Abstract
The cell envelope of mycobacteria, which include the causative agents of tuberculosis and leprosy, is crucial for their success as pathogens. Despite a continued strong emphasis on identifying the multiple chemical components of this envelope, it has proven difficult to combine its components into a comprehensive structural model, primarily because the available ultrastructural data rely on conventional electron microscopy embedding and sectioning, which are known to induce artifacts. The existence of an outer membrane bilayer has long been postulated but has never been directly observed by electron microscopy of ultrathin sections. Here we have used cryo-electron microscopy of vitreous sections (CEMOVIS) to perform a detailed ultrastructural analysis of three species belonging to the Corynebacterineae suborder, namely, Mycobacterium bovis BCG, Mycobacterium smegmatis, and Corynebacterium glutamicum, in their native state. We provide new information that accurately describes the different layers of the mycobacterial cell envelope and challenges current models of the organization of its components. We show a direct visualization of an outer membrane, analogous to that found in gram-negative bacteria, in the three bacterial species examined. Furthermore, we demonstrate that mycolic acids, the hallmark of mycobacteria and related genera, are essential for the formation of this outer membrane. In addition, a granular layer and a low-density zone typifying the periplasmic space of gram-positive bacteria are apparent in CEMOVIS images of mycobacteria and corynebacteria. Based on our observations, a model of the organization of the lipids in the outer membrane is proposed. The architecture we describe should serve as a reference for future studies to relate the structure of the mycobacterial cell envelope to its function.
The suborder of Corynebacterineae is a distinct group of gram-positive bacteria and comprises mycobacteria and other genera such as Corynebacterium, Rhodococcus, and Nocardia. The medical importance of the group is enormous; it includes the causative agents of human diseases such as tuberculosis and leprosy, Mycobacterium tuberculosis and Mycobacterium leprae, respectively. The structure of the cell envelope of these bacteria has been the subject of numerous studies because it is already clear that the powerful biological activities of known wall components contribute significantly to the disease process. Indeed, lipids isolated from the cell envelope can elicit responses by the host immune system very similar to the responses generated by M. tuberculosis infection (e.g., granuloma formation) (22).
Schematically, the envelope of this bacterial group is composed of a typical plasma membrane (PM) surrounded by a cell wall core, which, in turn, is surrounded by an outer layer (OL) called the capsule in the case of pathogenic mycobacterial species (Fig. 1). The cell wall core consists of peptidoglycan covalently bound to arabinogalactan, which itself is covalently bound to mycolic acids (very-long-chain, from C30 to C90, α-alkyl, β-hydroxy fatty acids) (9). This envelope is unusual in that it is very rich in lipids, and unlike other gram-positive microorganisms, Corynebacterineae possess an outer permeability barrier. It has been postulated that this barrier is formed by a lipid bilayer analogous to the outer membrane (OM) of gram-negative bacteria (33). The arrangement of the lipids in this hypothetical OM has been long debated (26, 32, 34, 43, 44). Based on the chemical structures of the main cell envelope constituents, several models of the cell envelope, in particular, the hypothetical OM bilayer of mycobacteria, were developed (26, 32, 33, 43, 44). According to these models, the innermost leaflet consists mainly of the mycolic acids, which are, at least in part, covalently linked to the cell wall arabinogalactan. The outermost leaflet is proposed to be composed of various glycolipids, including trehalose monomycolate and trehalose dimycolate; of phospholipids; and of species-specific lipids such as glycopeptidolipids (GPL), phthiocerol dimycocerosate, and sulfolipids (8, 26, 33, 44). The presence of pore-forming proteins in low numbers in mycobacteria relative to Escherichia coli may explain both the limited permeability of mycobacterial cell envelopes and the generally rather low susceptibility of these bacteria to toxic agents (6, 9, 36).
FIG. 1.
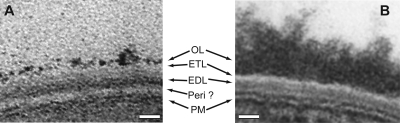
Thin-section transmission electron microscopy of chemically fixed and dehydrated Corynebacterineae. (A) M. smegmatis mc2155. (B) C. glutamicum (CGL2020). The cell envelope of M. smegmatis is composed of a PM; a thick, electron-transparent layer (Peri ?; interpreted as the postulated periplasmic space); a thick, internal, electron-dense layer (EDL; considered a complex of peptidoglycan and arabinogalactan); a thin, electron-transparent layer (ETL; assumed to be composed of mycolic acids and other lipids); and an electron-dense OL (a complex protein-carbohydrate matrix with some lipids) (15, 16). The EDL and the mycolic acids of the ETL form the cell wall core. The cell envelope of C. glutamicum looks similar, but it does not possess an obvious low-density hypothetical periplasmic space and the OL is thicker than in M. smegmatis (43). In both cases, the use of ruthenium red stain resulted in enhanced staining of the OL. Bars, 20 nm.
Nevertheless, not only is the precise organization of the OM bilayer debated, but even its existence can be questioned since there are no direct observations of this structure. On the one hand, the bilayer model is supported by data from freeze fracture electron microscopy which have clearly shown the occurrence of a major fracture plane in the outer part of the envelope, in addition to the expected plane that typifies the PM (5, 7, 33, 35, 43).
On the other hand, no sign of an OM bilayer was seen in ultrathin sections of Corynebacterineae (Fig. 1) (9, 11, 43), questioning its existence. Importantly, all electron microscopy analyses of ultrathin sections were done with specimens from which water had been removed, a prerequisite for electron microscopy observation at room temperature. We call this technique the conventional preparation method. Even when dehydration has been performed at low temperature by freeze-substitution in order to better preserve biological structures, no OM bilayer was seen in Corynebacterineae (9, 28, 39, 40). This is possibly due to the fact that, during dehydration, water-soluble molecules tend to aggregate and lipid molecules may be prone to extraction or rearrangement by organic solvents (14).
In an attempt to resolve the question of the existence of an OM bilayer in Corynebacterineae, we addressed the native structure of both mycobacteria and the closely related corynebacteria, whose cell envelope resembles that of mycobacteria after conventional electron microscopy (43). We focused our study on Mycobacterium smegmatis, Mycobacterium bovis BCG, and Corynebacterium glutamicum by cryo-electron microscopy (cryoEM) of vitreous sections (CEMOVIS). In the CEMOVIS technique, specimens are vitrified by high-pressure freezing (i.e., cooled to liquid nitrogen temperature without water crystallization). The vitreous specimens are then cryosectioned and imaged in a cryo-electron microscope in their fully hydrated, native state. The artifacts of aggregation and lipid extraction are therefore prevented (1). Importantly, this is the only sectioning technique for electron microscopy involving freezing where the vitreous state can be unambiguously confirmed, by electron diffraction. With this approach, we provide detailed insights into the structure of the mycobacterial cell envelope in its native state and a direct visualization of the mycobacterial OM. Furthermore, we demonstrate that mycolates are essential constituents of this structure through the use of a wild-type and a mycolate-free strain of C. glutamicum (41).
MATERIALS AND METHODS
Culture conditions.
M. smegmatis mc2155 (ATCC 700084) and M. smegmatis tmptB (49) were cultured in LB (Luria-Bertani broth; Difco, Basel, Switzerland) at 37°C with aeration. M. bovis BCG Pasteur (ATCC 35734) was cultured in 0.05% Tween 80, oleic acid-albumin-dextrose-catalase-enriched (oleic acid dextrose complex) Middlebrook 7H9 broth (Difco) at 37°C with aeration. Mycobacteria were harvested during the exponential growth phase (optical density at 600 nm of 3 for M. smegmatis and of 0.24 for M. bovis BCG). Wild-type C. glutamicum (ATCC 13032) was cultured on 3% agar-brain heart infusion medium (Difco) at 30°C. C. glutamicum Δpks13::km (41) was cultured on agar-brain heart infusion medium supplemented with 25 μg/liter kanamycin at 30°C. Corynebacterial colonies were harvested after overnight growth. E. coli B/r carrying pUC19 was cultured in 2 × YT in the presence of 100 μg/ml ampicillin (45). Cells were harvested during the exponential growth phase (optical density at 600 nm of 0.96). Streptococcus gordonii Challis was cultured and processed for CEMOVIS as previously described (58). Harvesting was done by centrifugation at 3,200 × g for 5 min.
Vitrification and cryosectioning.
For CEMOVIS, M. smegmatis and M. bovis BCG were washed twice in phosphate-buffered saline (PBS) supplemented with 20% dextran (20% dextran-PBS) (average molecular mass, 40 kDa; Sigma-Aldrich, Buchs, Switzerland). They were then introduced into copper tubes and vitrified with an EM PACT high-pressure freezer (Leica, Vienna, Austria). Corynebacterial colonies were scraped and resuspended in 20% dextran-PBS. E. coli was centrifuged and resuspended in 20% dextran-PBS. Corynebacteria and E. coli were introduced into membrane carriers (Leica) and vitrified with the same apparatus. Afterwards, tubes were mounted in the tube holder of an FC6/UC6 cryo-ultramicrotome (Leica) and trimmed to a pyramidal shape as previously described (51, 58). Membrane carriers were clamped in the flat specimen holder of the cryo-ultramicrotome. Copper was trimmed away with a diamond knife (Diatome, Bienne, Switzerland) on part of the specimen holder, and the specimen was trimmed to a pyramidal shape with the same knife. Forty- to 50-nm feed cryosections were cut with a 35 or 45° diamond knife (Diatome) under standard cutting conditions (58). They were collected on carbon-coated 1,000-mesh grids or noncoated lacey carbon grids (Agar Scientific, Essex, United Kingdom) and stored in liquid nitrogen or transferred immediately to the microscope.
For staining experiments, M. smegmatis mc2155 was washed three times in PBS and fixed for 2 h in 2.5% (wt/vol) glutaraldehyde in cacodylate buffer at room temperature. Cells were washed three times in cacodylate buffer and postfixed in 1% (wt/vol) OsO4 for 2 h at room temperature. They were washed three times in cacodylate buffer and then processed for high-pressure freezing as described above.
For cryoEM of whole-mount wild-type C. glutamicum, colonies were scraped and resuspended in 20 mM Tris, pH 7.5. A 4-μl sample volume was allowed to adsorb to a carbon-coated grid for 1 min, blotted with Whatman no. 4 filter paper (Merck, Zurich, Switzerland), and vitrified by plunging into liquid ethane at −178°C.
CryoEM.
For CEMOVIS, grids were transferred to a cryoholder (Gatan, Warrendale, PA) kept below −170°C and inserted into CM100, Tecnai 12, and Tecnai F30 cryo-electron microscopes (FEI, Eindhoven, Netherlands) equipped with a LaB6 cathode, a tungsten cathode, and a field emission gun, respectively. The accelerating voltages were 100, 120, and 200 kV, respectively. Specimens were irradiated with a low electron dose. Electron diffraction was used to check whether water was vitreous or crystalline. Crystalline sections were discarded. Images were recorded with a TemCam-F224HD charge-coupled device camera (Tietz Video and Image Processing Systems, Munich, Germany) at magnifications of ×22,500, ×33,000, and ×53,000. No image processing other than that described in the figure legends was performed.
Whole-mount plunge-frozen cells were transferred to a Gatan cryoholder and imaged at a magnification of ×50,000 in a CM200-FEG (FEI) operated at an accelerating voltage of 200 kV. Images were recorded on Kodak SO-163 plates and scanned at a pixel size of 0.5 nm. For conventional electron microscopy, C. glutamicum (CGL2020) and M. smegmatis mc2155 were grown, prepared, and imaged as previously described (16, 43).
Quantitative measurements.
Pixel size was calibrated by using a two-dimensional crystal of catalase (Agar Scientific). At magnifications of ×22,500, ×33,000, and ×53,000, the pixel sizes are 0.63, 0.50, and 0.31 nm, respectively. During cryosectioning, material is compressed along the cutting direction. Nevertheless, it has been shown that compression does not affect the dimensions measured perpendicularly to the cutting direction (13). Dimensions were measured accordingly on average density profiles calculated along rectangular selections with the software ImageJ (NIH, Bethesda, MD). The width of selection rectangles is specified in the figure legends.
The density of OsO4-stained cells was measured with the software EMMENU (Tietz Video and Image Processing Systems) in raw image files acquired at 120 kV at a defocus between −1.7 and −2.3 μm. The density of the OM was normalized to the density of the background.
RESULTS
Vitreous sections—general considerations.
We investigated the structure of the cell envelopes of two species of mycobacteria with CEMOVIS, namely, M. smegmatis, a fast-growing nonpathogenic species, and M. bovis BCG, a slow-growing vaccine strain belonging to the same complex as M. tuberculosis and showing more than 99.9% genome sequence identity with this pathogen (19). Henceforth, M. smegmatis refers to strain mc2155 unless mentioned differently. A low-magnification micrograph of M. smegmatis is shown in Fig. 2. Its quality is representative of the majority of the sections that we observed. It does not contain chatter or crevasses, which are cutting artifacts that complicate image interpretation because they produce irregular distortions (inhomogeneities) (2). Knife marks (arrows) and compression along the cutting direction cannot not be prevented but are homogeneous and therefore do not hinder image interpretation.
FIG. 2.
CEMOVIS of a cross-sectioned M. smegmatis mc2155 cell. Arrows, knife marks. Bar, 500 nm.
Structure of the cell envelopes of M. bovis BCG and M. smegmatis.
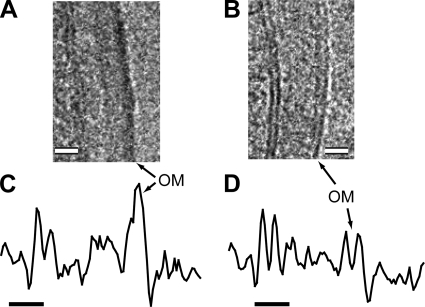
The cell envelopes of M. smegmatis and M. bovis BCG are structurally similar in CEMOVIS (Fig. 3A to F; Table 1). They are composed of a PM, a granular layer (GL), an inner wall zone (IWZ) of low density, a medial wall zone (MWZ) of intermediate density, and an OM of higher density. The bilayer aspect of the OM can be best visualized in micrographs recorded with a relatively small defocus value (Fig. 3A and D) and in the corresponding density profiles (Fig. 3C and F), whereas the organization of the GL, the IWZ, and the MWZ is best seen in micrographs recorded with a larger defocus value (Fig. 3B and E). The GL is found next to the PM, in the IWZ. The appearance of the mycobacterial GL and IWZ is similar to that of the GL and the IWZ of typical gram-positive bacteria (e.g., S. gordonii) visualized by CEMOVIS (58) (Fig. 3G and H; Table 1). The MWZ of mycobacteria is topologically identical to the gram-positive peptidoglycan layer (outer wall zone) but is thinner. On images recorded with a relatively large defocus, the MWZ seems separated from the OM by a low-density gap (Fig. 3F). However, on images recorded very close to focus, the gap is strongly reduced (Fig. 3C) or absent (data not shown), whereas the leaflets of the OM are distinct. The gap is thus a phase-contrast artifact that can lead to a slight underestimation of the MWZ thickness.
FIG. 3.
Cell envelope of mycobacteria and gram-positive and gram-negative bacteria by CEMOVIS. (A, B, and C) M. smegmatis mc2155. (D, E, and F) M. bovis BCG. (G and H) S. gordonii. (I and J) E. coli. Images were acquired at 100 kV and defocused by −2.5 μm (A, D, and I), −5 μm (B), −3.7 μm (E), and −4.5 μm (G). They were denoised by Gaussian filtering in Adobe Photoshop (radius of 0.6 pixel for panels A and D and of 1 pixel for panels B, E, G, and I). The density profiles in panels C, F, H, and J were obtained from nondenoised images corresponding to panels A, E, G, and I, respectively. They were averaged over a width of 70 pixels (C) and 50 pixels (F, H, and J). Note that the GL is in IWZ. OWZ, outer wall zone; PG, peptidoglycan layer. Bars, 20 nm (A, B, D, E, G, and I) and 10 nm (C, F, H, and J).
TABLE 1.
Dimension of cell envelope structures based on CEMOVIS analysis
| Organism | Avg position (nm) ± SD (no. of measurements)a
|
Avg thickness (nm) ± SD (no. of measurements)b
|
||||||
|---|---|---|---|---|---|---|---|---|
| GL | OM | PM | GL | IWZ | MWZ | OM | Cell envelope | |
| M. smegmatis | 8.4 ± 1.2 (8) | 35.2 ± 1.9 (8) | 7.1 ± 0.6 (8) | 3.6 ± 0.9 (8) | 16.6 ± 1.2 (8) | 7.3 ± 1.9 (8) | 7.1 ± 0.6 (8) | 42.4 ± 2.3 (8) |
| M. bovis BCG | 9.0 ± 1.3 (8) | 31.5 ± 1.9 (8) | 6.3 ± 0.7 (8) | 3.8 ± 0.8 (8) | 14.1 ± 1.6 (8) | 6.3 ± 1.0 (8) | 7.5 ± 0.8 (8) | 38.7 ± 2.4 (8) |
| Wild-type C. glutamicum | 8.7 ± 0.7 (9) | 44.5 ± 7.5 (9) | 6.7 ± 0.4 (9) | 4.3 ± 0.8 (9) | 18.0 ± 1.4 (8) | 20.9 ± 8.8 (8) | 4.7 ± 0.7 (9) | 48.6 ± 6.7 (8) |
| C. glutamicum Δpks13 | 8.4 ± 1.4 (6) | NAc | 5.5 ± 0.5 (6) | 3.6 ± 0.3 (6) | 15.4 ± 2.0 (6) | 18.7 ± 3.2 (6) | NA | 37.2 ± 4.1 (6) |
| E. coli K-12d | NA | 27.3 | 5.8 ± 0.4 (8) | NA | 11.5 | 6.4 ± 0.5 (8) | 6.9 ± 1.0 (8) | 33.7 |
| S. gordonii Challise | 8.3 ± 0.5 (10) | NA | 6.3 ± 0.3 (6) | 4.1 ± 0.9 (9) | 12.9 ± 1.0 (10) | 26.4 ± 3.6 (10) | NA | 45.5 ± 3.4 (10) |
Distance between the center of the PM and the center of the designated structure.
In the case of bilayers, the thickness corresponds to the total width of the structure and not to the peak-to-peak distance.
NA, not applicable.
There is no structure similar to the mycobacterial OM in classical gram-positive bacteria (e.g., S. gordonii), but the mycobacterial OM is structurally analogous to the OM of gram-negative bacteria (e.g., E. coli) (Fig. 3I and J). The thickness of the mycobacterial OM is similar to the published thickness of the gram-negative OM visualized by CEMOVIS (Table 1) (29). Even though an OM in mycobacteria has long been suggested and supported by freeze fracture electron microscopy, here we report a direct observation of such a bilayer in native mycobacteria. Together, these comparisons indicate the following three points: (i) the IWZ occupies the position shown to be a periplasmic space in other gram-positive bacteria (30, 31); (ii) the MWZ is likely formed, at least in part, of peptidoglycan; and (iii) the OM is made of molecules specific to mycobacteria and not ubiquitous in gram-positive bacteria. Because these molecules form a bilayer, they are likely to contain a hydrophobic moiety. OsO4 is considered to label predominantly lipids (56). We thus fixed M. smegmatis cells with glutaraldehyde, postfixed them with OsO4, and subsequently processed them for CEMOVIS. In a negative control, cells were processed for CEMOVIS directly after glutaraldehyde fixation. The density of the OM is, on average, 39% higher in OsO4-treated cells than in control cells (P < 0.01, Fig. 4). These data suggest that the OM is mainly made of lipids. Besides, the bilayer aspect of the OM is lost after OsO4 staining in most of the images, an observation that requires further investigation. Mycolic acids, which are the hallmark lipids of Corynebacterineae, of which a fraction is covalently bound to peptidoglycan via arabinogalactan, are likely to be a major constituent of the OM.
FIG. 4.
Staining of M. smegmatis mc2155 with OsO4. (A) Glutaraldehyde-fixed and OsO4-postfixed cell. (B) Glutaraldehyde-fixed cell. (C) Density profile of the cell envelope in panel A. (D) Density profile of the cell envelope in panel B. Images A and B were acquired at 120 kV. They were denoised by Gaussian filtering in Adobe Photoshop (radius of 0.6 pixel). They have the same intensity scale. Images were defocused by −1.9 μm (A) and −2.1 μm (B) Density profiles were obtained from nondenoised images. They were averaged over a width of 35 pixels, and both profiles are shown at the same scale. The apparent difference in the distance between the PM and the OM in panels C and D is due to the fact that the measured cell envelopes had a different orientation in relation to the cutting direction. Bars, 20 nm (A and B) and 10 nm (C and D).
In order to test our hypothesis that the OM of mycobacteria is made at least in part of mycolic acids, we wanted to observe mutant mycobacteria that would be devoid of mycolic acid. However, this was impossible because mycobacteria cannot survive in the absence of this compound (3, 18, 41, 42, 53). For the same reason, treating cells with drugs that inhibit the synthesis of mycolic acids (e.g., isoniazid) proved useless. Indeed, at the highest sublethal isoniazid concentration, the amount of mycolic acid per cell is only reduced by 20% (4). Furthermore, probes such as gold-coupled antibodies could not be used because they are too large to diffuse through the cell envelope. And since specimens must be kept frozen, immunolabeling of cryosections is impossible.
Nevertheless, we studied M. smegmatis tmptB, a mutant devoid of GPL and which forms very large cell aggregates (49). GPL are considered to be an important constituent of the OM outer leaflet in several mycobacterial species (26), and their disruption may therefore affect the OM. CEMOVIS revealed the structure of the contact zone between tmptB cells within an aggregate: all of the layers of the cell envelope are present, except the OM outer leaflet, and cells within the aggregate are interacting via their OM inner leaflet (see Fig. S1A in the supplemental material). This assertion is confirmed by images showing that, at the edge of the contact zone, the OM outer leaflet is highly curved and is continuous between the two cells involved in the contact, whereas this is not the case for the inner leaflet (see Fig. S1B in the supplemental material). Contacts between wild-type cells also exist, and their structure is identical to that of contacts seen between tmptB cells (see Fig. S1C in the supplemental material). However, the deletion of GPL generates an increase in the frequency of such contacts and, accordingly, the surface ratio of the OM outer leaflet versus the inner leaflet is reduced, which is in agreement with the expected localization of GPL in the OM outer leaflet and reinforces the hypothesis that the bilayer that we have observed is indeed the long-searched-for mycobacterial OM. The lipidic nature of the OM is further supported by the fact that the OM inner leaflet was never observed in direct contact with the aqueous environment but is always coated with either the OM outer or inner leaflet of another cell in contact.
Structures of the cell envelopes of wild-type and mycolic-acid-deficient C. glutamicum.
Corynebacteria are closely related to mycobacteria, and they possess a similar cell envelope. C. glutamicum has become widely used in the study of mycolic acids because, as opposed to mycobacteria, C. glutamicum can grow in the absence of mycolic acids (3, 18, 41, 42, 53). In order to further test our hypothesis that the OM of Corynebacterineae is made, in part, of mycolic acids, we studied C. glutamicum ATCC 13032 (henceforth referred to as wild-type C. glutamicum), which possesses mycolic acids, and C. glutamicum Δpks13::km (henceforth referred to as C. glutamicum Δpks13), a viable mutant strain that is deficient in the production of these fatty acids (41).
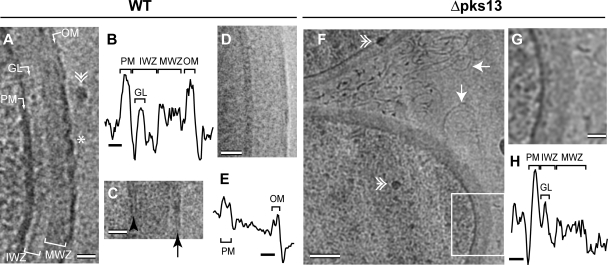
Our CEMOVIS micrographs show that the cell envelope of wild-type C. glutamicum is similar to that of mycobacteria; it consists of an IWZ, a GL, an MWZ, and an OM. Though similar, the cell envelopes are not identical; the MWZ of C. glutamicum is considerably thicker than the MWZ of mycobacteria, exhibiting values similar to those of classical gram-positive bacteria in CEMOVIS micrographs (Fig. 5A and C; Table 1) (58). This suggests that the amount of peptidoglycan/arabinogalactan could be more important in corynebacteria than in mycobacteria. On the other hand, the corynebacterial OM is thinner than the mycobacterial OM and the corynebacterial PM. For this reason, we expect that a higher resolution in micrographs is needed to visualize its bilayer aspect, which is more difficult to obtain. In addition, in order to be seen, the two dense layers of the OM need to be aligned along the viewing axis through most of the section thickness (see Fig. 2 in reference 59). However, the corynebacterial OM rarely appears smooth but shows many defects, which might thus hinder the bilayer aspect of the corynebacterial OM (Fig. 5A, asterisk). As a consequence, this aspect is visible only in local portions of micrographs recorded close to focus (Fig. 5C, arrow).
FIG. 5.
Cell envelope of corynebacteria visualized by CEMOVIS and whole-mount cryoEM. (A to E) Wild-type (WT) C. glutamicum. (F to G) C. glutamicum Δpks13. (A, C, F, and G) CEMOVIS. (D) Whole-mount cryoEM. (G) Higher magnification of the boxed area in panel F. Images A, C, F, and G were acquired at 100 kV, and image D was acquired at 200 kV. Images were defocused by −3.4 μm (A), −1.1 μm (C), −3 μm (D), and −5 μm (F and G). They were denoised by Gaussian filtering in Adobe Photoshop (radius of 1 pixel for panels A and D, 0.8 pixel for panel C, and 2.4 pixels for panels F and G). The density profiles in panels B and H were obtained from nondenoised images corresponding to panels A and G, respectively. The density profile in panel E was obtained from Gaussian filtered (radius of 1 pixel) image D. The density profiles were averaged over a width of 74 pixels (B), 65 pixels (E), and 70 pixels (H). The abbreviations are the same as those in Fig. 3. Double arrowhead, ice contamination; asterisk, cutting-induced defects in the OM; black arrowhead, PM; black arrow, OM; white arrow, filaments. Bars, 20 nm (A, C, D, and G), 10 nm (B, E, and H), and 100 nm (F).
In order to determine if these defects are a native feature of the corynebacterial OM or a cutting artifact, we examined whole-mount plunge-frozen wild-type C. glutamicum cells by cryoEM (13). The cytoplasm appears completely featureless due to the large thickness of the sample (data not shown), but both the PM and the OM can readily be observed. The OM appears smoother than in CEMOVIS micrographs, indicating that this structure is particularly prone to cutting artifacts (Fig. 5D). Furthermore, after Gaussian filtering of whole-mount cell micrographs, a bilayer aspect can be distinguished in many places of the OM but its dimensions are close to the resolution limit due to specimen thickness (Fig. 5D and E). Altogether, our data suggest that the corynebacterial OM is also a bilayer.
Importantly, the cell envelope of mycolate-free C. glutamicum Δpks13 lacks an OM (Fig. 5F to H). The MWZ thickness does not significantly differ between wild-type C. glutamicum and C. glutamicum Δpks13. This indicates that mycolic acids are not present in the MWZ and that the OM found in wild-type C. glutamicum is formed of mycolic acids and possibly of other lipids that interact with them. Furthermore, the extracellular medium of C. glutamicum Δpks13 contains a large amount of filaments. These are 3.9 ± 0.2 nm thick, which is thinner than the OM found in wild-type C. glutamicum. Thus, they possibly represent lipids that normally interact with mycolic acids to form the outer leaflet of the OM. In the absence of mycolic acid, they would still be exported but would not be linked to the cell wall anymore and would be released in the outer medium. This interpretation needs to be tested by further analysis of the released material.
DISCUSSION
Insights into the native structure of mycobacterial cell envelope and comparison with images of conventional preparations.
A variety of electron microscopy techniques have been used to decipher the unusual architecture of mycobacteria and related microorganisms. These include ultrathin sectioning of both conventionally processed and freeze-substituted samples, negative staining, and freeze fracture. Although the impression prevails from all of the methods that the cell envelope is layered, different techniques give different pictures of the layering. Consequently, the assignment of the known chemical components of the wall to ultrastructurally defined layers is not straightforward (9).
In ultrathin conventional sections, the cell envelope of mycobacteria is seen as being composed of an asymmetric PM with a thin inner leaflet and a thick outer leaflet (10, 47, 48); a thick, electron-dense layer (EDL); an electron-transparent layer (ETL); and an OL of variable density and thickness (Fig. 1). Because of the ETL's transparency and the disappearance of this layer after removal of lipids by alkaline hydrolysis (12), the ETL is assumed to contain lipids, mainly mycolic acids. The density of the EDL makes it likely to contain the peptidoglycan to which the arabinogalactan is attached at many sites. In mycobacteria, but not in corynebacteria, the PM is separated from the EDL by a space that may correspond to a periplasm (Fig. 1) (9, 43). Based on electron microscopy data, notably those from freeze fracture (5, 7, 33, 35, 43), and on the chemical structures of the main cell envelope constituents, a model of the envelope was developed in 1982 by Minnikin (33), followed by several improvements and modifications (26, 32, 34, 43, 44). Nevertheless, discrepancies remained between the images provided by various ultrastructural techniques and chemical knowledge.
Application of CEMOVIS to representatives of pathogenic and slow-growing, as well as saprophytic and rapid-growing, mycobacterial species, i.e., M. bovis BCG and M. smegmatis, respectively, resulted in images that differ from those previously obtained by conventional and freeze-substitution electron microscopy in the following important points. (i) The PM has its typical bilayer aspect, with a thickness similar to that observed in other bacterial cells (Table 1 and Fig. 6) (29-31, 58). The inner leaflet has the same density as the outer one, in contrast to the asymmetrical appearance often reported by thin conventional sections of mycobacteria, that was speculated to be due to the glycoconjugates in the outer leaflet (47, 48). This asymmetrical appearance of the membrane seen in conventional sections could be a consequence of the dehydration process involved in sample preparation, which could cause the collapse of the GL against the outer leaflet of the PM. (ii) A compartment similar to the periplasmic space of both gram-positive and gram-negative bacteria (29-31, 58) is apparent in CEMOVIS images not only in mycobacteria, as it was predicted from conventional electron microscopy (9), but also in corynebacteria. This could provide cells with a space where enzymatic reactions involved in cell envelope maintenance can take place. As mentioned, the GL lies in the mycobacterial and corynebacterial IWZ. This layer has been observed in gram-positive bacteria but not in gram-negative bacteria (58). Thus, its presence in both mycobacteria and corynebacteria is consistent with the fact that these bacteria belong to the gram-positive group. Recent cryo-electron tomography-based reports of a similar structure in cell wall-free Mycoplasma pneumoniae suggest that it could be made of membrane-bound proteins (20, 46). (iii) No ETL that is visible in conventional thin sections and that has been considered to be made up of cell wall-linked mycolates is seen in either M. bovis or M. smegmatis examined by CEMOVIS. The presence of this layer in conventional preparation of mycolate-free corynebacteria (i.e., Corynebacterium amycolatum [43]) indicates that the layer either represents a technical artifact or contains lipids other than mycolates. (iv) Most importantly, both M. bovis and M. smegmatis cells treated by CEMOVIS unambiguously show the presence of an outer bilayer, a key point of validation of the common basic feature of the current cell envelope models.
FIG. 6.
Schematic representation, at scale, of the cell envelopes of E. coli, M. smegmatis, C. glutamicum, and S. gordonii as seen with CEMOVIS. The abbreviations are the same as those in Fig. 3. The GL is drawn as bound to the PM. This hypothesis is based on the conclusion of our previous work with gram-positive bacteria (58). The IWZ is attributed, by analogy to other bacteria, to a periplasmic space. The MWZ represents the peptidoglycan layer in E. coli and S. gordonii and would correspond to the peptidoglycan-arabinogalactan layer in M. smegmatis and C. glutamicum. The OL of M. smegmatis and C. glutamicum is not depicted (see Discussion).
The density of the OM is strongly increased after fixation with OsO4, a chemical that stains mainly lipids (56). In cells devoid of GPL, which in the OM are considered to be mainly present in the outer leaflet (26), the OM inner leaflet has not been seen uncoated and in direct contact with the aqueous outer medium. Yet, the reduction in the surface area of the OM outer leaflet is correlated with an increase in the contact surface between the OM inner leaflets of two different cells forming an aggregate. These observations strongly support the hydrophobic and lipidic nature of the leaflets composing the OM.
To firmly establish the involvement of mycolates in the OM, we used corynebacteria as a surrogate model for studying essential mycobacterial compounds, such as mycolic acids, which are dispensable in corynebacteria (3, 18, 41, 42, 53). We compared the architecture of a wild-type strain and a mycolate-free mutant of C. glutamicum (41). Our data clearly show that whereas the wild-type strain had a distinct OM, the pks13 knockout mutant is devoid of this structure, making a compelling argument that the OM is a real structure that contains mycolic acids.
Challenges to the current models of the mycobacterial OM.
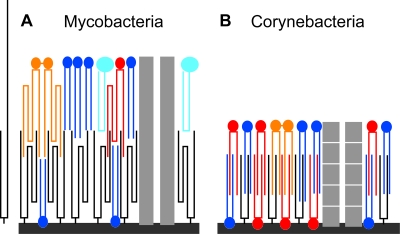
The thickness of the OM is 4 to 5 nm in C. glutamicum and 7 to 8 nm in mycobacteria. This observation is consistent with the presence of mycolyl residues in the structure and the shorter chain lengths of corynomycolic acids (32 to 36 carbons) compared to those of mycobacteria (70 to 90 carbons). The separation of the density peaks of the PM bilayer (3.9 ± 0.4 nm in M. bovis BCG) corresponds to the separation measured by cryoEM and X-ray scattering in liposomes made of phosphatidylcholine with acyl chain lengths of 16 to 18 carbons (25, 52). This is consistent with the lengths of the main fatty acid constituents of the PM (11). On the other hand, the mycobacterial OM is only slightly thicker than the PM (Table 1 and Fig. 6), whereas the main (meromycolic) chain of the mycolic acids is much longer (49 to 61 carbons in M. bovis BCG [50] and 35 to 58 carbons in M. smegmatis [57]). Similarly, both chains of C. glutamicum mycolic acids are made of 16 to18 carbons (8), like the main constituents of the PM, but the corynebacterial OM is considerably thinner than the PM. It therefore appears that to accommodate the limited thickness of the OM, the lipids facing the arabinogalactan-bound mycolic acids must be intercalated between mycolic acid chains, resulting in a zipper-like structure (Fig. 7). These lipids could be represented by extractable lipids (i.e., noncovalently bound to arabinogalactan) of a ubiquitous nature (e.g., trehalose mycolate, phospholipids) and of species-specific types (e.g., sulfolipid, phthiocerol dimycocerosate). Such a structure was originally proposed by Minnikin (33). The models proposed by Rastogi (44) and Liu et al. (26), in which the extractable lipids form a totally distinct monolayer rather than intercalating with the nonextractable mycolic acids, seems unlikely because they result in an OM that would be much thicker than what we see by CEMOVIS. Villeneuve et al. have recently proposed a novel conformational model of mycolic acids where the meromycolyl chain is folded upon itself to create a compact structure (54, 55). In this case, the thickness of a monolayer of mycolic acids corresponds to the length of mycolic acid short arm, which is unfolded (20 to 26 carbons). Although this model does not contradict our CEMOVIS data, it is not sufficient to fully explain them. Indeed, the short arm of M. smegmatis mycolic acids is 22 carbons long (57), i.e., 20 to 30% longer than phospholipids forming the PM, whereas the thickness of the PM equals that of the OM in our data. Likewise, both chains of corynebacterial mycolic acids should be unfolded, as suggested by the model of Villeneuve et al.; as mentioned above, these chains have the same length as the PM phospholipids (16 to 18 carbons), whereas the OM is thinner than the PM. Thus, even if the mycobacterial meromycolyl chain is folded and compact, both the mycobacterial and corynebacterial arabinogalactan-bound mycolic acids forming the inner leaflet of the OM have to be intercalated to a certain extent with the longest chains of free lipids (e.g., trehalose mycolate) forming the OM outer leaflet (Fig. 7).
FIG. 7.
Zipper model of the OM of Corynebacterineae. (A) Mycobacteria. (B) Corynebacteria. Hydrocarbon chains of the lipids are drawn to scale. Black, mycolic acid; dark blue, phospholipids (16- to 18-carbon-long chains); dark gray, peptidoglycan-arabinogalactan; light blue, GPL; light gray, porin; orange, trehalose dimycolate; red, trehalose monomycolate. Mycolic acids and trehalose mycolates are folded (54, 55). An unfolded mycolic acid is shown in panel A. It is too large to be accommodated in the OM. Porins are not drawn to scale. Of note, the porin of M. smegmatis MspA is expected to protrude out of the OM (27). The porin of corynebacteria has been proposed to be made by a stack of short proteins (6 kDa) (43). GPL are species-specific lipids found in M. smegmatis but not in M. bovis BCG (16). It has been suggested that the OM inner leaflet of corynebacteria contains a substantial amount of mycolic acids noncovalently bound to the peptidoglycan-arabinogalactan (43).
Since we submitted this work for publication, a different laboratory has published CEMOVIS images and cryo-electron tomograms of M. smegmatis, M. bovis BCG, and C. glutamicum (21). Their results are essentially similar to ours, but the models of the OM of mycobacteria that they propose significantly differ from ours in that the mycolic acids are unfolded. As we have explained above, we think that the unfolded meromycolic chain is too long to fit in the OM. Hoffmann and colleagues have not proposed any model for the OM of corynebacteria.
Future directions and conclusion.
In contrast to pictures from conventional (Fig. 1) and freeze-substitution (39, 40) techniques, no OL/capsule was seen at the surface of either mycobacteria or corynebacteria. This situation may be due to the fact that the components of such a layer would have the same density as the cryoprotection medium and would therefore be undistinguishable from this medium in CEMOVIS images. On the other hand, in mycobacteria, the capsular constituents are known to be loosely attached to the cell wall and most of them are found released in the culture fluids of in vitro-grown bacteria (23, 24, 37, 38), potentially explaining why they are not visible in our images. When mycobacteria grow intracellularly, these constituents are confined around the bacteria by the phagosomal membrane (9). Techniques were developed to maintain these constituents around in vitro-grown cells and visualize them by conventional transmission electron microscopy (17). Further studies are warranted to address the issue of the structure of the mycobacterial OL and capsule by CEMOVIS.
In conclusion, our study brings a new reference structure of the cell envelope of mycobacteria and corynebacteria. This should serve as a framework for building new models of the organization of chemical components in the cell envelope of mycobacteria. It also provides a basis for analyzing whether the cell envelope becomes significantly modified when mycobacteria are enclosed within phagosomes in macrophages, a question we are currently addressing.
Supplementary Material
Acknowledgments
We thank Jeanne Salje (Cambridge, United Kingdom) for providing the E. coli B/r strain and Gilles Etienne (Toulouse, France) for helpful discussions and for providing M. smegmatis tmptB. We are grateful to Pierre Gounon (Nice, France) for his technical help in conventional electron microscopy. We thank Nigel Unwin (Cambridge, United Kingdom) and Andreas Engel (Basel, Switzerland) for their support.
B.Z. is supported by an EMBO long-term fellowship.
Footnotes
Published ahead of print on 20 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Al-Amoudi, A., J. J. Chang, A. Leforestier, A. McDowall, L. M. Salamin, L. P. Norlen, K. Richter, N. S. Blanc, D. Studer, and J. Dubochet. 2004. Cryo-electron microscopy of vitreous sections. EMBO J. 233583-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Amoudi, A., D. Studer, and J. Dubochet. 2005. Cutting artefacts and cutting process in vitreous sections for cryo-electron microscopy. J. Struct. Biol. 150109-121. [DOI] [PubMed] [Google Scholar]
- 3.Alderwick, L. J., E. Radmacher, M. Seidel, R. Gande, P. G. Hitchen, H. R. Morris, A. Dell, H. Sahm, L. Eggeling, and G. S. Besra. 2005. Deletion of Cg-emb in Corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an arabinan-deficient mutant with a cell wall galactan core. J. Biol. Chem. 28032362-32371. [DOI] [PubMed] [Google Scholar]
- 4.Bardou, F., A. Quemard, M. A. Dupont, C. Horn, G. Marchal, and M. Daffé. 1996. Effects of isoniazid on ultrastructure of Mycobacterium aurum and Mycobacterium tuberculosis and on production of secreted proteins. Antimicrob. Agents Chemother. 402459-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barksdale, L., and K. S. Kim. 1977. Mycobacterium. Bacteriol. Rev. 41217-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 6429-63. [DOI] [PubMed] [Google Scholar]
- 7.Chami, M., N. Bayan, J. Dedieu, G. Leblon, E. Shechter, and T. Gulik-Krzywicki. 1995. Organization of the outer layers of the cell envelope of Corynebacterium glutamicum: a combined freeze-etch electron microscopy and biochemical study. Biol. Cell 83219-229. [DOI] [PubMed] [Google Scholar]
- 8.Daffé, M. 2005. The cell envelope of corynebacteria in Corynebacterium glutamicum, p. 121-148. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Inc., Boca Raton, FL.
- 9.Daffé, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39131-203. [DOI] [PubMed] [Google Scholar]
- 10.Daffé, M., M.-A. Dupont, and N. Gas. 1989. The cell envelope of Mycobacterium smegmatis: cytochemistry and architectural implications. FEMS Microbiol. Lett. 5289-93. [DOI] [PubMed] [Google Scholar]
- 11.Draper, P. 1998. The outer parts of the mycobacterial envelope as permeability barriers. Front. Biosci. 3D1253-D1261. [DOI] [PubMed] [Google Scholar]
- 12.Draper, P. 1971. The walls of Mycobacterium lepraemurium: chemistry and ultrastructure. J. Gen. Microbiol. 69313-324. [DOI] [PubMed] [Google Scholar]
- 13.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21129-228. [DOI] [PubMed] [Google Scholar]
- 14.Dubochet, J., and N. Sartori Blanc. 2001. The cell in absence of aggregation artifacts. Micron 3291-99. [DOI] [PubMed] [Google Scholar]
- 15.Etienne, G., F. Laval, C. Villeneuve, P. Dinadayala, A. Abouwarda, D. Zerbib, A. Galamba, and M. Daffé. 2005. The cell envelope structure and properties of Mycobacterium smegmatis mc2155: is there a clue for the unique transformability of the strain? Microbiology 1512075-2086. [DOI] [PubMed] [Google Scholar]
- 16.Etienne, G., C. Villeneuve, H. Billman-Jacobe, C. Astarie-Dequeker, M. A. Dupont, and M. Daffé. 2002. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 1483089-3100. [DOI] [PubMed] [Google Scholar]
- 17.Frehel, C., N. Rastogi, J.-C. Benichou, and A. Ryter. 1988. Do test tube-grown pathogenic mycobacteria possess a protective capsule? FEMS Microbiol. Lett. 56225-229. [Google Scholar]
- 18.Gande, R., K. J. Gibson, A. K. Brown, K. Krumbach, L. G. Dover, H. Sahm, S. Shioyama, T. Oikawa, G. S. Besra, and L. Eggeling. 2004. Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in Corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 27944847-44857. [DOI] [PubMed] [Google Scholar]
- 19.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 1007877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, G. P., and G. J. Jensen. 2006. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 60376-385. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann, C., A. Leis, M. Niederweis, J. M. Plitzko, and H. Engelhardt. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA 1053963-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karakousis, P. C., W. R. Bishai, and S. E. Dorman. 2004. Mycobacterium tuberculosis cell envelope lipids and the host immune response. Cell. Microbiol. 6105-116. [DOI] [PubMed] [Google Scholar]
- 23.Lemassu, A., and M. Daffé. 1994. Structural features of the exocellular polysaccharides of Mycobacterium tuberculosis. Biochem. J. 297351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemassu, A., A. Ortalo-Magne, F. Bardou, G. Silve, M. A. Laneelle, and M. Daffé. 1996. Extracellular and surface-exposed polysaccharides of non-tuberculous mycobacteria. Microbiology 1421513-1520. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, B. A., and D. M. Engelman. 1983. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J. Mol. Biol. 166211-217. [DOI] [PubMed] [Google Scholar]
- 26.Liu, J., E. Y. Rosenberg, and H. Nikaido. 1995. Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc. Natl. Acad. Sci. USA 9211254-11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahfoud, M., S. Sukumaran, P. Hulsmann, K. Grieger, and M. Niederweis. 2006. Topology of the porin MspA in the outer membrane of Mycobacterium smegmatis. J. Biol. Chem. 2815908-5915. [DOI] [PubMed] [Google Scholar]
- 28.Marienfeld, S., E. M. Uhlemann, R. Schmid, R. Kramer, and A. Burkovski. 1997. Ultrastructure of the Corynebacterium glutamicum cell wall. Antonie van Leeuwenhoek 72291-297. [DOI] [PubMed] [Google Scholar]
- 29.Matias, V. R., A. Al-Amoudi, J. Dubochet, and T. J. Beveridge. 2003. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 1856112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matias, V. R., and T. J. Beveridge. 2005. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol. 56240-251. [DOI] [PubMed] [Google Scholar]
- 31.Matias, V. R., and T. J. Beveridge. 2006. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 1881011-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNeil, M. R., and P. J. Brennan. 1991. Structure, function and biogenesis of the cell envelope of mycobacteria in relation to bacterial physiology, pathogenesis and drug resistance; some thoughts and possibilities arising from recent structural information. Res. Microbiol. 142451-463. [DOI] [PubMed] [Google Scholar]
- 33.Minnikin, D. E. 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles, p. 95-184. In C. Ratledge and J. Stanford (ed.), The biology of the mycobacteria, vol. 1. Physiology, identification and classification. Academic Press, Inc., New York, NY. [Google Scholar]
- 34.Minnikin, D. E., L. Kremer, L. G. Dover, and G. S. Besra. 2002. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 9545-553. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, H. T., D. D. Trach, N. V. Man, T. H. Ngoan, I. Dunia, M. A. Ludosky-Diawara, and E. L. Benedetti. 1979. Comparative ultrastructure of Mycobacterium leprae and Mycobacterium lepraemurium cell envelopes. J. Bacteriol. 138552-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niederweis, M. 2003. Mycobacterial porins—new channel proteins in unique outer membranes. Mol. Microbiol. 491167-1177. [DOI] [PubMed] [Google Scholar]
- 37.Ortalo-Magné, A., A. B. Andersen, and M. Daffé. 1996. The outermost capsular arabinomannans and other mannoconjugates of virulent and avirulent tubercle bacilli. Microbiology 142927-935. [DOI] [PubMed] [Google Scholar]
- 38.Ortalo-Magné, A., A. Lemassu, M. A. Laneelle, F. Bardou, G. Silve, P. Gounon, G. Marchal, and M. Daffé. 1996. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J. Bacteriol. 178456-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul, T. R., and T. J. Beveridge. 1994. Preservation of surface lipids and determination of ultrastructure of Mycobacterium kansasii by freeze-substitution. Infect. Immun. 621542-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul, T. R., and T. J. Beveridge. 1992. Reevaluation of envelope profiles and cytoplasmic ultrastructure of mycobacteria processed by conventional embedding and freeze-substitution protocols. J. Bacteriol. 1746508-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Portevin, D., C. De Sousa-D'Auria, C. Houssin, C. Grimaldi, M. Chami, M. Daffé, and C. Guilhot. 2004. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. USA 101314-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Portevin, D., C. de Sousa-D'Auria, H. Montrozier, C. Houssin, A. Stella, M. A. Laneelle, F. Bardou, C. Guilhot, and M. Daffé. 2005. The acyl-AMP ligase FadD32 and AccD4-containing acyl-CoA carboxylase are required for the synthesis of mycolic acids and essential for mycobacterial growth: identification of the carboxylation product and determination of the acyl-CoA carboxylase components. J. Biol. Chem. 2808862-8874. [DOI] [PubMed] [Google Scholar]
- 43.Puech, V., M. Chami, A. Lemassu, M. A. Laneelle, B. Schiffler, P. Gounon, N. Bayan, R. Benz, and M. Daffé. 2001. Structure of the cell envelope of corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology 1471365-1382. [DOI] [PubMed] [Google Scholar]
- 44.Rastogi, N. 1991. Recent observations concerning structure and function relationships in the mycobacterial cell envelope: elaboration of a model in terms of mycobacterial pathogenicity, virulence and drug-resistance. Res. Microbiol. 142464-476. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Seybert, A., R. Herrmann, and A. S. Frangakis. 2006. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J. Struct. Biol. 156342-354. [DOI] [PubMed] [Google Scholar]
- 47.Silva, M. T., and P. M. Macedo. 1983. A comparative ultrastructural-study of the membranes of Mycobacterium-Leprae and of cultivable Mycobacteria. Biol. Cell 47383-386. [Google Scholar]
- 48.Silva, M. T., and P. M. Macedo. 1984. Ultrastructural characterization of normal and damaged membranes of Mycobacterium leprae and of cultivable mycobacteria. J. Gen. Microbiol. 130369-380. [DOI] [PubMed] [Google Scholar]
- 49.Sondén, B., D. Kocincova, C. Deshayes, D. Euphrasie, L. Rhayat, F. Laval, C. Frehel, M. Daffé, G. Etienne, and J. M. Reyrat. 2005. Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol. Microbiol. 58426-440. [DOI] [PubMed] [Google Scholar]
- 50.Steck, P. A., B. A. Schwartz, M. S. Rosendahl, and G. R. Gray. 1978. Mycolic acids. A reinvestigation. J. Biol. Chem. 2535625-5629. [PubMed] [Google Scholar]
- 51.Studer, D., W. Graber, A. Al-Amoudi, and P. Eggli. 2001. A new approach for cryofixation by high-pressure freezing. J. Microsc. 203285-294. [DOI] [PubMed] [Google Scholar]
- 52.Tahara, Y., and Y. Fujiyoshi. 1994. A new method to measure bilayer thickness: cryo-electron microscopy of frozen hydrated liposomes and image simulation. Micron 25141-149. [DOI] [PubMed] [Google Scholar]
- 53.Tropis, M., X. Meniche, A. Wolf, H. Gebhardt, S. Strelkov, M. Chami, D. Schomburg, R. Kramer, S. Morbach, and M. Daffé. 2005. The crucial role of trehalose and structurally related oligosaccharides in the biosynthesis and transfer of mycolic acids in Corynebacterineae. J. Biol. Chem. 28026573-26585. [DOI] [PubMed] [Google Scholar]
- 54.Villeneuve, M., M. Kawai, H. Kanashima, M. Watanabe, D. E. Minnikin, and H. Nakahara. 2005. Temperature dependence of the Langmuir monolayer packing of mycolic acids from Mycobacterium tuberculosis. Biochim. Biophys. Acta 171571-80. [DOI] [PubMed] [Google Scholar]
- 55.Villeneuve, M., M. Kawai, M. Watanabe, Y. Aoyagi, Y. Hitotsuyanagi, K. Takeya, H. Gouda, S. Hirono, D. E. Minnikin, and H. Nakahara. 2007. Conformational behavior of oxygenated mycobacterial mycolic acids from Mycobacterium bovis BCG. Biochim. Biophys. Acta 17681717-1726. [DOI] [PubMed] [Google Scholar]
- 56.White, D. L., S. B. Andrews, J. W. Faller, and R. J. Barrnett. 1976. The chemical nature of osmium tetroxide fixation and staining of membranes by X-ray photoelectron spectroscopy. Biochim. Biophys. Acta 436577-592. [DOI] [PubMed] [Google Scholar]
- 57.Wong, M. Y. H., P. A. Steck, and G. R. Gray. 1979. The major mycolic acids of Mycobacterium smegmatis: characterization of their homologous series. J. Biol. Chem. 2545734-5740. [PubMed] [Google Scholar]
- 58.Zuber, B., M. Haenni, T. Ribeiro, K. Minnig, F. Lopes, P. Moreillon, and J. Dubochet. 2006. Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J. Bacteriol. 1886652-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuber, B., I. Nikonenko, P. Klauser, D. Muller, and J. Dubochet. 2005. The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes. Proc. Natl. Acad. Sci. USA 10219192-19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.