Abstract
The pfam04002 annotation describes RadC as a bacterial DNA repair protein. Although the radC gene is expressed specifically during competence for genetic transformation in Streptococcus pneumoniae, we report that radC mutants exhibit normal uptake and processing of transforming DNA. They also display normal sensitivity to DNA-damaging agents, providing no support for the rad epithet.
RadC is an ∼220-residue widespread bacterial protein (COG2003). A conserved domain corresponding to the C-terminal moiety of the protein is called pfam04002 (or RadC; for an explanation of the truncation of pfam04002 compared to COG2003, see the supplemental material). Rad serves as a mnemonic for radiation, and pfam04002 is annotated as “a DNA repair protein. RadC plays a role in repair of DNA damage after UV and X-ray irradiation in prokaryotes. The Escherichia coli radC gene encodes a RecG-like DNA recombination/repair function. RadC may function specifically in recombinational repair that is associated with the replication fork.” However, this annotation is solely based on the study of the radC102 mutation of E. coli. The mutation was reported to cause mild UV and X-ray sensitivity and was initially mapped near to pyrE and recG (5). The study of an E. coli strain carrying the very same mutation led to the conclusion that RadC was required to prevent deletions at chromosomal tandem repeats induced by replication fork defects (21). However, the radC102 mutation was then unambiguously demonstrated to be an allele of recG by Lombardo and Rosenberg (11), who concluded that the function of the E. coli RadC protein and of its many bacterial homologs remains to be determined. Since then, no publication describing the inactivation of E. coli radC and its phenotypic consequence(s) has appeared.
Intriguingly, radC is expressed specifically in cells competent for genetic transformation in four divergent naturally transformable species: Streptococcus pneumoniae (14, 19), Streptococcus gordonii (24), Haemophilus influenzae (18), and Bacillus subtilis (3, 7, 13). In S. pneumoniae, the radC gene (spd_0975 in D39, spr0996 in R6, and sp1088 in TIGR4) was concluded to be essential in two independent studies (20, 22). However, in a third report, radC was inactivated through replacement of the entire coding sequence by a kanamycin resistance cassette, and the protein appeared individually dispensable for chromosomal transformation (14). In B. subtilis, radC (ysxA) is not essential, and its inactivation did not affect chromosomal transformation (13). These puzzling observations prompted us to determine whether S. pneumoniae radC could be inactivated and to investigate the possible role(s) of RadC in transformation, as well as in the repair of DNA damage. All of the strains and plasmids used in the present study are listed, together with the primers, in Table S1 in the supplemental material.
Inactivation of radC.
In vitro mariner mutagenesis (16) was used to generate minitransposon insertions in the radC1-radC2 PCR fragment (Fig. 1). Insertions were readily obtained. Of 10 randomly selected insertions analyzed and found to be evenly distributed in the targeted region, 9 were localized within radC, demonstrating that this gene is not essential, at least in the S. pneumoniae D39-R6 lineage. This result contradicts two previous reports describing radC as an essential gene of S. pneumoniae (20, 22) but is consistent with the data of Peterson et al. (14).
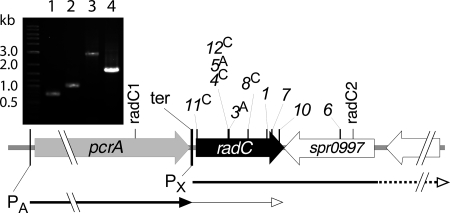
FIG. 1.
Genetic organization of the pcrA-radC chromosomal region and location of mariner minitransposon insertion mutants of radC. Transcripts previously identified in the region are shown by horizontal arrows. Expression from PA (σA promoter) is much lower for radC than for pcrA, most likely because of the presence of a terminator sequence, ter (20); only radC is induced at competence (14, 19). The dotted line indicates that transcripts initiated at PX (σX competence-specific promoter) extend into the inversely oriented flanking genes (14). The location and orientation of spc cassette insertions were determined as previously described (16) through PCRs using primers MP127 or MP128 (see Table S1 in the supplemental material) in combination with either one of the two primers used to generate the radC1-radC2 PCR fragment. Cassette-chromosome junctions were determined by DNA sequencing at position +9 (with respect to the the first nucleotide of radC taken as +1) for spc11C, +248 for spc12C, spc5A, and spc4C, and + 255 for spc3A. The inset shows the control PCR of insertion spc4C (strain R1966) retained for the present study, confirming the loss of the wild-type radC1-radC2 fragment (shown in lane 4) and its replacement by a fragment (lane 3), the size of which was fully consistent with a simple minitransposon insertion in radC, and the spc4C location with the primer pairs MP127-radC1 (lane 1) and MP127-radC2 (lane 2).
A transformation-dedicated function of RadC?
We first investigated whether transformation with chromosomal point mutations was affected in the absence of RadC by using standard procedures for transformation (6). In full agreement with a previous report (14), chromosomal transformation frequency appeared to be similar to that in wild-type cells (Table 1). We then compared the frequency of integration of pR290, a nonreplicative recombinant plasmid carrying a 1,596-bp insert of pneumococcal DNA (see Table S1 in the supplemental material). This recombination event involves integration of a long stretch of heterologous DNA (the vector moiety). Integration of pR290 occurred at a normal frequency in the absence of RadC (Table 1). Altogether, these data suggest that RadC is not required (or is individually dispensable) for the integration of homologous DNA, as well as for homology-dependent integration of nonreplicative plasmids.
TABLE 1.
Comparison of chromosomal transformation, mismatch repair, plasmid installation, and gene conversion in wild-type and radC mutant cells
| Strain (genotype) | Transforming DNA
|
Gene conversiond (Smr cells/ 10,000 cells ± SD) | |||||
|---|---|---|---|---|---|---|---|
| Chromosomal
|
Plasmid
|
||||||
| str41 (%) | rif23 (%) | Rif/Str ± SD | pR290 (%)a | pLS1 (%)b | pLS70 (%)c | ||
| R1502 (wild type) | 4.94 | 0.59 | 0.119 ± 0.034 | 0.70 | 0.046 | 0.43 | |
| R990 (rpsL+-rpsL1, Sms) | 0.117 ± 0.076 | ||||||
| R1969 (radC) | 2.67 | 0.23 | 0.088 ± 0.029 | 0.71 | 0.041 | 0.42 | |
| R2327 (rpsL+-rpsL1, Sms; radC) | 0.298 ± 0.088 | ||||||
Nonreplicative plasmid carrying a 1,596-bp chromosomal insert (see Table S1 in the supplemental material).
Replicative plasmid (rolling-circle type; pMV158 derivative) (see Table S1 in the supplemental material).
Replicative plasmid (pLS1 derivative) carrying a 3,486-bp chromosomal insert (see Table S1 in the supplemental material).
Calculated from four to five independent cultures inoculated from individual colonies resuspended in 2 ml of C+Y medium and grown to an OD550 of ∼0.4 before plating.
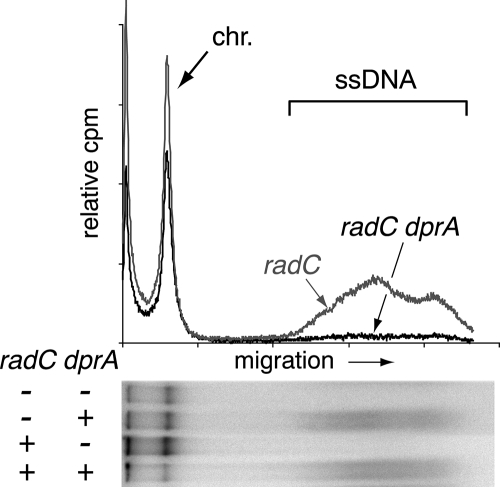
We also examined whether RadC is important for the stability of incoming single-stranded DNA (ssDNA). The fate of transforming DNA was compared in wild-type and radC mutant cells using as the donor a 7,771-bp S. pneumoniae fragment uniformly labeled with 32P. After 3 min of contact between competent cells and DNA and a further incubation of cells for 1 min in the presence of DNase I to terminate uptake, ssDNA was recovered from transformed cells by using a previously described method (2), with only minor modifications (see the supplemental material). No effect of the inactivation of radC on the amount of intact ssDNA recovered was detected (Fig. 2), suggesting that RadC does not affect the stability of incoming ssDNA. Inactivation of dprA (or recA) was previously shown to destabilize completely incoming ssDNA (1). To determine whether RadC could be responsible for ssDNA degradation when DprA is missing, we compared the fate of internalized transforming DNA in dprA and radC dprA mutant cells (Fig. 2). No stabilization of incoming ssDNA was detected in the latter, ruling out the possibility that RadC is a nuclease responsible for ssDNA degradation in the absence of DprA (or RecA).
FIG. 2.
Inactivation of radC has no detectable effect on the stability of internalized ssDNA in both wild-type and dprA mutant genetic backgrounds. Total DNA extracted from cells transformed with 32P-labeled DNA was fractionated by gel electrophoresis (see the supplemental material). The top of the figure shows densitometer tracings of electropherograms used to calculate the amount of donor label recovered in each lane (expressed as relative cpm). The bottom of the figure shows electropherograms from radC dprA double mutant (R1939), radC mutant (R1933), dprA mutant (R1750), and wild-type (R1928) cell extracts. The positions of chromosomal DNA (chr.) and of internalized ssDNA are indicated by an arrow and a bracket, respectively.
A role for RadC in competence regulation?
In the experiments described above, transformation was induced with synthetic CSP (8), and competence was monitored using an ssbB::luc fusion (17). The kinetics of ssbB expression of wild-type and radC mutant cells appeared indistinguishable, suggesting that RadC plays no role in the response to CSP and in the decay of CSP-induced competence (see Fig. S1 in the supplemental material). In addition, a comparison of the spontaneous induction of competence in wild-type and radC mutant cultures revealed no significant difference (see Fig. S2 in the supplemental material). Altogether, these data indicate that RadC is either individually dispensable or plays no role in the regulation of competence.
A role in mismatch repair?
Mismatch repair proficiency of radC mutant cells was also analyzed both during and outside competence. During chromosomal transformation, mismatch repair is known to be responsible for the low efficiency of transformation of the rif23 marker compared to the str41 marker. The Hex system (4) rejects potential rifampin-resistant transformants following recognition of the rif23/rif+ mismatch at the heteroduplex stage in transformation. Mismatch repair was unaffected in radC mutant cells (Table 1).
To evaluate mismatch repair proficiency outside competence, we took advantage of a previously constructed duplication of the rpsL gene with the two alleles, rpsL1 and rpsL+ (23). Gene conversion can occur between rpsL1, which confers streptomycin resistance (Smr), and rpsL+, which confers Sm sensitivity (Sms). Since the sensitive allele is dominant over the resistant allele, conversion of the former allele is revealed by the appearance of Smr colonies. This process was previously shown to depend on RecA and to be susceptible to mismatch repair (23) because the transient heteroduplex structure formed between the two rpsL genes creates a mismatch (rpsL+/rpsL1) recognized by the Hex system. Hex-dependent mismatch correction reduces spontaneous conversion to Smr by ∼18-fold (23). The conversion frequency measured with radC mutant cells did not differ significantly from that of the wild-type parent (Table 1). We concluded that RadC plays no role in Hex-dependent generalized mismatch repair.
Sensitivity to DNA damages.
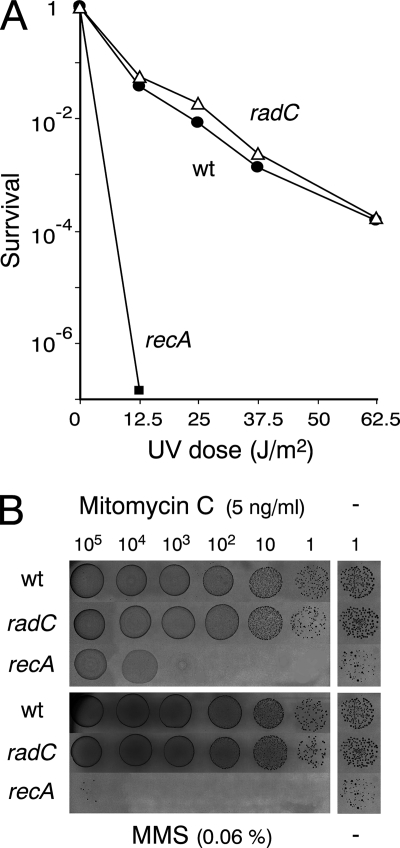
To assess a possible involvement of RadC in the repair of DNA damages outside competence, we compared the sensitivity to UV irradiation, to mitomycin C, and to methyl methanesulfonate (MMS) of a wild-type and a radC mutant strain (as well as a recA mutant as a control) as follows. Stocks of bacteria grown in THY medium (or C+Y medium for the recA mutant) to an optical density at 550 nm (OD550) of 0.4 were diluted 100-fold in C+Y medium (pH 6.8 to 7.0) and incubated at 37°C to an OD550 of 0.2. Then, 20 μl of each culture and of 10-fold serial dilutions were spotted on 1-day-old d-agar plates containing horse blood and catalase (500 U/ml). To assay UV sensitivity, plates were then UV irradiated at 2.5 J/m/s for 5, 10, 15, or 25 s. To measure sensitivity to mitomycin C and to MMS, plates contained variable concentrations of mitomycin C (5, 10, 15, or 20 ng/ml) or MMS (0.02, 0.04, 0.06, or 0.08%). In all cases, plates were incubated overnight at 37°C. The radC mutant strain exhibited sensitivity to UV (Fig. 3A) and to mitomycin C or MMS (Fig. 3B) similar to that of the wild-type strain, providing no support to the hypothesis that RadC plays a role in the repair of DNA damages in S. pneumoniae.
FIG. 3.
Inactivation of radC has no detectable effect on resistance to UV irradiation (A) and mitomycin C and MMS (B). The strains were the wild type (R1501), the radC mutant strain (R1966), and the recA mutant strain (R1409).
A helicase-related function of RadC?
radC was previously shown to be coexpressed with pcrA (20) (Fig. 1). PcrA is a DNA helicase involved in the replication of rolling-circle replicating plasmids (20). This coexpression prompted us to investigate whether RadC could be required for the establishment of a rolling-circle replicon plasmid (pLS1; see Table S1 in the supplemental material). Plasmid establishment frequencies were indistinguishable in radC mutant and wild-type cells (Table 1). We also investigated homology-facilitated plasmid establishment (12) using as donor plasmid pLS70, a pLS1 derivative carrying a 3,486-bp long chromosomal insert (see Table S1 in the supplemental material). The presence of a chromosomal insert is known to increase 10-fold or more the frequency of plasmid establishment by transformation in S. pneumoniae (12). The facilitation mechanism is likely to depend on synapsis of the internalized ssDNA plasmid fragment with the recipient chromosome, which favors reconstitution of an intact replicon. The frequency of pLS70 establishment was 10-fold higher than that of pLS1 in both wild-type and radC mutant cells (Table 1), indicating that RadC plays no role in the establishment of rolling-circle replicons via transformation. These data provide no support to the hypothesis that RadC has a helicase-related function in connection with PcrA.
Concluding remarks.
In the facultative phototrophic bacterium Rhodobacter capsulatus, radC was shown to be relatively highly expressed under both chemotrophic and phototrophic growth conditions (10). The expression of radC was reported to increase about fivefold after UV irradiation. However, the survival rate of radC mutant cells to mild UV treatment was not significantly different from that of the wild-type parent (10). In B. subtilis, radC belongs to the σM-responsive genes (9). As such, it is induced in response to environmental stresses, for example, stress generated by cationic antimicrobial peptides (15). During competence radC is specifically coinduced with six flanking genes: maf-(radC)-mreBCD-minCD. These genes encode proteins involved in cell shape determination and septum placement, suggesting that RadC could have a function related to cell envelope metabolism. However, neither of these observations provides a real clue as to the role of RadC since the genomic context of radC orthologues varies between species (data not shown). Thus, both the function of RadC proteins in nontransformable bacteria and their specific role during competence in naturally transformable species remain to be determined.
In view of the observations concerning radC102 of E. coli (11) and those on radC of S. pneumoniae (the present study), the pfam Group (http://pfam.sanger.ac.uk/) has followed our suggestion to rename the RadC family (to DUF2466; DUF stands for domain of unknown function), thereby avoiding the systematic annotation of newly sequenced bacterial genomes with a potentially misleading gene name.
Supplementary Material
Acknowledgments
We thank Nathalie Campo and Patrice Polard for critical reading of the manuscript.
This study was supported in part by a grant from the Agence Nationale de la Recherche (projet n_ BLAN06-3_141806).
Footnotes
Published ahead of print on 13 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bergé, M., I. Mortier-Barrière, B. Martin, and J. P. Claverys. 2003. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming single strands. Mol. Microbiol. 50527-536. [DOI] [PubMed] [Google Scholar]
- 2.Bergé, M., M. Moscoso, M. Prudhomme, B. Martin, and J. P. Claverys. 2002. Uptake of transforming DNA in gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45411-421. [DOI] [PubMed] [Google Scholar]
- 3.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. J. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 431331-1345. [DOI] [PubMed] [Google Scholar]
- 4.Claverys, J. P., and S. A. Lacks. 1986. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol. Rev. 50133-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felzenszwalb, I., N. J. Sargentini, and K. C. Smith. 1984. Characterization of a new radiation-sensitive mutant, Escherichia coli K-12 radC102. Radiat. Res. 97615-625. [PubMed] [Google Scholar]
- 6.Guiral, S., V. Hénard, C. Granadel, B. Martin, and J. P. Claverys. 2006. Inhibition of competence development in Streptococcus pneumoniae by increased basal-level expression of the ComDE two-component regulatory system. Microbiology 152323-331. [DOI] [PubMed] [Google Scholar]
- 7.Hamoen, L. W., W. K. Smits, A. De Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 2025517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 9211140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jervis, A. J., P. D. Thackray, C. W. Houston, M. J. Horsburgh, and A. Moir. 2007. SigM-responsive genes of Bacillus subtilis and their promoters. J. Bacteriol. 1894534-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katsiou, E., C. M. Nickel, A. F. Garcia, and M. H. Tadros. 1999. Molecular analysis and identification of the radC gene from the phototrophic bacterium Rhodobacter capsulatus B10. Microbiol. Res. 154233-239. [DOI] [PubMed] [Google Scholar]
- 11.Lombardo, M. J., and S. M. Rosenberg. 2000. radC102 of Escherichia coli is an allele of recG. J. Bacteriol. 1826287-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López, P., M. Espinosa, D. Stassi, and S. A. Lacks. 1982. Facilitation of plasmid transfer in Streptococcus pneumoniae by chromosomal homology. J. Bacteriol. 150692-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogura, M., H. Yamaguchi, K. Kobayashi, N. Ogasawara, Y. Fujita, and T. Tanaka. 2002. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 1842344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson, S., C. K. Sung, R. Cline, B. V. Desai, E. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. Burr, Y. Do, S. Ahn, J. Gilbert, R. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae. Mol. Microbiol. 511051-1070. [DOI] [PubMed] [Google Scholar]
- 15.Pietiainen, M., M. Gardemeister, M. Mecklin, S. Leskela, M. Sarvas, and V. P. Kontinen. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 1511577-1592. [DOI] [PubMed] [Google Scholar]
- 16.Prudhomme, M., A. Camilli, and J. P. Claverys. 2007. In vitro mariner mutagenesis of Streptococcus pneumoniae: tools and traps, p. 511-518. In R. Hakenbeck and G. S. Chhatwal (ed.), The molecular biology of streptococci. Horizon Scientific Press, Norfolk, United Kingdom.
- 17.Prudhomme, M., and J. P. Claverys. 2007. There will be a light: the use of luc transcriptional fusions in living pneumococcal cells, p. 519-524. In R. Hakenbeck and G. S. Chhatwal (ed.), The molecular biology of streptococci. Horizon Scientific Press, Norfolk, United Kingdom.
- 18.Redfield, R. J., A. D. Cameron, Q. Qian, J. Hinds, T. R. Ali, J. S. Kroll, and P. R. Langford. 2005. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae. J. Mol. Biol. 347735-747. [DOI] [PubMed] [Google Scholar]
- 19.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae. Mol. Microbiol. 361279-1292. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz-Maso, J. A., S. P. Anand, M. Espinosa, S. A. Khan, and G. del Solar. 2006. Genetic and biochemical characterization of the Streptococcus pneumoniae PcrA helicase and its role in plasmid rolling circle replication. J. Bacteriol. 1887416-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saveson, C. J., and S. T. Lovett. 1999. Tandem repeat recombination induced by replication fork defects in Escherichia coli requires a novel factor, RadC. Genetics 1525-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song, J. H., K. S. Ko, J. Y. Lee, J. Y. Baek, W. S. Oh, H. S. Yoon, J. Y. Jeong, and J. Chun. 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells 19365-374. [PubMed] [Google Scholar]
- 23.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 675190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vickerman, M. M., S. Iobst, A. M. Jesionowski, and S. R. Gill. 2007. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J. Bacteriol. 1897799-7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.