Abstract
Comamonas testosteroni TA441 degrades steroids such as testosterone via aromatization of the A ring, followed by meta-cleavage of the ring. In the DNA region upstream of the meta-cleavage enzyme gene tesB, two genes required during cholic acid degradation for the inversion of an α-oriented hydroxyl group on C-12 were identified. A dehydrogenase, SteA, converts 7α,12α-dihydroxyandrosta-1,4-diene-3,17-dione to 7α-hydroxyandrosta-1,4-diene-3,12,17-trione, and a hydrogenase, SteB, converts the latter to 7α,12β-dihydroxyandrosta-1,4-diene-3,17-dione. Both enzymes are members of the short-chain dehydrogenase/reductase superfamily. The transformation of 7α,12α-dihydroxyandrosta-1,4-diene-3,17-dione to 7α,12β-dihydroxyandrosta-1,4-diene-3,17-dione is carried out far more effectively when both SteA and SteB are involved together. These two enzymes are encoded by two adjacent genes and are presumed to be expressed together. Inversion of the hydroxyl group at C-12 is indispensable for the subsequent effective B-ring cleavage of the androstane compound. In addition to the compounds already mentioned, 12α-hydroxyandrosta-1,4,6-triene-3,17-dione and 12β-hydroxyandrosta-1,4,6-triene-3,17-dione were identified as minor intermediate compounds in cholic acid degradation by C. testosteroni TA441.
Rhodococcus equi and Comamonas testosteroni (formerly Nocardia restrictus and Pseudomonas testosteroni) are known for the ability to utilize testosterone and various other steroids, such as the cholic acid analogs. In the 1950s and 1960s, the mechanism by which testosterone is degraded in these bacteria was extensively studied, and the main intermediate compounds in the degradation pathway were determined (2-6, 16-19). In our previous work, we simultaneously identified the genes and the intermediate compounds that are accumulated by gene disruption mutants and revealed the testosterone degradation pathway and degradation genes of C. testosteroni TA441 (7-13). Two steroid degradation gene clusters were identified in TA441; one contains the meta-cleavage enzyme gene tesB, 16 ORFs, and the positive regulator of the steroid degradation gene tesR, and the other consists of ORF18, ORF17, and tesIHA2A1DEFG. TA441 degrades testosterone via aromatization of the A ring, followed by cleavage of the ring by enzymes encoded by these genes. Most of the enzymes involved in the transformation of testosterone to 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid and (2Z,4Z)-2-hydroxyhexa-2,4-dienoic acid are encoded by the ORF18-tesG gene cluster. ORF18 has been reported to encode a CoA transferase which adds CoA to 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid, and most of the putative ORFs in the region downstream of tesB have been suggested to be involved in 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid degradation, which is thought to be similar to the β-oxidation pathway of fatty acids. However, the region upstream of tesB was not clear. In this report, we describe newly isolated genes essential for the degradation of 12α-hydroxylated steroids in the region upstream of tesB.
MATERIALS AND METHODS
Abbreviations.
The following abbreviations are used in this report: ORF, open reading frame; CoA, coenzyme A; HPLC, high-performance liquid chromatography; TFA, trifluoroacetic acid; RT, retention time; FAB-MS, fast atomic bombardment-mass spectrometry; PFG, pulsed-field gradient; DQF-COSY, double quantum filtered chemical shift correlation spectroscopy; HMBC, heteronuclear multiple bond correlation; NOE, nuclear Overhauser effect; DMSO, dimethyl sulfoxide; 2D, two-dimensional; NMR, nuclear magnetic resonance.
Culture conditions.
C. testosteroni TA441 and mutant strains were grown at 30°C in LB medium, C medium (13), or 1/2 LB + 1/2 C medium (a mixture of equal volumes of LB and C media) with suitable carbon sources when necessary. Testosterone and cholic acid analogs were added as filter-sterilized DMSO solutions with a final concentration of 0.1% (wt/vol).
Cloning of genes in the upstream region of tesB and nucleotide sequence determination.
Total DNA of strain TA441 was digested with EcoT22I and ligated to the pUC19 vector, which had been digested with PstI and treated with alkaline phosphatase (shrimp alkaline phosphatase; Roche Molecular Biochemicals, Mannheim, Germany), and the objective transformant was selected on the basis of colony hybridization with tesB as a probe. Plasmids for the DNA sequence were constructed by using the Genome Priming System (New England BioLabs, Ipswich, MA), and the plasmid solution for PCR was prepared with the Wizard Plus Minipreps DNA Purification System (Promega, Madison, WI). DNA sequence determination and analysis were performed with an ABI model 373A automated DNA sequencer and dye terminator sequencing protocols (Perkin-Elmer Japan, Chiba).
Construction of gene disruption mutants, plasmids, and complement mutants.
ORF9 was disrupted by the insertion of a kanamycin resistance gene into the SalI site. The resultant plasmid, pORF9-Kmr, was used for inactivation of ORF9 in TA441 by homologous recombination. Insertion of the kanamycin resistance gene into ORF9 was confirmed by Southern hybridization. Gene disruption of ORF8, -7, and -6 and both ORF9 and ORF8 (ORF9,8) was performed in the same manner, by the insertion of kanamycin resistance genes into ClaI, ApaI, BglII, and HincII sites, respectively. As a result of HincII treatment, a DNA fragment of about 750 bp, containing parts of ORF9 and ORF8, was dropped, which resulted in the disruption of both ORFs. PCR-amplified ORF9, ORF8, and ORF9,8 were transferred into pUC19 to construct pUCORF9, pUCORF8, and pUCORF9,8 and into broad-host-range plasmid pMFY42 (14), which can be maintained in Pseudomonas and its relatives and gives them tetracycline resistance, to construct pMFYORF9, pMFYORF8, and pMFYORF9,8, respectively. The broad-host-range plasmid carrying each gene was introduced into the gene disruption mutant of TA441 by electroporation; a kanamycin- and tetracycline-resistant TA441 mutant was selected. Retention of the plasmids by the gene disruption mutants and transformants was confirmed by Southern hybridization with suitable probes.
Growth of TA441 and mutant strains on testosterone and cholic acid.
Growth was monitored in terms of CFU counts. The mutants were grown in C medium containing each steroid at 0.1% (wt/vol), and growth was monitored by counting colonies that appeared on LB plates on which appropriate dilutions of the culture had been spread, with incubation at 30°C, as described previously (13).
Northern analysis.
The total RNA of TA441 incubated in 1/2 LB + 1/2 C medium with testosterone, cholic acid, or succinate (negative control) was purified 6 h after the start of incubation. Northern analysis was carried out with the purified RNA with ORF6 as the probe.
HPLC analysis.
After the addition of a double volume of methanol to the culture, the mixture was centrifuged and the supernatant was directly injected into an HPLC apparatus. An HPLC apparatus (Alliance 2695 with a UV detector and a 996 photodiode array detector; Nihon Waters, Tokyo, Japan) equipped with an Inertsil ODS-3 column (4.6 by 250 mm; GL Sciences Inc., Tokyo, Japan) was used, and elution was carried out with a linear gradient of 20% solution A (CH3CN-CH3OH-TFA ratio = 95:5:0.05) and 80% solution B (H2O-CH3OH-TFA ratio = 95:5:0.05) to 65% solution A and 35% solution B over 10 min; this was maintained for 3 min and then changed to 20% solution A. The flow rate was 1.0 ml/min.
General experimental procedures.
FAB-MS (positive-ion mode) data were recorded on a JEOL JMS-700 mass spectrometer (JEOL Ltd., Tokyo, Japan) with a glycerin matrix. One- and 2D NMR spectra were recorded on a JNM-ECP500 or a JNM-ECA600 spectrometer (JEOL Ltd., Tokyo, Japan). Tetramethylsilane at 0 ppm in CDCl3 solution and residual proton signal at 2.49 ppm in a DMSO-d6 solution were used as internal references for 1H chemical shifts. 13C chemical shifts were obtained with reference to DMSO-d6 (39.5 ppm) or CDCl3 (77.0 ppm) at 25°C.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession number AB063577.
RESULTS
Isolation and DNA sequence of the gene region upstream of the meta-cleavage enzyme gene tesB.
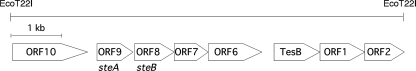
C. testosteroni TA441 degrades certain steroids, such as testosterone, via aromatization of the A ring, followed by meta-cleavage. The main steroid degradation genes are in two clusters, one containing the meta-cleavage enzyme gene tesB, 16 ORFs, and the positive regulator of steroid degradation genes tesR and the other containing ORF18, ORF17, and tesIHA2A1DEFG (7-13). Since the DNA sequence in the region upstream of tesB has not been identified, an Escherichia coli library of the total DNA of TA441 digested with EcoT22I was hybridized with a tesB probe and the DNA sequence of the 8-kb fragment cloned in the positive colony was determined. The fragment contained five putative ORFs (ORF10 to ORF6), tesB, ORF1, and ORF2 (Fig. 1). The results of a database search are shown in Table 1. The deduced amino acid sequences of ORF9 to -6 showed the highest similarity to corresponding proteins in Pseudoalteromonas haloplanktis TAC125. The deduced amino acid sequences of ORF9 and ORF8 showed significant similarity to short-chain dehydrogenases, and ORF6 was suggested to be an oxidoreductase. The putative amino acid sequences of ORF7 and ORF6 showed similarity to the corresponding putative ORFs in uncultured bacterium 442. Many ORFs in the region downstream of tesB also showed similarity to the corresponding putative ORFs in uncultured bacterium 442 (12).
FIG. 1.
Isolated 8.0-kb EcoT22I fragment of C. testosteroni TA441 containing tesB. tesB, ORF1, and ORF2 were identified in our previous study (13).
TABLE 1.
Comparison of the putative amino acid sequences of the isolated genes to the most similar proteins found in databases
| ORF (bp) | Intergenic space (bp) | Putative function or family | Similar protein or putative protein | % Amino acid identity | Organism | Accession no. |
|---|---|---|---|---|---|---|
| ORF10 (1,473) | Transcriptional regulator, XRE family protein | SKA58_11855 | 55 | Sphingomonas sp. strain SKA58 | AAQG01000024 | |
| ORF9 (732) | ORF10-9 (235) | 3-Oxoacyl-(acyl carrier protein) reductase, short-chain dehydrogenase/reductase family | PSHAa0894 | 51 | Pseudoalteromonas haloplanktis TAC125 | YP_339418 |
| ORF8 | 39 | C. testosteroni TA441 | AB063577 | |||
| ORF8 (768) | ORF9-8 (13) | 3-Oxoacyl-(acyl carrier protein) reductase, short-chain dehydrogenase/reductase family | PSHAa0879 | 66 | P. haloplanktis TAC125 | YP_339403 |
| ORF7 (648) | ORF8-7 (29) | ? | PSHAa0895 | 78 | P. haloplanktis TAC125 | YP_339419 |
| Hypothetical protein | 71 | Uncultured bacterium 442 | AY458639 | |||
| BaiE: bile acid 7-α-dehydratase | 27 | Lactobacillus plantarum WCFS1 | AL935261 | |||
| ORF6 (1,083) | ORF7-6 (10) | NADH-dependent flavin oxidoreductase, Oye family | PSHAa0880 | 59 | P. haloplanktis TAC125 | YP_339404 |
| Oxidoreductase, FAD/FMNa binding | 42 | Uncultured bacterium 442 | AY458639 |
FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide.
Induction of ORF6 during growth on steroids.
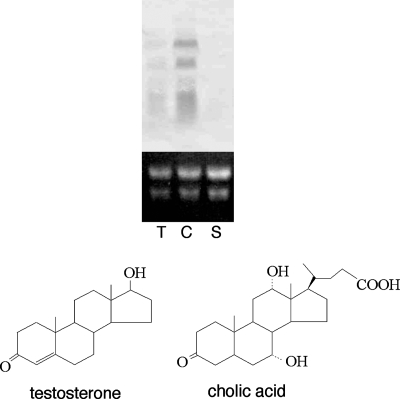
To examine whether ORF9 to ORF6 are involved in steroid degradation, total RNA of TA441 incubated in 1/2 LB + 1/2 C medium with testosterone, cholic acid, or succinate (negative control) was purified 6 h after the start of incubation and subjected to Northern analysis with an ORF6 probe. ORF9 to ORF6 are thought to form an operon because a possible terminator was not found in ORF9 to ORF6 and the genes were organized tightly with little intervals (Table 1). ORF9 to ORF6 were induced in TA441 during growth on testosterone and cholic acid but not induced during growth on succinate, indicating that these ORFs are involved in steroid degradation (Fig. 2).
FIG. 2.
Induction of ORF6 in C. testosteroni TA441 incubated with testosterone (T) and cholic acid (C). Total RNA was purified 6 h after the start of incubation of TA441 in 1/2 LB + 1/2 C medium with each compound at 0.1% (wt/vol) and analyzed by Northern hybridization. Succinate (S) was used as a negative control.
Growth of mutants with ORF9 to -6 disrupted on steroids.
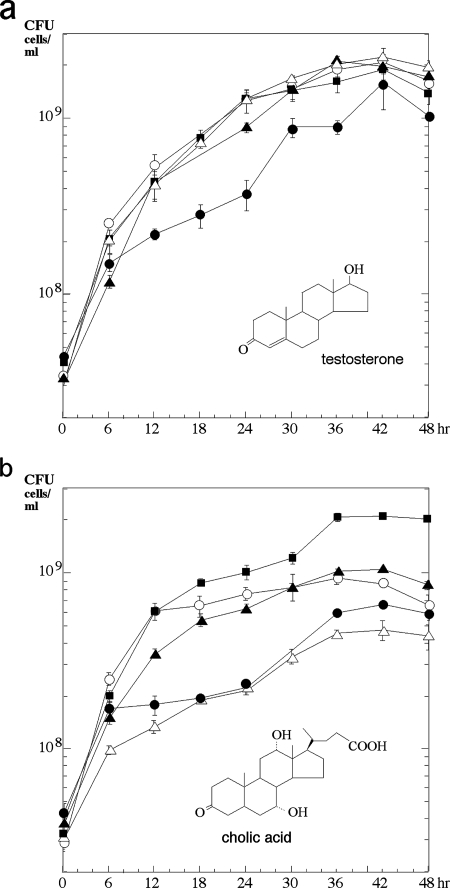
Gene mutants carrying disruptions of ORF9, ORF8, ORF7, and ORF6—in which the target ORF is disrupted by a kanamycin resistance gene without a terminator—were constructed, and their growth on testosterone as a sole carbon source was monitored; it was found that these mutants were able to grow on testosterone (Fig. 3a). The growth of each strain until around 6 h after the start of the incubation was probably caused by the nutrients carried by the cells from preculture on LB medium. The reason for the delay in the growth of the ORF8 disruption mutant is not clear. The same experiment was then performed with cholic acid, which contains a different A ring from testosterone and the side chain portion at C-17, together with hydroxyl groups at C-7 and C-12, as the sole carbon source (Fig. 3b). As the gene disruption mutants apparently showed less growth on cholic acid than did TA441, these ORFs are thought to be involved in cholic acid degradation.
FIG. 3.
Growth of TA441 and gene disruption mutants with testosterone (a) or cholic acid (b) as the sole carbon source. The genes were disrupted by the insertion of a kanamycin resistance-encoding gene. Each strain was grown in 10 ml of C medium supplemented with 0.1% (wt/vol) testosterone (TA441, ▪; ORF6 disruption mutant, ▵; ORF7 disruption mutant, ▴; ORF8 disruption mutant, •; ORF9 disruption mutant, ○). Growth is represented by CFU counts. The data shown are averages of more than three experiments.
HPLC analysis of a culture of mutants with ORF9 to -6 disrupted that were incubated with cholic acid.
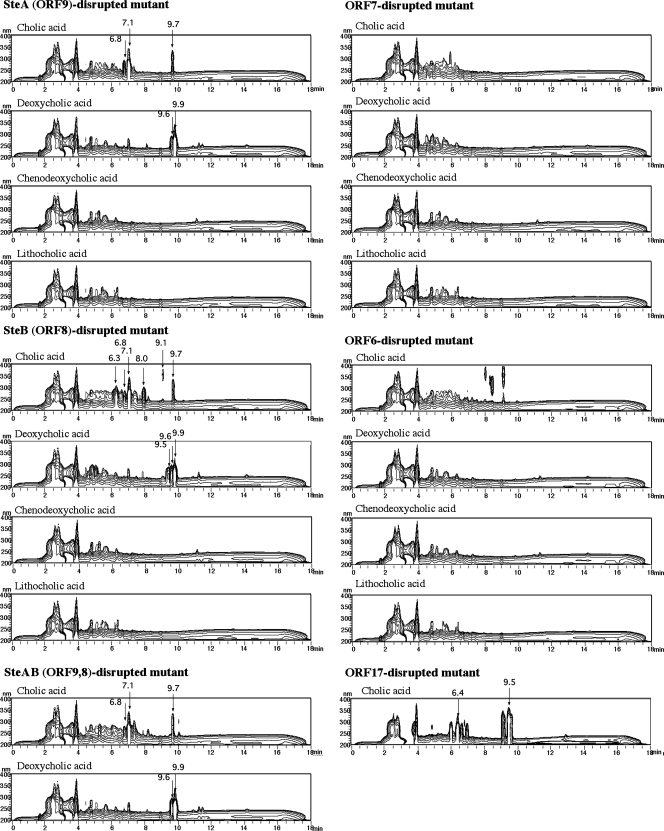
Cholic acid has a C24 carbon skeleton with a side chain at C-17 and three hydroxyl groups at C-3, C-7, and C-12. In a comparison between testosterone and cholic acid, the presence of the side chain on C-17 and two hydroxyl groups at C-7 and C-12 might be major structural differences, so it was thought that ORF9 to ORF6 might be involved in the conversion of one of these functional groups. ORF9, ORF8, ORF7, and ORF6 gene disruption mutants were incubated in 1/2 LB + 1/2 C medium with a cholic acid analog (cholic acid, chenodeoxycholic acid, deoxycholic acid, or lithocholic acid), and the cultures were analyzed by HPLC at suitable intervals. Chenodeoxycholic acid and deoxycholic acid contain a hydroxyl group at C-7 and C-12, respectively, while lithocholic acid does not contain any hydroxyl groups at either of these sites. Charts of the HPLC analysis conducted 3 days after the start of incubation are shown in Fig. 4. Any added cholic acid analogs were not detected under the conditions used. Peaks eluted before an RT of 4 min are due to the contents of the medium. Characteristic peaks considered to represent intermediate compounds were detected in the cultures of mutants with ORF9 or ORF8 disrupted that were incubated with cholic acid or deoxycholic acid, indicating that ORF9 and ORF8 are involved in the conversion of a hydroxyl group at C-12. ORF9 and -8 were named steA and -B, respectively. Compound I (RT = 7.1 min) is the major intermediate compound in the culture of an SteA disruption mutant incubated with cholic acid, and compound II (RT = 9.7 min) was the most conspicuous possible intermediate compound among the rest of the compounds detected. In the culture of the SteB disruption mutant incubated with cholic acid, compound III (RT = 6.3 min) was detected as a major compound in addition to these. The mutant with both the steA and steB genes disrupted that was incubated with cholic acid showed the same HPLC chromatogram as the SteA disruption mutant (Fig. 4). These results indicate that SteA acts in advance of SteB and that compounds I and II are intermediates upstream of the steroid degradation pathway compared to compound III.
FIG. 4.
Three-dimensional HPLC analysis of cultures of the ORF9 disruption mutant (ORF9−), the ORF8 disruption mutant (ORF8−), the ORF7 disruption mutant (ORF7−), the ORF6 disruption mutant (ORF6−), the ORF9 and ORF8 disruption mutant (ORF9,8−), and the ORF17 disruption mutant (ORF17−) incubated in 1/2 LB + 1/2 C medium with 0.1% (wt/vol) cholic acid, deoxycholic acid, chenodeoxycholic acid, and lithocholic acid for 3 days. The vertical axis indicates wavelength (nanometers), and the horizontal axis indicates RT; the UV absorbance of each compound is represented in contours. Possible intermediate compounds are indicated by arrows. The values above the arrows are RTs.
Isolation and identification of compounds accumulated by an SteB disruption mutant incubated with cholic acid.
The culture of an SteB disruption mutant incubated with cholic acid (500 mg/500 ml) was extracted three times with the same volume of ethyl acetate under neutral conditions. The ethyl acetate layer was treated with Na2SO4 and concentrated in vacuo; a crystal substance was precipitated during concentration. This crystal substance was collected by filtration, washed, and dissolved in a small amount of methanol for further study. This compound was detected at an RT of 7.1 min, as shown in Fig. 4. The filtrate of the concentrated acetate layer was dissolved in a small amount of methanol and subjected to HPLC analysis, which revealed the presence of compounds I, II, and III and a compound detected at an RT of 6.8 min (Fig. 4). Fractions containing each of these were collected from the eluent. The water layer was extracted twice with the same volume of ethyl acetate under acidic conditions, and compounds contained in this layer were isolated in the same manner. A compound detected at an RT of 8.0 min in Fig. 4 was contained in this layer. Compounds were isolated from each fraction of the HPLC eluent, resolved in CD3OD immediately, and analyzed by NMR and MS.
The molecular formula of compound I was determined to be C19H24O4 by high-resolution FAB-MS data [m/z 317.1746 (M+H)+]. The 13C NMR spectrum confirmed the presence of 19 carbon signals, including two ketone carbonyl carbons, four sp2 carbons, and two oxygenated methine carbons. Analysis of PFG-DQF-COSY, PFG-heteronuclear single quantum coherence, and PFG-HMBC spectra established complete assignments of all 1H and 13C signals (Table 2) and suggested the 7,12-dihydroxyandrosta-1,4-diene-3,17-dione structure. The stereochemistry of C-7 and C-12 was determined to be an α configuration for both hydroxyl groups because methine signals of H-7 and H-12 in the 1H NMR spectrum showed a broad, singlet-like pattern having no large vicinal coupling constant values, respectively (1). NOE differential spectral data with irradiation of H-7, H-12, and also two singlet methyl H-18 and H-19 signals supported the stereochemistry. Based on these data, compound I was identified as 7α,12α-dihydroxyandrosta-1,4-diene-3,17-dione (20).
TABLE 2.
NMR data for compounds accumulated by ORF8 disruption mutant (compounds I to III) and ORF17 disruption mutant (compounds IV and V) incubated with cholic acida
| No. | Compound Ib
|
Compound IIc
|
Compound IIId
|
Compound IVe
|
Compound Vf
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 13C NMR δ in ppm | 1H NMR δ in ppm (J in Hz) | 13C NMR δ in ppm | 1H NMR δ in ppm (J in Hz) | 13C NMR δ in ppm | 1H NMR δ in ppm (J in Hz) | 13C NMR δ in ppm | 1H NMR δ in ppm (J in Hz) | 13C NMR δ in ppm | 1H NMR δ in ppm (J in Hz) | |
| 1 | 156.20 | 7.11 d (9.6) | 152.85 | 7.05 d (10.3) | 154.86 | 7.05 d (10.1) | 155.85 | 7.14 d (9.6) | 152.07 | 7.04 d (10.3) |
| 2 | 126.29 | 6.09 dd (9.6, 1.4) | 128.20 | 6.30 dd (10.3, 1.4) | 127.03 | 6.11 dd (10.1, 2.3) | 126.85 | 6.10 dd (9.6, 1.4) | 128.47 | 6.28 dd (10.3, 1.4) |
| 3 | 184.63 | 186.65 | 184.41 | 184.52 | 186.08 | |||||
| 4 | 125.75 | 5.92 br.s | 124.11 | 6.08 br.s | 126.35 | 5.97 br.s | 125.96 | 5.92 br.s | 124.54 | 6.05 br.s |
| 5 | 166.73 | 162.45 | 165.05 | 166.13 | 161.21 | |||||
| 6 | 40.56 | 2.69 br.d (13.8) | 128.65 | 6.34 dd (9.6, 2.8) | 40.16 | 2.80 ddd (13.8, 2.8, 1.8) | 40.43 | 2.69 br.d (13.1) | 128.88 | 6.33 dd (9.6, 2.8) |
| 2.37 dd (13.8, 3.4) | 2.43 dd (13.8, 2.8) | 2.38 m | ||||||||
| 7 | 67.09 | 3.99 m | 135.78 | 6.14 dd (9.6, 1.4) | 66.09 | 4.07 m | 66.43 | 3.96 m | 134.76 | 6.10 dd (9.6, 2.1) |
| (7-OH) | 4.56 d (3.4) | 4.76 d (4.1) | 4.57 br.s | |||||||
| 8 | 38.62 | 1.83 ddd (11.0, 11.0, 2.1) | 37.42 | 2.47 m | 37.53 | 2.25 m | 37.58 | 1.77 m | 36.58 | 2.45 m |
| 9 | 38.55 | 1.90 m | 42.18 | 1.98 m | 44.30 | 1.91 m | 41.99 | 1.53 m | 46.12 | 1.54 ddd (13.1, 9.6, 3.4) |
| 10 | 42.91 | 40.84 | 43.12 | 42.87 | 40.90 | |||||
| 11 | 29.48 | 1.74 m | 27.98 | 2.00 m | 38.65 | 2.88 dd (14.0, 14.0) | 30.82 | 1.77 m | 28.76 | 2.06 ddd (13.1, 4.8, 3.4) |
| 1.71 m | 1.84 m | 2.26 m | 1.47 m | 1.65 ddd (13.1, 13.1, 11.0) | ||||||
| 12 | 67.54 | 3.85 m | 68.92 | 4.21 dd (2.8, 2.8) | 204.95 | 69.74 | 3.48 dd (10.3, 4.8) | 72.10 | 3.81 dd (11.0, 4.8) | |
| (12-OH) | 4.56 d (3.4) | 4.35 br.s | ||||||||
| 13 | 51.85 | 53.37 | 56.88 | 50.84 | 51.98 | |||||
| 14 | 37.34 | 2.29 m | 41.28 | 2.21 m | 45.94 | 1.93 m | 43.18 | 1.48 m | 46.74 | 1.50 ddd (12.7, 12.4, 12.5) |
| 15 | 20.47 | 1.97 m | 20.58 | 2.19 m | 20.04 | 2.03 m | 20.34 | 1.95 m | 21.27 | 2.18 m |
| 1.47 m | 1.77 m | 1.65 m | 1.58 m | 1.86 m | ||||||
| 16 | 36.40 | 2.28 m | 35.81 | 2.53 m | 36.35 | 2.30 m | 35.12 | 2.38 m | 35.50 | 2.55 m |
| 1.92 m | 2.15 m | 2.11 ddd (18.8, 9.0, 9.0) | 1.95 m | 2.18 m | ||||||
| 17 | 217.80 | 218.66 | 211.33 | 218.90 | 220.83 | |||||
| 18 | 14.02 | 0.77 s | 13.59 | 0.99 s | 14.17 | 1.16 s | 7.76 | 0.87 s | 8.26 | 1.08 s |
| 19 | 18.14 | 1.16 s | 20.47 | 1.21 s | 17.79 | 1.27 s | 18.18 | 1.17 s | 20.75 | 1.23 s |
Abbreviations for NMR signals: s, singlet; d, doublet; m, multiplet; br, broad.
RT = 7.1 min in DMSO-d6.
RT = 9.7 min in CDCl3.
RT = 6.3 min in DMSO-d6.
RT = 6.4 min in DMSO-d6.
RT = 9.5 min in CDCl3.
The 19 resonances in the 13C NMR spectrum of compound II, together with the result obtained from high-resolution FAB-MS measurement of the [M+H]+ pseudomolecular ion m/z 299.1643 in the positive mode suggested that the molecular formula is C19H22O3. In the 1H NMR spectrum, five conjugated olefinic proton signals were observed. PFG-DQF-COSY spectral data suggested that compound II possessed the double bonds in the C-1 and C-4 positions of the A ring and the C-6 position of the B ring. H-6 at 6.34 ppm and H-7 at 6.14 ppm had vicinal coupling with J = 9.6 Hz typical for a Z configuration and showed a correlation to H-8 at 2.47 ppm. These assignments were supported by heteronuclear long-range correlations in the PFG-HMBC spectrum; i.e., H-6 showed correlations to C-8 and C-10 and H-7 showed correlations to C-5, C-8, and C-9. The PFG-HMBC data revealed the presence of ketone groups at C-3 and C-17 and also a hydroxyl group at C-12. Complete NMR assignments were established by detailed analysis of 2D spectral data and are summarized in Table 2. Stereochemistry of C-12 was determined from small vicinal coupling constant values of H-12 at 4.21 ppm (dd, J = 2.8, 2.8 Hz) and NOE between the H-12 and H-18 methyl signals at 0.99 ppm. Compound II was therefore elucidated as 12α-hydroxyandrosta-1,4,6-triene-3,17-dione, which is a new steroidal metabolite with an androstane skeleton.
As dehydration of the C-7 hydroxyl group is often observed among intermediate compounds in cholic acid degradation by TA441 (unpublished data) and because the amount of 12α-hydroxyandrosta-1,4,6-triene-3,17-dione was very small compared to those of compounds I and III, we concluded that this compound is not a major intermediate.
The molecular formula of compound III was determined to be C19H22O4 from high-resolution FAB-MS data [m/z 315.1596 (M+H)+]. In the 13C NMR spectrum, three ketone carbonyl carbon signals were observed at 184.41 (C-3), 204.95 (C-12), and 211.33 (C-17) ppm. The last two of these had long-range correlation from methyl protons H-18 in the PFG-HMBC spectrum, suggesting that C-12 and C-17 were oxidized to ketone groups. One hydroxyl group-attached carbon signal was observed at 66.09 ppm, and its proton signal H-7 was observed at 4.07 ppm as a singlet-like broad signal with small vicinal coupling constant values, which were quite similar to the signals of compound I. Detailed analyses of several 2D NMR spectra confirmed the complete 1H and 13C NMR assignments and confirmed the chemical structure to be 7α-hydroxyandrosta-1,4-diene-3,12,17-trione. Compound III is also a new steroidal metabolite.
ORF17, an ORF in another major steroid degradation gene cluster of TA441, is a putative ferredoxin reductase component of a hydroxylase at C-9. As the ORF17 disruption mutant was found to accumulate androsta-1,4-diene-3,17-dione when incubated with testosterone, 7α,12α-hydroxyandrosta-1,4-diene-3,17-dione and 12α-hydroxyandrosta-1,4,6-triene-3,17-dione might be accumulated by the ORF17 disruption mutant when it is incubated with cholic acid. To investigate this, the ORF17 disruption mutant was incubated with cholic acid and the resulting culture was analyzed by HPLC (Fig. 4). The RTs of the detected compounds were different from those detected in the culture of the SteB disruption mutant incubated with cholic acid. The compounds that accumulated in the largest amounts were detected at RTs of 6.4 min (compound IV) and 9.5 min (compound V). These compounds were purified and identified in the same manner as compounds accumulated by the SteB disruption mutant.
The molecular formula C19H24O4 of compound IV was identical to that of compound I. The 1H and 13C NMR spectral data were also similar to those of compound I, which suggested that compound IV should be a stereoisomer of compound I. In a comparison of the 13C NMR data of compounds I and IV, relatively large differences were observed for carbon signals of C-9, C-14, and C-18. C-9 and C-14 were shifted to low field and C-18 was shifted to high field in compound IV because of a stereochemical change of a hydroxyl group attached to C-12 from α of compound I to a β configuration. In the 1H NMR spectrum, H-12 at 3.48 (dd, J = 10.3, 4.8 Hz) ppm showed a large vicinal coupling constant value between H-11β and H-12α. The 1H and 13C NMR data (Table 2) indicated that the stereochemistry of C-7 should be the same as that of compound I. Therefore, compound IV was identified as 7α,12β-dihydroxyandrosta-1,4-diene-3,17-dione (20).
The molecular formula of compound V was the same as that of compound II, and its UV spectral data were quite similar to those of compound II, indicating that compound V was an isomer of compound II. 1H and 13C NMR data (Table 2) indicated that compound V had the same planar structure as compound II, but the stereochemistry of the C-12 hydroxyl group was a β configuration because chemical shifts of carbon signals for the ring C portion were quite similar to those of compound IV. Therefore, compound V was identified as 12β-hydroxyandrosta-1,4,6-triene-3,17-dione (15).
A number of further possible intermediate or shunt products were detected by HPLC in addition to the main intermediate compounds. However, characterization of these compounds was not successful as their quantities were insufficient for identification. These compounds may have been produced by side reactions of TA441 enzymes acting on the excess accumulation of main intermediate compounds.
Compounds I and IV in cholic acid degradation by TA441.
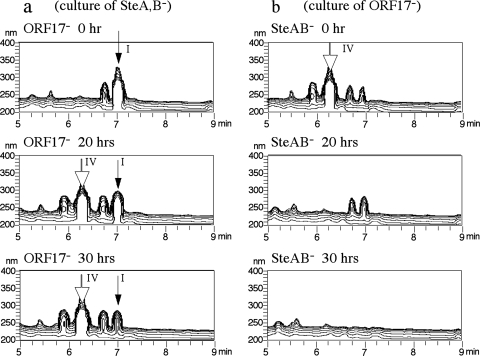
Compound I, accumulated by the SteA disruption mutant, and compound IV, accumulated by the ORF17 disruption mutant, are diastereoisomers; compound I has the same configuration of C-12 as that of cholic acid, which may suggest that compound I is produced prior to compound IV during cholic acid degradation. To confirm this speculation, an SteAB disruption mutant was incubated with cholic acid, the culture was sterilized by filtration, and the resultant culture was treated with the ORF17 disruption mutant while a culture of the ORF17 disruption mutant incubated with cholic acid was treated with the SteAB disruption mutant in the same manner. The cultures were analyzed by HPLC at suitable intervals. In the culture of the SteAB disruption mutant treated with the ORF17 disruption mutant, compound I was converted to compound IV, which accumulated because of the lack of ORF17 (Fig. 5a). In the culture of the ORF17 disruption mutant treated with the SteAB disruption mutant, compound IV disappeared quickly and no product was detected (Fig. 5b). These results indicated that compound I was produced early and then converted to compound IV during cholic acid degradation. The slow degradation of compound I in the culture of the ORF17 disruption mutant treated with the SteAB disruption mutant might be due to a side reaction of TA441 enzymes, as all of the genes except steA and steB are available in the SteAB disruption mutant.
FIG. 5.
(a) Conversion of compound I by the ORF17 disruption mutant (ORF17−). The ORF9,8 disruption mutant (ORF9,8−) was incubated with cholic acid for 3 days, centrifuged, and filter sterilized to remove ORF9,8− mutant cells. Three-dimensional HPLC analyses of the culture incubated with the ORF17− mutant after 20 and 30 h are shown. (b) Conversion of compound IV by the ORF9,8− mutant. The ORF17− mutant was incubated with cholic acid for 3 days, centrifuged, and filter sterilized to remove ORF17− mutant cells. Three-dimensional HPLC analyses of the culture incubated with the ORF9,8− mutant after 20 and 30 h are shown.
Complementation experiment with steA and steB.
For the complementation experiment, a range of mutants were constructed and incubated with cholic acid to examine the function of SteA and SteB. These mutants were an SteA disruption mutant with broad-host-range plasmid pMFY42 (SteA− mutant with pMFY42, negative control) and with pMFY42 carrying steA (SteA− mutant with pMFYSteA), an SteB disruption mutant with pMFY42 (SteB− mutant with pMFY42, negative control) and with pMFY42 carrying steB (SteB− mutant with pMFYSteB), and an SteA and -B disruption mutant with pMFY42 (SteAB− mutant with pMFY42, negative control), with pMFYSteA (SteAB− mutant with pMFYSteA), with pMFYSteB (SteAB− mutant with pMFYSteB), and with pMFY42 carrying steAB (SteAB− mutant with pMFYSteAB). Tetracycline was added to the culture to maintain the plasmid. The culture was analyzed by HPLC every 24 h after the start of incubation (data not shown). The period of cultivation was longer in this experiment than in the others because the growth of the mutants was slower than that of other TA441 mutants due to the presence of tetracycline. The broad-host-range plasmid gives TA441 tetracycline resistance, but growth was slower with tetracycline than without it. In the culture of the SteA− mutant with pMFYSteA, compound I, which was accumulated in the culture of the SteA− mutant with pMFY42, was not detected, indicating that SteA is necessary for the conversion of compound I. The SteAB− mutant with pMFY42 gave HPLC results that were similar to those of the SteA− mutant with pMFY42 and the SteAB− mutant with pMFYSteB, indicating that SteA catalyzes the reaction prior to the reaction catalyzed by SteB. The SteB− mutant with pMFY42 accumulated both compounds III and I. In the culture of the SteB− mutant with pMFYSteB, neither of these compounds was detected, indicating that SteB is necessary for the conversion of compound III. These results led to the presumption that SteA converts compound I to compound III, which is followed by the conversion of compound III to compound IV by SteB.
Conversion of compounds I and III by SteA and SteB, respectively.
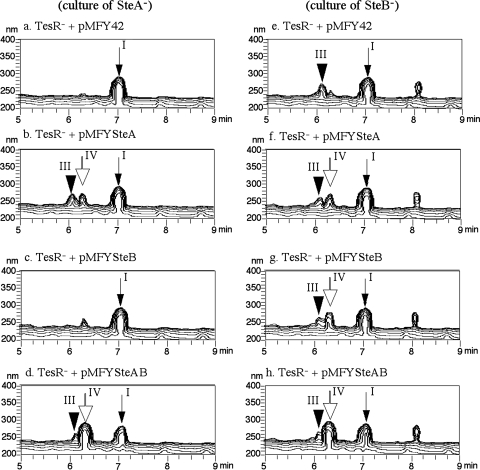
To confirm the functions of SteA and SteB, each of these enzymes was cloned into E. coli and the transformation of compounds I and III in the resultant E. coli cells was examined. However, we could not detect transformation in either the induced E. coli cells or the cell extracts. We then introduced the broad-host-range plasmids constructed as described in the previous section into the TesR disruption (TesR−) mutant (TesR− mutant with pMFY42, pMFYSteA, pMFYSteB, or pMFYSteAB). TesR is a positive regulator of steroid degradation genes in TA441; without TesR, steroid degradation genes are not induced (12). The SteA disruption mutant and the SteB disruption mutant were each incubated in 1/2 LB + 1/2 C medium with 0.1% cholic acid for 3 days. Because the culture, especially that of the SteB disruption mutant, seemed to cause cell lysis of the TesR− mutants, the culture was diluted 10 times with 1/2 LB + 1/2 C medium after centrifugation and filter sterilization to remove SteA disruption and SteB disruption mutant cells. The resultant solution was used for a transformation experiment. The TesR− mutants were incubated in LB medium with kanamycin and tetracycline until there was enough growth for the experiment (usually 30 to 40 h), and the cells were collected by centrifugation. Each mutant was inoculated into the prepared reaction solution and incubated at 30°C; the cultures were analyzed by HPLC every 24 h. Figure 6 shows HPLC charts of the cultures 2 days after the start of incubation. Compound I was converted to compound III by the TesR− mutant with pMFYSteA (Fig. 6b), and compound III was converted to compound IV by the TesR− mutant with pMFYSteB (Fig. 6g). In the reaction solution of the TesR− mutant with pMFYSteA, a small amount of compound IV was detected (Fig. 6b and f). Repetition of the experiments gave the same results, implying that the SteA enzyme might have a slight activity on compound III to produce compound IV. Both compounds I and III were converted very effectively by the TesR− mutant with pMFYSteAB (Fig. 6d and h). The amount of compound IV detected in the culture treated with this mutant was about 10 times greater than the amount of compound IV produced by the TesR− mutant with pMFYSteA. The data showed that SteB is most important for the conversion of compound III into compound IV and the conversion of compound I to compound IV proceeds effectively only when both SteA and SteB are involved.
FIG. 6.
Transformation of compounds I and III by the TesR disruption (TesR−) mutant expressing each ORF. Cultures of the ORF9 disruption mutant (a to d) and the ORF8 disruption mutant (e to h) were used as reaction solutions after filter sterilization and dilution 10 times with 1/2 LB + 1/2 C medium. Each reaction solution was treated with the TesR− mutant carrying pMFY42 (negative control) (a, e), pMFYORF9 (b, f), pMFYORF8 (c, g), or pMFYORF9,8 (d, h) for 2 days, and the reaction solution was analyzed by HPLC 2 days after the start of the incubation. The TesR− strain is a mutant of TA441 with a mutation in TesR, a positive regulator of steroid degradation genes.
DISCUSSION
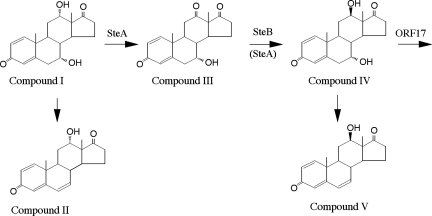
Five ORFs (ORF10 to ORF6) were identified in the upstream DNA region of a steroid degradation gene cluster consisting of tesB to tesR in C. testosteroni TA441. We showed that SteA is a dehydrogenase which converts 7α,12α-dihydroxyandrosta-1,4-diene-3,17-dione (compound I) to 7α-hydroxyandrosta-1,4-diene-3,12,17-trione (compound III), and SteB is a hydrogenase which converts compound III to 7α,12β-dihydroxyandrosta-1,4-diene-3,17-dione (compound IV) (Fig. 7). SteA converts only a small portion of compound I into compound III when only SteA is expressed, but the conversion of compound I to compound IV proceeds quite effectively when both SteA and SteB are involved. steA to ORF6 are thought to form an operon because a possible terminator was not found in tesA to ORF6 and the genes were organized tightly with little interval. As members of an operon are expressed together, TA441 is presumed to convert compound I to compound IV effectively and does not accumulate compound III without a gene disruption. This is supported by that the lack of previous reports for compound III, while compounds I and IV were reported from Pseudomonas sp. strain NCIB 10590 (15, 20). Bacterial degradation of cholic acid analogs was studied extensively in the 1970s and 1980s. From a culture of Pseudomonas sp. strain NCIB 10590 incubated with cholic acid, compounds I, IV, and V were identified as intermediate compounds in cholic acid degradation (19, 20). In those reports, transformation of a 12α-hydroxyl group of cholic acid to a 12β-hydroxyl group was predicted, but the detailed mechanism was not clear as an androstane with ketone moiety at C-12 was not successfully isolated.
FIG. 7.
Proposed mechanism of conversion of compound I to compound IV during cholic acid degradation by C. testosteroni TA441. Compounds: I, 7α,12α-dihydroxyandrosta-1,4-diene-3,17-dione; II, 12α-hydroxyandrosta-1,4,6-triene-3,17-dione; III, 7α-hydroxyandrosta-1,4-diene-3,12,17-trione; IV, 7α,12β-dihydroxyandrosta-1,4-diene-3,17-dione; V, 12β-hydroxyandrosta-1,4,6-triene-3,17-dione.
Since the ORF9 disruption mutant converted most of the added cholic acid into compounds I and II, it is suggested that a steroid compound with a 12α-hydroxyl group is hydroxylated at the 9 position far less effectively than one containing a 12β-hydroxyl group. The ORF18-encoded enzyme is thought to add CoA to 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid in steroid degradation by TA441, and a gene disruption-carrying ORF18 mutant accumulates 7α,12β-dihydroxy-9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid when it is incubated with cholic acid (7). This compound was the only intermediate compound of which a large amount was accumulated in the culture of an ORF18 disruption mutant incubated with cholic acid, indicating that a 12α-hydroxyl group is converted into a 12β-hydroxyl group before hydroxylation at the 9 position and the following degradation proceeds with intermediate compounds with a 12β-hydroxyl group in cholic acid degradation by TA441. The culture of the ORF17 disruption mutant containing compound IV was incubated with a TesR disruption mutant with pMFY42 (negative control), pMFYTesB, pMFYTesA, or pMFYTesAB in the same manner as the conversion experiment presented in Fig. 6, but production of compounds I and III was not observed (data not shown). This result indicates that reverse reactions from compound IV to compound III and then to compound I do not occur or that the efficiency is quite low compared to the reactions leading from compound I to compound IV.
To the best of our knowledge, compounds II and III identified in this study are new substances and have not previously been reported. A gene disruption mutant enables us to isolate compounds which rarely accumulate in the culture of an intact strain together with major intermediate compounds in the degradation pathway. It will be important to identify these minor and major intermediate compounds and clarify the whole mechanism of bacterial steroid degradation in detail.
Acknowledgments
This work was partly supported by a grant from the Eco Molecular Science Research Program from RIKEN. M.H. was supported by a grant from the Special Postdoctoral Researchers Program from RIKEN.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Barnes, P. J., J. D. Baty, R. F. Bilton, and A. N. Mason. 1976. Degradation of deoxycholic acid by Pseiidontionias species NCIB 10590. Tetrahedron 3289-93. [Google Scholar]
- 2.Coulter, A. W., and P. Talalay. 1968. Studies on the microbial degradation of steroid ring A. J. Biol. Chem. 2433238-3247. [PubMed] [Google Scholar]
- 3.Dodson, R. M., and R. D. Muir. 1958. Microbiological transformations. II. Microbiological aromatization of steroids. J. Am. Chem. Soc. 805004-5005. [Google Scholar]
- 4.Dodson, R. M., and R. D. Muir. 1958. Microbiological transformations. III. The hydroxylation of steroids at C-9. J. Am. Chem. Soc. 806148. [Google Scholar]
- 5.Dodson, R. M., and R. D. Muir. 1961. Microbiological transformations VI. The microbiological aromatization of steroids. J. Am. Chem. Soc. 834627-4631. [Google Scholar]
- 6.Gibson, D. T., K. C. Wang, C. J. Sih, and H. Whitlock, Jr. 1966. Mechanisms of steroid oxidation by microorganisms. IX. On the mechanism of ring A cleavage in the degradation of 9,10-seco steroids by microorganisms. J. Biol. Chem. 241551-559. [PubMed] [Google Scholar]
- 7.Horinouchi, M., T. Hayashi, H. Koshino, and T. Kudo. 2006. ORF18-disrupted mutant of Comamonas testosteroni TA441 accumulates significant amounts of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid and its derivatives after incubation with steroids. J. Steroid Biochem. Mol. Biol. 10178-84. [DOI] [PubMed] [Google Scholar]
- 8.Horinouchi, M., T. Hayashi, H. Koshino, T. Kurita, and T. Kudo. 2005. Identification of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid, 4-hydroxy-2-oxohexanoic acid, and 2-hydroxyhexa-2,4-dienoic acid and related enzymes involved in testosterone degradation in Comamonas testosteroni TA441. Appl. Environ. Microbiol. 715275-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horinouchi, M., T. Hayashi, H. Koshino, T. Yamamoto, and T. Kudo. 2003. Gene encoding the hydrolase for the product of the meta-cleavage reaction in testosterone degradation by Comamonas testosteroni Appl. Environ. Microbiol. 692139-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horinouchi, M., T. Hayashi, and T. Kudo. 2004. The genes encoding the hydroxylase of 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione in steroid degradation in Comamonas testosteroni TA441. J. Steroid Biochem. Mol. Biol. 92143-154. [DOI] [PubMed] [Google Scholar]
- 11.Horinouchi, M., T. Hayashi, T. Yamamoto, and T. Kudo. 2003. A new bacterial steroid degradation gene cluster which consists of aromatic compound degradation genes for seco-steroids and 3-ketosteroid dehydrogenase genes in Comamonas testosteroni TA441. Appl. Environ. Microbiol. 694421-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horinouchi, M., T. Kurita, T. Yamamoto, E. Hatori, T. Hayashi, and T. Kudo. 2004. Steroid degradation gene cluster of Comamonas testosteroni consisting of 18 putative genes from meta-cleavage enzyme gene tesB to regulator gene tesR. Biochem. Biophys. Res. Commun. 324597-604. [DOI] [PubMed] [Google Scholar]
- 13.Horinouchi, M., T. Yamamoto, K. Taguchi, H. Arai, and T. Kudo. 2001. meta-cleavage enzyme gene tesB is necessary for testosterone degradation in Comamonas testosteroni TA441. Microbiology 1473367-3375. [DOI] [PubMed] [Google Scholar]
- 14.Nagata, Y., T. Nariya, R. Ohtomo, M. Fukuda, K. Yano, and T. M. 1993. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of γ-hexachlorocyclohexane (γ-HCH) in Pseudomonas paucimobilis. J. Bacteriol. 1756403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen, R. W., and R. F. Bilton. 1983. The degradation of cholic acid by Pseudomonas sp. N.C.I.B. 10590 under anaerobic conditions. Biochem. J. 216641-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sih, C. J., S. S. Lee, Y. K. Tsong, and K. C. Wang. 1965. 3,4-Dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione. An intermediate in the microbiological degradation of ring A of androst-4-ene-3,17-dione. J. Am. Chem. Soc. 871385-1386. [PubMed] [Google Scholar]
- 17.Sih, C. J., S. S. Lee, Y. K. Tsong, and K. C. Wang. 1966. Mechanisms of steroid oxidation by microorganisms. VIII. 3,4-Dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione, an intermediate in the microbiological degradation of ring A of androst-4-ene-3,17-dione. J. Biol. Chem. 241540-550. [PubMed] [Google Scholar]
- 18.Sih, C. J., and K. C. Wang. 1963. Mechanisms of steroid oxidation by microorganisms. II. Isolation and characterization of 3aα-H-4α-[3′-propionic acid]-7aβ-methylhezahydro-1,5-indanedione. J. Am. Chem. Soc. 852135-2137. [Google Scholar]
- 19.Sih, C. J., K. C. Wang, D. T. Gibson, and H. W. J. Whitlock. 1965. On the mechanism of ring A cleavage in the degradation of 9,10-seco steroids by microorganisms. J. Am. Chem. Soc. 871386-1387. [DOI] [PubMed] [Google Scholar]
- 20.Tenneson, M. E., J. D. Baty, R. F. Bilton, and A. N. Mason. 1979. The degradation of cholic acid by Pseudomonas sp. N.C.I.B. 10590. Biochem. J. 184613-618. [DOI] [PMC free article] [PubMed] [Google Scholar]