate that the diversity within this superfamily might allow distinguishing up to eight subfamilies (16).
The cbb3-type cytochrome oxidases (cbb3-Cox) are the second most abundant oxidases after the A subfamily and constitute for more than 20% of the heme oxidase superfamily genome sequences (8, 16, 34). Despite the relative abundance and their likely importance for the life cycle of many pathogenic bacteria extending from Vibrio cholerae to Helicobacter pylori (26), no structural data are thus far available, and the information about the assembly and subunit interactions of these enzymes is limited. cbb3-Cox is composed of four subunits; the catalytic subunit I (CcoN/FixN) contains the diagnostic six histidine residues which ligate the low-spin heme b and a high-spin heme b3-CuB binuclear center, where oxygen reduction takes place (13). The oxygen affinity of cbb3-Cox is about fivefold higher than that of aa3-type cytochrome oxidase (aa3-Cox), which is probably important for respiration under lower oxygen concentrations (33). The subunits II (CcoO/FixO) and III (CcoP/FixP) are membrane-bound c-type cytochromes (cyt c), required for transferring electrons from the donor cyt c to the catalytic binuclear center within the subunit I (13, 19, 49). Subunit IV (CcoQ/FixQ) is suggested to be a small, single-spanning membrane protein of unknown function. In contrast to subunits I to III, CcoQ is apparently not essential for the stability or activity of cbb3-Cox in Rhodobacter sphaeroides (29) and Bradyrhizobium japonicum (55). Nevertheless, CcoQ appears to be a bona fide subunit of this enzyme, because it is detectable in purified enzyme preparations of B. japonicum (56) and because it forms, together with CcoN, CcoP, and CcoO, a biologically active, 230-kDa complex in R. capsulatus membranes, as revealed by blue-native-polyacrylamide gel electrophoresis (BN-PAGE) (21). Based on initial studies with a CcoQ deletion strain in R. sphaeroides, it has been suggested that CcoQ monitors electron flow through cbb3-Cox (30, 31) and transmits a thus-far-uncharacterized signal to the RegB/RegA (PrrB/PrrA) two-component system (10, 11), which is a global regulator of multiple energy-generating and energy-consuming processes in Rhodobacter species (9). Alternatively, it has also been proposed that under aerobic conditions, CcoP would be highly susceptible toward proteolytic degradation and that CcoQ might be required for protecting CcoP (31). The necessity to protect cbb3-Cox against high oxygen concentrations by recruiting CcoQ as an additional subunit (31) was suggested to be related to the evolution of terminal oxidases (8, 32, 40). cbb3-Cox is predicted to be a “primitive” form of oxidase, which probably has evolved from the anaerobic enzyme NO reductase (40, 57) and as such might be more susceptible to oxidative damage.
The occurrence of a fourth subunit associated with the core subunits of the terminal oxidases is observed not only for cbb3-Cox but also for other members of the heme copper oxidase superfamily, such as the aa3-Cox of Paracoccus denitrificans (53) and R. sphaeroides (45) or the bo3-type ubiquinol oxidase of Escherichia coli (3, 27, 38). However, whether the fourth subunits in different terminal oxidases execute comparable functions, e.g., monitoring electron flow or protecting individual subunits, is currently unknown.
An intrinsic advantage of using R. capsulatus over R. sphaeroides is that in the former species cbb3-Cox is the only cyt c oxidase (13, 18). Thus, the determination of the cyt c oxidase activities is not complicated by additional related enzymes with similar activities. In the present study, we analyzed the role of CcoQ on the stability and activity of R. capsulatus cbb3-Cox. By combining activity measurements, BN-PAGE, and chemical cross-linking, we demonstrated that CcoQ specifically interacts with the CcoP subunit of cbb3-Cox and that this interaction is important for the stability of cbb3-Cox. In the absence of CcoQ, the amount of the active 230-kDa cbb3-Cox complex was significantly reduced, which is in line with the reduced cbb3-Cox activity detected in the CcoQ-null (ΔccoQ) strain. In contrast to what was observed with R. sphaeroides, the cbb3-Cox activity in R. capsulatus was impaired by the lack of CcoQ subunit under all of the growth conditions tested and not limited to aerobic conditions only (31). This indicates that in R. capsulatus the primary role of CcoQ is not to protect CcoP against proteolytic degradation under aerobic conditions but rather to stabilize the interaction of the CcoP subunit with a preassembled CcoNO complex.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmid used are listed in Table 1. E. coli strains harboring plasmids were grown in LB medium supplemented with appropriate antibiotics (100, 50, and 12.5 μg/ml for ampicillin, kanamycin, and tetracycline, respectively). R. capsulatus strains were grown in Sistrom's minimal medium A (44) or in enriched medium MPYE (4) at 35°C in liquid cultures in the dark. For semiaerobic growth, 500-ml cultures were grown in 1,000-ml flasks and were shaken at 100 rpm. For aerobic growth, 500-ml cultures were grown in 5,000-ml flasks and were shaken at 200 rpm. Anaerobic-photosynthetic growth was achieved by culturing the cells in completely filled 1,000 ml Duran bottles that were incubated at 35°C under continuous illumination of 20 W/m2 using Osram light bulbs. Light intensity was measured with a LI-250A light meter (Li-COR Biosciences, Lincoln, NE).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description and relevant phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| HB101 | F−proA2 hsdS20(rB− mB−) recA13 ara14 lacY1 galK2 rpsL20 supE44 xyl-5 mtl-1 | 39 |
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 39 |
| R. capsulatus | ||
| 37b4 | Wild-type strain for the preparation of cell extracts for in vitro transcription-translation assays | 21 |
| MT1131 | crtD121, Rifr, cbb3-Cox wild type | 4 |
| GK32 | ΔccoNO::Kan; cbb3-Cox minus | 18 |
| CW2 | ΔccoI::Spec, cbb3-Cox minus | 20 |
| M7G | ccoP269 | 23 |
| Plasmids | ||
| pRK415 | Tetr | 6 |
| pRK2013 | Kanr | 6 |
| pOX15 | ccoNOQP in pRK415 | 18 |
| pAP4 | ccoNOP in pRK415 | This study |
| pET22b-CcoQ | ccoQ under T7 promoter control | This study |
| pC2P2.71 | cycA under lac promoter control | 7 |
Tetr, tetracycline resistance; Ampr, ampicillin resistance; Rifr, rifampin resistance; Kanr, kanamycin resistance.
Molecular genetic techniques.
Standard molecular genetic techniques were performed as described by Sambrook et al. (39). For in vitro synthesis of CcoQ, the corresponding gene was PCR amplified by using plasmid pOX15 as a template and cloned into the pET22b expression vector (Novagen, Bad Schwalbach, Germany), yielding pET22b-CcoQ. For constructing the plasmid pAP4, the 1.3-kb BamHI fragment of pOX15 was cloned into pBluescript SK(+), resulting in plasmid pAP1. This plasmid was used as a template for inverse PCR with the primer CcoQ-1 (5′-ACG CAG GAT ATG ATA GTC CAT CCC CAG CTC C-3′) and CcoQ-2 (GGC TGA GAC CCC GGA CAC GAC GAC GGC GCC A-3′). The PCR amplified plasmid (pAP2) lacking ccoQ was ligated and transformed into E. coli DH5α. The 1.15-kb BamHI fragment of pAP2 was isolated and used to replace the 1.3-kb BamHI fragment of pOX15, to yield pAP4, carrying an in-frame deletion of 49 amino acids within CcoQ.
Preparation of cell extracts and ICM.
High-speed supernatants (S-135 extract) of cell homogenates for efficient in vitro transcription/translation of Rhodobacter proteins were prepared from R. capsulatus strain 37b4. Intracytoplasmic membranes (ICM) were prepared as described previously (14, 21, 52) from R. capsulatus strain MT1131, which exhibits wild-type cbb3-Cox activity and from the different MT1131 derivatives listed in Table 1.
In vitro protein synthesis, protease protection assay, and carbonate resistance.
T7-RNA polymerase dependent in vitro expression of CcoQ was achieved using the plasmid pET22b-CcoQ. In vitro synthesis of cyt c2 was performed using plasmid pC2P2.71, which carries the cycA gene under the control of both its own and the lac promoter. Cell-free protein synthesis using [35S]methionine with R. capsulatus S-135 extracts was carried out for 30 min at 35°C as described previously (14, 21, 52). For cotranslational integration of in vitro-synthesized proteins into membranes, ICM were added after 5 min of synthesis, and the reaction mix was incubated for 25 min at 35°C. For protease treatment, samples were incubated with 0.5 mg of proteinase K/ml for 20 min at 25°C. Subsequently, 1 volume of 10% trichloroacetic acid (TCA) was added, and the sample was incubated for 10 min at 56°C. After centrifugation for 10 min at 20,000 × g, the TCA pellet was resuspended in sodium dodecyl sulfate (SDS) loading buffer and loaded onto a 22% urea-SDS-polyacrylamide gel. For analyzing carbonate resistance, freshly prepared Na2CO3 (pH 11.3) was added to the in vitro reaction mix (final concentration, 0.18 M), and the samples were incubated on ice for 30 min. After centrifugation in a Beckmann TLA 100.3 rotor for 15 min at 70,000 rpm, the pellet thus obtained was directly dissolved in SDS loading buffer. The supernatant was neutralized with glacial acetic acid, TCA precipitated, and centrifuged for 10 min at 20,000 × g, and the pellet obtained was dissolved in SDS loading buffer. CcoQ samples in SDS loading buffer were loaded onto a 22% urea-SDS-polyacrylamide gel, and the cyt c2 samples were loaded on a 16.5% Tris-Tricine SDS-polyacrylamide gel (41). Radiolabeled proteins were visualized by phosphorimaging using a Molecular Dynamics PhosphorImager and quantified by using the ImageQuant software from Molecular Dynamics.
Chemical cross-linking and immunoprecipitation.
Chemical cross-linking using disuccinimidyl suberate (DSS; Pierce Chemical Co., Rockford, IL) was performed as described previously (28). The in vitro reactions for subsequent cross-linking experiments were performed in the presence of HEPES-NaOH instead of triethanolamine acetate. Immunoprecipitations were performed using 4- to 10-fold scaled-up reactions with either polyclonal rabbit antibodies to CcoP or CcoN (18) or monoclonal anti-His tag antibodies. For immunoprecipitation, antibodies were covalently linked to protein A-Sepharose matrix (28). The anti-His tag monoclonal antibody was purchased from Novagen (Bad Schwalbach, Germany).
Enzyme assays.
TMPD (N,N,N′,N′-tetramethyl-p-phenylenediamine) oxidase activity was measured at 28°C in a closed reaction chamber (1-ml volume) with a fiber optic oxygen meter (Fibox 3; PreSens GmbH, Regensburg, Germany). R. capsulatus membranes were dissolved in ICM buffer (21) to a final concentration of approximately 0.1 mg/ml. Oxygen consumption was initiated by the addition of 10 μl of 1 M sodium ascorbate (final concentration, 10 mM) and 5 μl of 24 mM TMPD (final concentration, 0.2 mM). Oxygen consumption was recorded at 28°C using OxyView 3.5.1 software (PreSens GmbH) and terminated after several minutes of recording by the addition of 100 μM NaCN (final concentration). Net TMPD oxidase activity was determined by subtraction of the endogenous respiration rate from that induced by ascorbate. Alternatively, cyt c oxidase activity was measured spectrophotometrically by monitoring the oxidation of reduced horse heart cyt c (Sigma, St. Louis, MO) at 550 nm and 25°C using a TIDAS II Spectrophotometer (Spectralytics GmbH, Essingen, Germany). R. capsulatus ICM were dissolved in assay buffer (10 mM Tris-HCl [pH 7.0], 120 mM KCl) to a final protein concentration of approximately 0.02 mg/ml. Then, 1 ml of horse heart cyt c (0.4 mM final concentration) was reduced by adding 100 μl of sodium dithionate from a stock solution of 100 mM, and dithionate was subsequently removed by gel filtration using a PD MiniTrap G-25 column (GE Healthcare, Munich, Germany). The standard assay mixture (2 ml) was composed of 50 μl of reduced cyt c (final concentration, 5 μM) and the appropriate volume (1 to 5 μl) of ICM in the assay buffer.
BN-PAGE.
For blue native-polyacrylamide gel electrophoresis (BN-PAGE) analyses, ICM (300 μg of protein total) were resuspended in 25 μl of lysis buffer (50 mM NaCl, 5 mM 6-aminohexanoic acid, 50 mM imidazole/HCl [pH 7.0]) and solubilized with n-dodecylmaltoside (Roche Diagnostics, Mannheim, Germany) at a 1:1 (wt/wt) ICM protein/detergent ratio from a 10% dodecylmaltoside stock solution in lysis buffer. After incubation for 10 min at 20°C, the samples were centrifuged for 30 min at 55,000 rpm in a TLA100.3 rotor. After centrifugation, the supernatant was supplemented with 6 μl of loading buffer (5% Coomassie blue in 500 mM 6-aminohexanoic acid) and 5 μl of 50% glycerol and then loaded onto a 5 to 20% BN-polyacrylamide gel (42).
In-gel heme staining, in-gel α-naphtol and dimethylphenylenediamine (NADI) staining and immunoblotting.
SDS-Tris-Tricine polyacrylamide gels were treated with 3,3′,5,5′-tetramethylbenzidine (TMBZ) to reveal the c-type cytochromes according to the method of Thomas et al. (48). In-gel activity staining for the cyt cbb3-Cox was performed by incubating BN-polyacrylamide gels with a 1:1 (vol/vol) mixture of 35 mM α-naphthol dissolved in ethanol and 30 mM N,N-dimethyl-p-phenylenediamine in water, which produced a blue color in the presence of active enzyme. For immunoblot analyses, proteins were electroblotted onto Immobilon-P transfer membranes, and polyclonal antibodies against CcoP and CcoN (18) were used with horseradish peroxidase-conjugated goat anti-rabbit antibodies from Caltag Laboratories (Burlingame, CA) as secondary antibodies and ECL (GE Healthcare, Munich, Germany) as the detection substrate.
RESULTS
CcoQ is a type I membrane protein.
The ccoQ gene of R. capsulatus encodes a small, putative membrane protein of 58 amino acids. Computer-aided topology predictions suggest that it has a single transmembrane domain encompassing amino acid residues 12 to 30 and an 11-amino-acid long N-terminal portion facing the periplasm (Fig. 1A). In order to experimentally determine the topology of CcoQ, we used an in vitro transcription-translation system of R. capsulatus, which allows the in vitro synthesis of a C-terminally His-tagged CcoQ derivative and its subsequent integration into R. capsulatus inside-out ICM (21). In vitro synthesis of CcoQ-His6 resulted in single radioactively labeled band of about 7 kDa, which is in agreement with its predicted size (Fig. 1B). In order to analyze whether CcoQ was integrated into the ICM in vitro, alkaline carbonate extraction, which is a routinely used method to differentiate between membrane-integral and soluble or membrane-associated proteins, was performed. In the absence of ICM, the vast majority of in vitro-synthesized CcoQ was recovered in the supernatant after carbonate extraction and centrifugation. Only a small amount of CcoQ was found in the pellet after carbonate extraction in the absence of membranes, which is probably due to aggregation of the rather hydrophobic CcoQ. Importantly, in the presence of ICM most of the in vitro-synthesized CcoQ was found to be carbonate resistant, e.g., found in the pellet fraction, indicating that CcoQ was integrated into the R. capsulatus ICM. As a control, we also analyzed carbonate resistance of the soluble cyt c2. Unlike CcoQ, cyt c2 was exclusively found in the supernatant after carbonate extraction, independently of whether ICM were present or not.
FIG. 1.
CcoQ is a type I integral membrane protein. (A) Predicted topology of CcoQ in R. capsulatus and sequence alignment of CcoQ from the closely related species R. capsulatus (R.c.), R. sphaeroides (R.s.), and P. denitrificans (P.d.). The alignment was performed by using CLUSTAL; an asterisk (*) in the sequence indicates identical amino acids, and a colon (:) indicates similar amino acids. The putative transmembrane domain of CcoQ in R. capsulatus is underlined. (B) CcoQ and cyt c2 were in vitro synthesized in the presence or absence of R. capsulatus inverted inner membrane vesicles (ICM) using a coupled R. capsulatus in vitro transcription-translation system. After in vitro synthesis the reaction mixture was extracted with alkaline Na2CO3, pH 11.3 and centrifuged. The supernatant (S) reflecting soluble proteins and the pellet (P) reflecting membrane-integral proteins were loaded on a 22% urea-SDS-PAGE (CcoQ) or on a 16.5% Tris-Tricine (cyt c2) gel. Radioactively labeled proteins were visualized by phosphorimaging. For quantification, the amount of soluble material and that of the pellet fraction was set as 100%, and the material present in the individual fractions was quantified. The quantification is based on at least three independent experiments. (C) For proteinase K protection, the in vitro reaction mixture was split, and one-half was directly precipitated for 30 min at 4°C with ice-cold TCA; the other half was incubated with proteinase K (0.5 mg/ml, final concentration) for 20 min at 25°C. The proteinase K-treated sample was also then TCA precipitated. After centrifugation of the TCA-precipitated samples, the pellets were resuspended in SDS-loading dye, heat denatured, and loaded onto a 22% urea-SDS-PAGE gel. Immunoprecipitation experiments were performed with α-His antibodies covalently bound to protein A-Sepharose beads. For proteinase K-treated samples, the protease inhibitor phenylmethylsulfonyl fluoride was added before the addition of the antibody beads.
In order to further verify the membrane integration of CcoQ and to determine its topology, in vitro-synthesized CcoQ-His6 was subjected to proteinase K treatment. In the absence of ICM, the 7-kDa band was completely degraded by proteinase K (Fig. 1C). However, in the presence of ICM, an ∼3-kDa membrane protected fragment of CcoQ was observed (Fig. 1C; CcoQ-MPF), confirming its integration into the membrane. Considering that CcoQ is exclusively labeled via its N-terminal initiating methionine, the detection of a radioactively labeled protease protected fragment of CcoQ also indicated that the initiating methionine was not accessible to proteinase K. This observation suggests that the N terminus of CcoQ is probably located on the periplasmic side of the membrane, i.e., inside the ICM vesicles. In order to confirm this topology, we performed immunoprecipitation experiments with antibodies directed against the C-terminal His tag of CcoQ. The antibodies precipitated the 7-kDa band but not the membrane protected 3-kDa band, indicating that the C-terminal tail of CcoQ was cleaved off by proteinase K (Fig. 1C). These data experimentally confirmed the prediction that CcoQ is an integral membrane protein with an Nout-Cin topology (Fig. 1A).
The lack of CcoQ impairs cbb3 oxidase activity in R. capsulatus.
Although the CcoQ subunit appears to be present in most, if not all, cbb3-Cox containing species (34), the analyses of ccoQ deletions in R. sphaeroides (29) and B. japonicum (55) indicated that CcoQ is not essential for their cbb3-Cox activity. In R. sphaeroides, cbb3-Cox is the predominant cytochrome oxidase only under semiaerobic conditions, while in the presence of high oxygen concentrations this bacterium uses its mitochondrion-like aa3-Cox (30). Interestingly, the lack of CcoQ has apparently the most pronounced effect on R. sphaeroides cbb3-Cox at high oxygen concentrations, which has led to the hypothesis that CcoQ might be required for protecting cbb3-Cox against oxidative damage (31). Unlike R. sphaeroides, cbb3-Cox is the only cytochrome oxidase present in R. capsulatus and is involved in respiration under both aerobic and semiaerobic conditions (46, 47). Thus, to examine the role of CcoQ in R. capsulatus cbb3-Cox under different growth conditions, we constructed the plasmid pAP4, which carries an in-frame ccoQ deletion within the ccoNOQP operon. This low-copy plasmid was transferred into the ccoNOQP deletion strain GK32 (18), and the cbb3-Cox oxidase activity of the merodiploid thus obtained was determined in whole cells using the NADI staining (19). In the presence of an active cytochrome oxidase, NADI produces a blue dye, indophenol blue, which is easily detected in whole cells. No significant difference was seen on the NADI staining of aerobically grown cells of GK32/pOX15, which carries the complete ccoNOQP operon or GK32/pAP4, which encodes only ccoNOP and lacks an intact ccoQ. Colonies of both strains turned blue within less than 1 min (data not shown), while those from GK32/pRK415 (carrying the empty vector pRK415) did not respond to the NADI stain even after 30 min, a finding in agreement with the absence of the cbb3-Cox activity in this strain. In order to exclude that the lack of ccoQ had a polar effect on the downstream ccoP, pAP4 was also transferred into the R. capsulatus cbb3-Cox mutant M7G (23). This mutant assembles a cbb3-Cox subcomplex lacking the CcoP subunit (18, 21). Like the wild type, M7G/pAP4 turned blue within less than 1 min upon NADI staining, indicating that the lack of ccoQ did not significantly impair the expression of the downstream ccoP.
Because the NADI staining of intact R. capsulatus cells does not allow for a quantitative assessment of the cbb3-Cox activity, we used both oxygen uptake as well as horse heart cyt c oxidation assays for determining the specific cbb3-Cox activity in purified membranes from strains grown under aerobic, semiaerobic, or anaerobic-photosynthetic conditions. In wild-type membranes, the highest cbb3-Cox activity was observed under semiaerobic conditions, and the lowest activity was under anaerobic-photosynthetic conditions (Table 2). Independently of the growth conditions, no oxygen uptake activity or cyt c oxidation was observed with the cbb3-Cox deletion strain GK32 or with GK32 carrying the empty vector pRK415. In GK32 carrying the ccoNOQP genes on the low-copy vector pOX15, cbb3-Cox activity was ca. 2.5 to 3 times higher than in wild-type membranes, which is expected considering a copy number of three to five copies/cell for pRK415 derived plasmids. In GK32/pAP4, which encodes only ccoNOP but lacks ccoQ, the cbb3-Cox activity was reduced to ca. 20% of what was observed in GK32/pOX15 and to ca. 60% of the wild-type activity (Table 2). Importantly, a comparable reduction in cbb3-Cox activity was observed in membranes from aerobically, semiaerobically, or anaerobic-photosynthetically grown cells, which indicated that in R. capsulatus the lack of CcoQ impaired the cbb3-Cox activity, irrespective of the oxygen concentration (Table 2). The important contribution of CcoQ for cbb3-Cox activity was further confirmed by analyzing M7G/pAP4. The ccoP mutant M7G regained cbb3-Cox activity after crossing in pAP4, but the activity was significantly lower than in GK32 pOX15.
TABLE 2.
Cytochrome cbb3 oxidase activity in R. capsulatus strains grown under different conditionsa
| Strain | % Cytochrome cbb3 oxidase activity
|
|||||
|---|---|---|---|---|---|---|
| Aerobic
|
Semiaerobic
|
Anaerobic-photosynthetic
|
||||
| O2 uptake | cyt c oxidation | O2 uptake | cyt c oxidation | O2 uptake | cyt c oxidation | |
| MT1131 (wild type) | 100 | 100 | 100 | 100 | 100 | 100 |
| GK32 (ΔccoNO) | <1 | <1 | <1 | <1 | <1 | <1 |
| GK32/pRK415 (empty vector) | <1 | <1 | <1 | <1 | <1 | <1 |
| GK32/pOX15 (ccoNOQP in pRK415) | 262 | 240 | 290 | 270 | 315 | 254 |
| GK32/pAP4 (ccoNOP in pRK415) | 61 | 68 | 53 | 65 | 59 | 47 |
| M7G (ccoP269) | NT | NT | 5 | 3 | NT | NT |
| M7G/pAP4 (ccoNOP in pRK415) | NT | NT | 80 | 90 | NT | NT |
Cytochrome cbb3 oxidase activity in R. capsulatus membranes (ICM) was analyzed by either measuring oxygen uptake or measuring horse heart cyt c oxidation. At least three independent measurements were performed. The activity of wild-type MT1131 membranes grown under the indicated conditions was set as 100%. For O2 uptake measurements, this corresponds to 680 nmol of O2/mg of protein·h (aerobic conditions), to 1,322 nmol of O2/mg of protein·h (semiaerobic conditions), and to 115 nmol of O2/mg of protein·h (anaerobic-photosynthetic conditions). For cyt c oxidations, 100% activity corresponds to 1,500 nmol of cyt c/mg of protein·h (aerobic conditions), to 2,745 nmol of cyt c/mg of protein·h (semiaerobic conditions), and to 140 nmol of cyt c/mg of protein·h (anaerobic-photosynthetic conditions). NT, not tested.
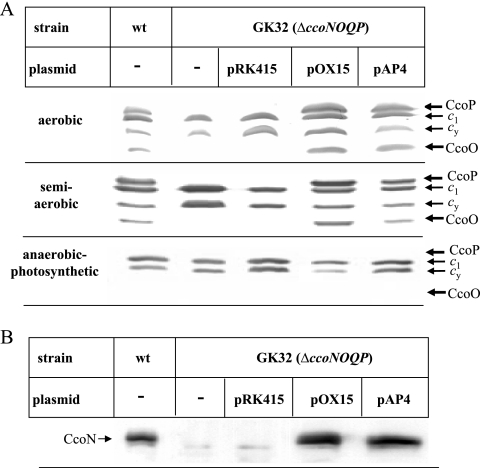
We next analyzed whether the decreased cbb3-Cox activity observed in the absence of CcoQ was correlated with reduced steady-state amounts of the cbb3-Cox subunits. Heme staining using TMBZ revealed four membrane-bound c-type cytochromes in wild-type membranes (19): CcoP (32 kDa) and CcoO (28 kDa) are subunits of cbb3-Cox, while cyt c1 (31 kDa) is part of the cyt bc1 complex, and cyt cy (29 kDa) is a membrane-bound electron carrier (25) (Fig. 2). In the cbb3-Cox knockout strain GK32 carrying either no plasmid or the empty vector pRK415, only the cytochromes c1 and cy were detectable. However, in GK32 cells carrying either pOX15 or pAP4 all four membrane-bound cytochromes were equally detectable under both aerobic and semiaerobic conditions. In membranes from anaerobic-photosynthetically grown cells, however, CcoP and CcoO were not detectable by the heme staining procedure, which is in agreement with the low cbb3-Cox activity under anaerobic-photosynthetic growth conditions (Table 2). By Western blotting we also determined the steady-state amounts of the catalytic subunit CcoN. CcoN was detectable in wild-type cells grown under aerobic conditions but not in GK32 or GK32/pRK415 (Fig. 2B). Importantly, there was no significant difference in the amount of CcoN between GK32/pOX15 and GK32/pAP4, indicating that the lack of CcoQ had also no considerable effect on the stability of CcoN. Similar results were also observed for cells grown under aerobic or anaerobic-photosynthetic conditions (data not shown), although the amount of CcoN was significantly lower in cells grown under anaerobic-photosynthetic conditions (see Fig. 6 below). In summary, these data indicated that the lack of CcoQ reduced the activity of cbb3-Cox without significantly reducing the steady-state amounts of the CcoN, CcoO, and CcoP subunits in R. capsulatus membranes.
FIG. 2.
The lack of CcoQ does not impair the steady-state stability of the cbb3-Cox subunits. (A) Membranes of wild-type R. capsulatus (wt), the cbb3-Cox deletion strain GK32, and GK32 carrying the empty plasmid pRK415, a plasmid-borne copy of ccoNOQP (pOX15), or a plasmid-borne copy of ccoNOP (pAP4), grown under aerobic, semiaerobic, or anaerobic-photosynthetic conditions on MPYE medium were isolated and separated on 16.5% Tris-Tricine-SDS gels. In each lane, 200 μg of protein was loaded. The membrane-bound c-type cytochrome profile was then revealed by TMBZ staining. CcoP and CcoO correspond to the cyt c subunits of cbb3 Cox, c1 is a subunit of the cyt bc1 complex, and cy corresponds to the membrane-bound cyt cy. (B) Immune detection of the catalytic subunit CcoN in membranes as described in panel A, grown under aerobic conditions. Membranes were separated on a 16.5% Tris-Tricine-SDS gels, blotted onto a polyvinylidene difluoride (PVDF) membrane, and decorated with polyclonal antibodies to CcoN.
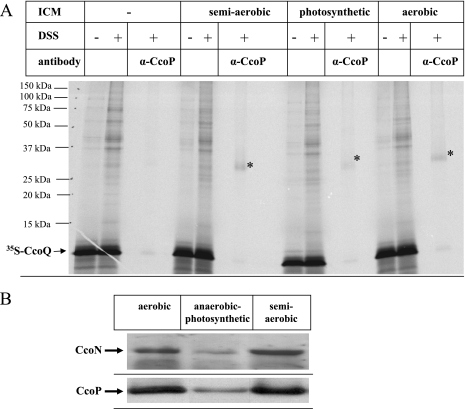
FIG. 6.
The ability of CcoQ to interact with the CcoP subunit is not influenced by the growth conditions. (A) Chemical cross-linking of in vitro-synthesized CcoQ was performed as described in Fig. 4 using wild-type membranes grown either under semiaerobic, anaerobic-photosynthetic, or aerobic conditions on MPYE medium. (B) Immunodetection of CcoN and CcoP in wild-type membranes grown either under semiaerobic, anaerobic-photosynthetic, or aerobic conditions on MPYE medium.
The amount of the active 230-kDa cbb3-Cox complex is reduced in the absence of CcoQ.
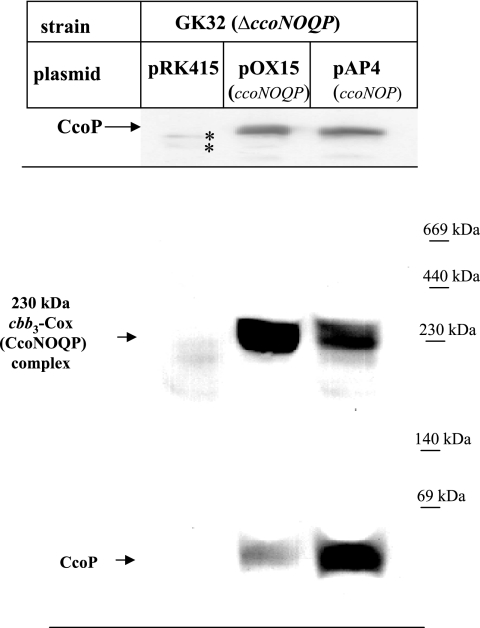
Because the reduced cbb3-Cox activity in the absence of the CcoQ subunit was not correlated with significantly reduced steady-state amounts of the cbb3-Cox subunits, we analyzed whether their functional assembly into an active cbb3-Cox complex was impaired. We have recently demonstrated by BN-PAGE analyses that cbb3-Cox forms an active 230-kDa complex that contains all four subunits, including CcoQ (21) (Fig. 3). In addition, CcoP is also detectable as a low-molecular-weight band, running below the 69-kDa size marker band on BN-PAGE (Fig. 3). Membranes from strains GK32/pRK415, GK32/pOX15, and GK32/pAP4 were solubilized and analyzed by BN-PAGE to examine whether the active 230-kDa cbb3-Cox complex formation was affected by the lack of CcoQ. Immunoblots with antibodies to CcoP revealed in GK32/pOX15 the presence of the 230-kDa cbb3-Cox complex (CcoNOQP-complex) and the low-molecular-weight CcoP band (Fig. 3, bottom), which were both missing in membranes from GK32/pRK415. Both bands were also detectable in GK32/pAP4, but CcoP was found predominantly in the low-molecular-weight band, and only a small portion was fully assembled into the active 230-kDa cbb3-Cox complex. Importantly, when these membranes were separated on SDS-PAGE and probed with the CcoP antibodies, no significant difference in the total amount of CcoP was observed (Fig. 3, top), in agreement with the heme staining data shown in Fig. 2. In summary, these data suggest that the lack of CcoQ impaired the formation of the active 230-kDa cbb3 complex, apparently without changing much the steady-state levels of the CcoN, CcoO, and CcoP subunits. The inability to form a stable cbb3-Cox complex in the absence of CcoQ also nicely explains why the cbb3-Cox activity is reduced by ca. 80% in the absence of CcoQ (Table 2).
FIG. 3.
The lack of the CcoQ subunit impairs the functional assembly of cbb3-Cox. Membranes (200 μg of protein) of GK32 grown under aerobic conditions on MPYE medium and carrying either the empty plasmid pRK415, pOX15 or pAP4 were separated on a 16.5% Tris-Tricine SDS-PAGE gel and, after Western transfer, decorated with α-CcoP antibodies (upper panel). An asterisk (*) indicates cross-reacting bands. The same membranes (300 μg of protein) were also separated after solubilization with dodecyl maltoside (1 mg/mg of protein) on a 5 to 20% BN-PAGE gradient gel. After Western transfer onto a PVDF membrane, immune detection with α-CcoP antibodies was performed. Indicated are the 230-kDa active cbb3-Cox complex (CcoNOQP complex) and a low-molecular-weight CcoP complex.
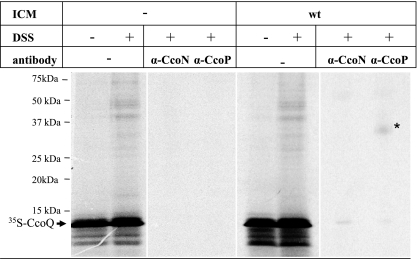
In vitro cross-linking indicates that CcoQ is in close proximity of the CcoP subunit of cyt cbb3 oxidase.
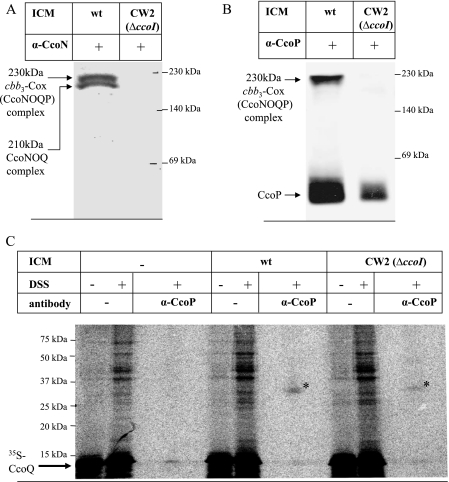
The observed assembly defect of cbb3-Cox in the absence of CcoQ suggested that a function of CcoQ might be to assist or to stabilize binding of CcoP to a preformed CcoNO subcomplex. This would probably require a direct molecular contact between CcoP and CcoQ. Chemical cross-linking of in vitro-synthesized membrane proteins provides a powerful tool for revealing such protein-protein interactions. Radiolabeled CcoQ was synthesized in vitro in the presence of membranes and incubated with the membrane permeable cross-linker DSS. Then, cross-linked products were immunoprecipitated with antibodies directed against either the catalytic subunit CcoN or the 32-kDa cyt subunit CcoP. The CcoN antibodies failed to precipitate a specific CcoQ cross-linked product; however, antibodies against CcoP immunoprecipitated a specific 36-kDa cross-linked band, which was only observed in the presence of membranes but not in their absence (Fig. 4). Although these data strongly suggest that CcoQ indeed interacts with the 32-kDa CcoP subunit of cbb3-Cox, this cross-linking approach did not allow us to determine whether CcoQ interacts with CcoP that is a part of the active 230-kDa cbb3-Cox complex or with the low-molecular-weight CcoP band that is detectable in BN-PAGE (Fig. 3). In order to differentiate between the two possibilities, we made use of the ccoI deletion mutant CW2. In this mutant, the putative Cu-transporting ATPase CcoI is deleted (20), which prevents the formation of the active 230-kDa cbb3 oxidase complex (21). Nevertheless, although in CW2 membranes the 230-kDa CcoNOQP and the 210-kDa CcoNOQ complexes are not detectable (Fig. 5A), it still contains a small amount of the low-molecular-weight CcoP band (21) (Fig. 5B). In agreement with the lack of the active 230-kDa CcoNOQP complex, no NADI-stainable band was detectable on BN-PAGE (data not shown). Although it is currently unknown why in this particular mutant a small amount of the low-molecular-weight CcoP band is detectable even in the absence of a stable CcoNO core complex, the use of this mutant allowed us to determine whether CcoQ would also cross-link to the low-molecular-weight CcoP band. When CcoQ was in vitro synthesized in the presence of wild-type membranes and subjected to DSS cross-linking with subsequent immunoprecipitation, we again detected the 36-kDa CcoQ-CcoP cross-linking product (Fig. 5C). Importantly, a weak CcoP-CcoQ cross-linking product was also detected in CW2 membranes, which indicates that CcoQ interacts with CcoP that is part of the active 230-kDa cbb3-Cox complex, as well as with the low-molecular-weight CcoP band. Thus, the low-molecular-weight CcoP band most likely reflects a protein complex consisting of at least CcoP and CcoQ. This finding is also supported by our observation that, in the absence of CcoQ, the low-molecular-weight CcoP band was running slightly lower on BN-PAGE than in CcoQ-containing membranes (Fig. 3). Although this shift in migration was only minimal, it was reproducibly observed.
FIG. 4.
Chemical cross-linking reveals a CcoQ-CcoP interaction. CcoQ was in vitro synthesized as described in Fig. 1 and incubated with homo-bifunctional cross-linker DSS in the presence or absence of R. capsulatus wild-type membranes, grown aerobically on MPYE medium. Immunoprecipitations were performed with 150-μl in vitro reaction mixtures using protein A-Sepharose-coupled antibodies directed against CcoP and CcoN.
FIG. 5.
CcoQ interacts with the low-molecular-weight CcoP. (A) BN-PAGE analyses of wild-type membranes and membranes derived from the ΔccoI mutant CW2, both grown under aerobic conditions on MPYE medium. After Western transfer onto a PVDF membrane, immunodetection with α-CcoN antibodies was performed. Indicated are the 230-kDa active cbb3-Cox complex (CcoNOQP complex) and the 210-kDa CcoNOQ complex, which presumably reflects an assembly intermediate (20). (B) BN-PAGE analyses as described in panel A but with antibodies to the CcoP subunit. Indicated are the active 230-kDa cbb3-Cox complex and a low-molecular-weight CcoP band. (C) Chemical cross-linking was performed as described in Fig. 4 using in vitro-synthesized CcoQ and wild-type or CW2 membranes. wt, Wild type.
In R. sphaeroides, CcoQ has been implicated in stabilizing or regulating the cbb3-Cox activity in response to oxygen (31); we therefore also tested the ability of CcoQ to interact with CcoP in membranes derived from either aerobically, semiaerobically, or anaerobic-photosynthetically grown cells. When these membranes were incubated with in vitro-synthesized CcoQ and treated with DSS, CcoP antibodies precipitated comparable amounts of the 36-kDa CcoP-CcoQ cross-linking product in both aerobic and semiaerobic membranes (Fig. 6A). The CcoP-CcoQ cross-linking product was, however, significantly weaker in membranes from photosynthetically grown cells. Considering that photosynthetically grown membranes contain significantly less cbb3-Cox (Fig. 2), the lower amount of the CcoP-CcoQ cross-link probably reflects the lower CcoP concentration in these membranes (Fig. 6B), rather than growth-condition-dependent differences in the CcoQ-CcoP interaction.
DISCUSSION
Our study provides the first analyses of the molecular contact sites and the function of the conserved CcoQ subunit in R. capsulatus cbb3-Cox. The ΔccoQ mutant exhibited significantly reduced cbb3-Cox activity, although the steady-state amounts of CcoN, CcoP, and CcoO as revealed by Western blotting and heme staining were not significantly different from a wild-type strain. Chemical cross-linking and BN-PAGE analyses demonstrated a specific CcoQ-CcoP interaction, which appears to be required for the stable association of CcoP with the CcoN and CcoO subunits. The different experimental approaches provided no indication that the ΔccoQ phenotype in R. capsulatus was significantly influenced by growth conditions. This is different from the closely related species R. sphaeroides, in which the lack of CcoQ appears to predominantly impair cbb3-Cox activity under aerobic conditions (31).
Subunit interactions and assembly of cbb3-Cox in R. capsulatus.
Due to the lack of a three-dimensional structure of cbb3-Cox, no information about the molecular contacts between its four subunits has been available thus far. Our study indicates that CcoQ is in close contact with the CcoP subunit. Using in vitro labeling, we showed earlier that in vitro-synthesized CcoQ is able to radioactively label not only the 230-kDa cbb3-Cox holo-complex but also a 210-kDa subcomplex, which contains CcoN and CcoO but lacks CcoP (21). Although these data do not necessarily indicate that CcoQ is a stable constituent of both complexes, they indicate that CcoQ can bind to a CcoNO subcomplex even in the absence of CcoP, suggesting that CcoP is not the only contact site for CcoQ. Molecular modeling approaches have recently provided the first tentative atomic models for cbb3-Cox (15, 43). In one model, it is suggested that CcoQ might be in close contact to the catalytically important helix 7 of the CcoN subunit (43). The corresponding position appears to be also occupied in aa3-Cox, either by subunit IV as in the case of the aa3-Cox of P. denitrificans (17) or by lipids as in the aa3-Cox of R. sphaeroides (45) and B. taurus (50). In our experiments, using the primary amine-directed cross-linker DSS, we were unable to identify a CcoN-CcoQ cross-linking product (Fig. 4). CcoQ contains three cross-linkable amines corresponding to those of the initiator methionine and the lysine residues at position 36 and 51 (Fig. 1C). Considering that the predicted helix 7 of R. capsulatus CcoN also contains a lysine residue at position 295, the lack of a CcoN-CcoQ cross-linking product cannot be explained by the absence of DSS reacting side chains. A likely possibility might be either that the spacer length of DSS (11.4Å) is too short or that CcoQ and CcoN are not in stable enough contact to each other to covalently link the two proteins. It is also possible that in the 210-kDa complex, CcoQ can bind via the CcoO subunit. Due to lack of antibodies against the CcoO subunit, this possibility awaits further verification.
The interaction of CcoQ with the CcoP subunit is important for the assembly process of cbb3-Cox. Previous data suggested that the CcoN and CcoO subunits form a stable subcomplex to which the CcoP subunit is recruited (5, 21, 55). In BN-PAGE, the subcomplex formed by CcoO and CcoN is visible as a 210-kDa complex, while CcoP is detectable in the active 230-kDa cbb3-Cox complex, as well as in a low-molecular-weight complex (Fig. 5A, 21). Our cross-linking data now reveal that CcoQ is also able to interact with this low-molecular-weight CcoP band (Fig. 5), which could indicate that the low-molecular-weight CcoP band does not represent monomeric CcoP as we had originally suggested (21) but instead a CcoP-CcoQ complex. This would further suggest that CcoQ can obviously bind to CcoP before CcoP associates with the CcoNO core complex to form the active 230-kDa cbb3-Cox complex (CcoNOQP complex). In the absence of CcoQ, the ability of CcoP to form a stable complex with CcoNO is drastically reduced (Fig. 3), which supports the idea that CcoQ is either an important determinant for the stability of the cbb3-Cox holo-complex or it is involved in recruiting CcoP into the CcoNO subcomplex. A model in which a preformed CcoNO complex interacts with a preformed CcoQP complex to yield the functional cbb3-Cox complex would suggest that CcoQ is most likely not a stable component of the 210-kDa CcoNO subcomplex. Although this is apparently in conflict with the labeling of the 210-kDa complex by in vitro-synthesized CcoQ (21), it should be noted that the in vitro labeling experiments primarily reflects binding to the membranes but not necessarily that binding occurs by exchanging with endogenous CcoQ. Nevertheless, we can currently not exclude the possibility that CcoQ is a stable component of the 210-kDa CcoNO subcomplex. It is also possible that the complex detected at 210-kDa in BN-PAGE is a mixture of a CcoNO subcomplex and a CcoNOQ subcomplex that has lost the CcoP subunit.
Although many terminal oxidases in bacteria contain a fourth subunit, no general function has been assigned thus far to these subunits. CcoQ and CtaH, the fourth subunit of P. denitrificans aa3-Cox (53), are both single-spanning membrane proteins but lack significant sequence conservation. They also appear to have a different topology in the membrane: while the N terminus of CtaH is facing the cytoplasm, as determined by X-ray crystallography (17), our data suggest that the N terminus of CcoQ is located in the periplasm (Fig. 1). Finally, deleting CtaH has no detectable effect on the aa3-Cox assembly or activity (53), which is clearly different from what is observed here in the case of R. capsulatus cbb3-Cox, where the assembly, stability, and activity are perturbed in the absence of CcoQ. Still different from CtaH and CcoQ, subunit IV (CyoD) of E. coli bo3-type ubiquinol oxidase spans the membrane three times (3), and its absence results in a perturbed redox metal center in the subunit I, although no general assembly defect is seen (27, 38). A similar phenotype is also observed for the B. subtilis aa3-type menaquinol oxidase. Subunit IV (QoxD) contains three transmembrane domains, and in its absence the aa3-menaquinol oxidase activity is reduced, although no general assembly defect is seen (51). Thus, even though many respiratory terminal oxidases contain a small, poorly conserved membrane-integral fourth subunit, the available data do not allow assigning them a general function in respect to the assembly or activity of these oxidases.
The lack of CcoQ impairs cbb3-Cox activity in R. capsulatus independently of the growth conditions.
Although cbb3-Cox is characterized by a higher oxygen affinity than the mitochondrion-like aa3-Cox (33, 34), its expression is not exclusively linked to low oxygen concentrations. In B. japonicum, cbb3-Cox is primarily expressed under semiaerobic conditions, while under high oxygen concentrations this species uses aa3-Cox (35, 36). The same oxygen-dependent switch is observed in R. sphaeroides: aa3-Cox is the predominant oxidase under aerobic conditions, while cbb3-Cox is mainly expressed at low oxygen concentrations or during the transition from anaerobic to aerobic growth (1, 24). Also, many pathogenic bacteria, such as Brucella suis (22) or Campylobacter jejuni (54), use cbb3-Cox mainly at low oxygen concentrations. On the other hand, in Thiobacillus denitrificans cbb3-Cox is highly upregulated under aerobic conditions (2). In contrast to R. sphaeroides, R. capsulatus lacks aa3-Cox (13) but instead contains in addition to cbb3-Cox a less-well-characterized CydAB-type quinol oxidase (23). This quinol oxidase is predicted to exhibit an even higher oxygen affinity than cbb3-Cox (9). The expression and activity of cbb3-Cox in R. capsulatus is highest under semiaerobic conditions (Table 2) (47). Nevertheless, the R. capsulatus enzyme exhibits significant activity also under aerobic conditions (Table 2) (47). Thus, in R. capsulatus cbb3 Cox appears to be active under both aerobic and semiaerobic conditions, while the CydAB quinol oxidase would facilitate respiration under very low oxygen concentrations. This is also reflected by the differential effects of multiple regulators such as RegA, FnrL, and HvrA on cbb3-Cox and cydAB expression (9).
It has been observed that under aerobic conditions that the CcoP subunit of R. sphaeroides cbb3-Cox becomes susceptible to proteolytic degradation in the absence of CcoQ (31). This has led to the hypothesis that in the absence of CcoQ high oxygen concentrations cause a selective loss of the heme groups of CcoP, which then undergoes a conformational change that converts it into a target for an unknown serine metalloprotease (31). In R. capsulatus, the lack of CcoQ does not seem to impair the stability or the heme content of the CcoP subunit, independently of the growth conditions, as revealed by heme staining and immunoblot analyses (Fig. 2 and 6). Although our cross-linking data support the idea that CcoQ interacts with CcoP, in R. capsulatus this interaction does not seem to be required specifically for the stabilization of the CcoP subunit. It rather appears to be required for facilitating or stabilizing the assembly of the CcoPQ subcomplex with the CcoNO subcomplex to yield the active 230-kDa cbb3-Cox complex. Thus, the available data indicate that CcoQ apparently plays very distinct and unrelated roles for cbb3-Cox in the closely related species R. capsulatus and R. sphaeroides. The physiological and biochemical significance of these differences remain to be determined in future studies.
Acknowledgments
This study was supported by the German Science Foundation (DFG grants SFB 388 (project A12) and GRK 1478 (project P5) to H.-G.K., the French-German University (DFH) to H.-G.K., the German Academic Exchange Service (DAAD) to F.D., the National Institutes of Health (GM 38237) and the U.S. Department of Energy (91ER 20052) to F.D., and the Erasmus Program of the European Union to G.P.
Footnotes
Published ahead of print on 13 June 2008.
REFERENCES
- 1.Arai, H., J. H. Roh, and S. Kaplan. 2008. Transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 190286-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beller, H. R., T. E. Letain, A. Chakicherla, S. R. Kane, T. C. Legler, and M. A. Coleman. 2006. Whole genome transcriptional analysis of chemolithoautotrophic thiosulfate oxidation by Thiobacillus denitrificans under aerobic versus denitrifying conditions. J. Bacteriol. 1887006-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chepuri, V., and R. B. Gennis. 1990. The use of gene fusions to determine the topology of all of the subunits of the cytochrome o terminal oxidase complex of Escherichia coli. J. Biol. Chem. 26512978-12986. [PubMed] [Google Scholar]
- 4.Daldal, F., S. Cheng, J. Applebaum, E. Davidson, and R. C. Prince. 1986. Cytochrome c2 is not essential for photosynthetic growth of Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 832012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Gier, J.-W., M. Schepper, W. N. M. Reijnders, S. J. van Dyck, D. J. Slotboom, A. Warne, M. Saraste, K. Krab, M. Finel, A. H. Stouthamer, R. J. M. van Spanning, and J. van der Oost. 1996. Structural and functional analysis of aa3-type and cbb3-type cytochrome c oxidases of Paracoccus denitrificans reveals significant differences in proton-pump design. Mol. Microbiol. 201247-1260. [DOI] [PubMed] [Google Scholar]
- 6.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 777347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donohue, T. J., A. G. McEwan, and S. Kaplan. 1986. Cloning, DNA sequence and expression of the Rhodobacter sphaeroides cytochrome c2 gene. J. Bacteriol. 168962-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ducluzeau, A.-L., S. Ouchane, and W. Nietschke. 2008. The cbb3 oxidases are an ancient innovation of the domain bacteria. Mol. Biol. Evol. 251158-1166. [DOI] [PubMed] [Google Scholar]
- 9.Elsen, S., L. R. Swem, D. L. Swem, and C. E. Bauer. 2004. RegB/RegA, a highly conserved redox responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eraso, J. M., and S. Kaplan. 2000. From redox flow to gene regulation: role of PrrC protein of Rhodobacter sphaeroides 2.4.1. Biochemistry 392052-2062. [DOI] [PubMed] [Google Scholar]
- 11.Eraso, J. M., and S. Kaplan. 2002. Redox flow as an instrument of gene regulation. Methods Enzymol. 348216-221. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Horsman, A., B. Barquera, J. Rumbley, J. Ma, and R. B. Gennis. 1994. The superfamily of heme-copper respiratory oxidase. J. Bacteriol. 1765587-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray, K. A., M. Grooms, H. Myllykallio, C. Moomaw, C. Slaughter, and F. Daldal. 1994. Rhodobacter capsulatus contains a novel cb-type cytochrome c oxidase without a CuA center. Biochemistry 333120-3127. [DOI] [PubMed] [Google Scholar]
- 14.Helde, R., B. Wiesler, E. Wachter, A. Neubüser, H. K. Hoffschulte, T. Hengelage, K. L. Schimz, R. A. Stuart, and M. Müller. 1997. Comparative characterization of SecA from the alpha-subclass purple bacterium Rhodobacter capsulatus and Escherichia coli reveals differences in membrane and precursor specificity. J. Bacteriol. 1794003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemp, J., C. Christian, B. Barquera, R. B. Gennis, and T. J. Martinez. 2005. Helix switching of a key active site residue in the cytochrome cbb3 oxidases. Biochemistry 4410766-10775. [DOI] [PubMed] [Google Scholar]
- 16.Hemp, J., H. Han, J. H. Roh, S. Kaplan, T. J. Martinez, and R. B. Gennis. 2007. Comparative genomics and site-directed mutagenesis support the existence of only one input channel for protons in the C-family (cbb3 Oxidase) of heme-copper oxygen reductases. Biochemistry 469963-9972. [DOI] [PubMed] [Google Scholar]
- 17.Iwata, S., C. Ostermeier, B. Ludwig, and H. Michel. 1995. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature 376660-669. [DOI] [PubMed] [Google Scholar]
- 18.Koch, H.-G., O. Hwang, and F. Daldal. 1998. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J. Bacteriol. 180969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koch, H.-G., H. Myllykallio, and F. Daldal. 1998. Using genetics to explore cytochrome function and structure in Rhodobacter. Methods Enzymol. 29781-94. [Google Scholar]
- 20.Koch, H.-G., C. Winterstein, A. S. Saribas, J. O. Alben, and F. Daldal. 2000. Roles of the ccoGHIS gene products in the biogenesis of the cbb3-type cytochrome c oxidase. J. Mol. Biol. 29749-65. [DOI] [PubMed] [Google Scholar]
- 21.Kulajta, C., J. O. Thumfart, S. Haid, F. Daldal, and H.-G. Koch. 2006. Multi-step assembly pathway of the cbb3 type cytochrome c oxidase complex. J. Mol. Biol. 355989-1004. [DOI] [PubMed] [Google Scholar]
- 22.Loisel-Meyer, S., M. P. Jimenez de Bagües, S. Köhler, J. P. Liautard, and V. Jubier-Maurin. 2005. Differential use of the two high-oxygen-affinity terminal oxidases of Brucella suis for in vitro and intramacrophagic multiplication. Infect. Immun. 737768-7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marrs, B. L., and H. Gest. 1973. Genetic mutations affecting the respiratory electron-transport system of the photosynthetic bacterium Rhodopseudomonas capsulata. J. Bacteriol. 1141045-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouncey, N. J., and S. Kaplan. 1998. Oxygen regulation of the ccoN gene encoding a component of the cbb3 oxidase in Rhodobacter sphaeroides 2.4.1: involvement of the FnrL protein. J. Bacteriol. 1802228-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myllykallio, H., D. Zannoni, and F. Daldal. 1999. The membrane-attached electron carrier cytochrome cy from Rhodobacter sphaeroides is functional in respiratory but not in photosynthetic electron transfer. Proc. Natl. Acad. Sci. USA 964348-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myllykallio, H., and U. Liebl. 2000. Dual role for cytochrome cbb3 oxidase in clinically relevant proteobacteria? Trends Microbiol. 8542-543. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura, H., K. Saiki, T. Mogi, and Y. Anraku. 1997. Assignment and functional role of the cyoABCDE gene products required for the Escherichia coli bo-type quinol oxidase. J. Biochem. 122415-421. [DOI] [PubMed] [Google Scholar]
- 28.Neumann-Haefelin, C., U. Schäfer, M. Müller, and H.-G. Koch. 2000. SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 196419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh, J.-I., and S. Kaplan. 1999. The cbb3 terminal oxidase of Rhodobacter sphaeroides: structural and functional implications for the regulation of spectral complex formation. Biochemistry 382688-2696. [DOI] [PubMed] [Google Scholar]
- 30.Oh, J.-I., and S. Kaplan. 2001. Generalized approach to the regulation and integration of gene expression. Mol. Microbiol. 391116-1123. [DOI] [PubMed] [Google Scholar]
- 31.Oh, J.-I., and S. Kaplan. 2002. Oxygen adaptation: the role of the CcoQ subunit of the cbb3 cytochrome c oxidase of Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 27716220-16228. [DOI] [PubMed] [Google Scholar]
- 32.Pereira, M. M., M. Santana, and M. Teixera. 2001. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta 1505185-208. [DOI] [PubMed] [Google Scholar]
- 33.Pitcher, R. S., T. Brittain, and N. J. Watmough. 2002. Cytochrome cbb3 oxidase and bacterial microaerobic metabolism. Biochem. Soc. Trans. 30653-658. [DOI] [PubMed] [Google Scholar]
- 34.Pitcher, R. S., and N. J. Watmough. 2004. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta 1655388-399. [DOI] [PubMed] [Google Scholar]
- 35.Preisig, O., D. Anthamatten, and H. Hennecke. 1993. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc. Natl. Acad. Sc. USA 903309-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preisig, O., R. Zufferey, L. Thöny-Meyer, C. Appleby, and H. Hennecke. 1996. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 1781532-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richter, O.-M. H., and B. Ludwig. 2003. Cytochrome c oxidase: structure, function, and physiology of a redox-driven molecular machine. Rev. Physiol. Biochem. Pharmacol. 14747-74. [DOI] [PubMed] [Google Scholar]
- 38.Saiki, K., H. Nakamura, T. Mogi, and Y. Anraku. 1996. Probing a role of subunit IV of the Escherichia coli bo-type ubiquinol oxidase by deletion and cross-linking analyses. J. Biol. Chem. 27115336-15344. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Saraste, M., and J. Castresana. 1994. Cytochrome oxidase evolved by tinkering with denitrification enzymes. FEBS Lett. 3411-4. [DOI] [PubMed] [Google Scholar]
- 41.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 42.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199223-231. [DOI] [PubMed] [Google Scholar]
- 43.Sharma, V., A. Puustinen, M. Wikström, and L. Laakkonen. 2006. Sequence analysis of the cbb3 oxidases and an atomic model for the Rhodobacter sphaeroides enzyme. Biochemistry 455754-5765. [DOI] [PubMed] [Google Scholar]
- 44.Sistrom, W. R. 1960. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 22778-785. [DOI] [PubMed] [Google Scholar]
- 45.Svensson-Ek, M., J. Abramson, G. Larsson, S. Törnroth, P. Brzezinski, and S. Iwata. 2002. The X-ray crystal structure of wild type and EQ(I-286) mutant cytochrome c oxidase from Rhodobacter sphaeroides. J. Mol. Biol. 321329-339. [DOI] [PubMed] [Google Scholar]
- 46.Swem, L. R., S. Elsen, T. H. Bird, D. L. Swem, H.-G. Koch, H. Myllykallio, F. Daldal, and C. E. Bauer. 2001. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J. Mol. Biol. 309121-138. [DOI] [PubMed] [Google Scholar]
- 47.Swem, D. L., and C. E. Bauer. 2002. Coordination of ubiquinol oxidase and cytochrome cbb3 oxidase expression by multiple regulators in Rhodobacter capsulatus. J. Bacteriol. 1842815-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75168-176. [DOI] [PubMed] [Google Scholar]
- 49.Thöny-Meyer, L., C. Beck, O. Preisig, and H. Hennecke. 1994. The ccoNOQP gene cluster codes for a cb-type cytochrome oxidase that functions in aerobic respiration of Rhodobacter capsulatus. Mol. Microbiol. 174705-716. [DOI] [PubMed] [Google Scholar]
- 50.Tsukhara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, and S. Yoshikawa. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 2721136-1144. [DOI] [PubMed] [Google Scholar]
- 51.Villani, G., M. Tattoli, N. Capitanio, P. Glaser, S. Papa, and A. Danchin. 1995. Functional analysis of subunits III and IV of Bacillus subtilis aa3-600 quinol oxidase by in vitro mutagenesis and gene replacement. Biochim. Biophys. Acta 123267-74. [DOI] [PubMed] [Google Scholar]
- 52.Wieseler, B., and M. Müller. 1993. Translocation of precytochrome c2 into intracytoplasmic membrane vesicles of Rhodobacter capsulatus requires a peripheral membrane protein. Mol. Microbiol. 7167-176. [DOI] [PubMed] [Google Scholar]
- 53.Witt, H., and B. Ludwig. 1997. Isolation, analysis and deletion of the gene coding for subunit IV of cytochrome c oxidase in Paracoccus denitrificans. J. Biol. Chem. 2725514-5517. [DOI] [PubMed] [Google Scholar]
- 54.Woodall., C. A., M. A. Jones, P. A. Barrow, J. Hinds, G. L. Marsden, D. J. Kelly, N. Dorrell, B. W. Wren, and D. J. Maskell. 2005. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low oxygen concentration. Infect. Immun. 735278-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zufferey, R., O. Preisig, H. Hennecke, and L. Thöny-Meyer. 1996. Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum. J. Biol. Chem. 2719114-9119. [DOI] [PubMed] [Google Scholar]
- 56.Zufferey, R., E. Arslan, L. Thöny-Meyer, and H. Hennecke. 1998. How replacement of the 12 conserved histidines of subunit I affect assembly, cofactor binding, and enzymatic activity of the Bradyrhizobium japonicum cbb3-type oxidase. J. Biol. Chem. 2736452-6459. [DOI] [PubMed] [Google Scholar]
- 57.Zumft, W. G. 2005. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J. Inorg. Biochem. 99194-215. [DOI] [PubMed] [Google Scholar]