Abstract
The gene for the Campylobacter ferric receptor (CfrA), a putative iron-siderophore transporter in the enteric food-borne pathogen Campylobacter jejuni, was cloned, and the membrane protein was expressed in Escherichia coli, affinity purified, and then reconstituted into model lipid membranes. Fourier transform infrared spectra recorded from the membrane-reconstituted CfrA are similar to spectra that have been recorded from other iron-siderophore transporters and are highly characteristic of a β-sheet protein (∼44% β-sheet and ∼10% α-helix). CfrA undergoes relatively extensive peptide hydrogen-deuterium exchange upon exposure to 2H2O and yet is resistant to thermal denaturation at temperatures up to 95°C. The secondary structure, relatively high aqueous solvent exposure, and high thermal stability are all consistent with a transmembrane β-barrel structure containing a plug domain. Sequence alignments indicate that CfrA contains many of the structural motifs conserved in other iron-siderophore transporters, including the Ton box, PGV, IRG, RP, and LIDG motifs of the plug domain. Surprisingly, a homology model reveals that regions of CfrA that are expected to play a role in enterobactin binding exhibit sequences that differ substantially from the sequences of the corresponding regions that play an essential role in binding/transport by the E. coli enterobactin transporter, FepA. The sequence variations suggest that there are differences in the mechanisms used by CfrA and FepA to interact with bacterial siderophores. It may be possible to exploit these structural differences to develop CfrA-specific therapeutics.
Campylobacter jejuni is a gram-negative food-borne pathogen that commonly populates the gastrointestinal tract of birds and mammals. In humans, C. jejuni infection leads to severe abdominal cramping, headache, diarrhea and, in extreme cases, death (34, 44). C. jejuni infects 1% of the U.S. population annually. Of those affected, 0.1% of the patients develop Guillain-Barré syndrome, an autoimmune disorder of the peripheral nervous system that results in weakening of muscles and, in some cases, neurological problems (9, 31, 41). Both C. jejuni infection and Guillain-Barré syndrome are a considerable burden on the public healthcare system (9, 41).
Most bacteria, including C. jejuni, have an obligatory requirement for iron and have thus developed sophisticated mechanisms for scavenging iron from their hosts (45). One key mechanism involves the production of low-molecular-weight molecules (500 to 1,000 Da), called siderophores, that bind ferric ions with high affinity (1, 46). Secreted siderophores chelate iron from surrounding tissues and are then bound to iron-siderophore receptors/transporters located in the bacterial outer membrane. An energy-dependent TonB-mediated conformational change in the ferric-siderophore transporter translocates the ferric-siderophore into the periplasm. Periplasmic binding proteins then transport the iron-bound siderophore to an inner membrane ATP binding cassette transporter that moves the iron-bound siderophore directly into the cell (1, 12, 15, 38, 47).
Although the mechanisms used by C. jejuni to scavenge iron appear complex, considerable evidence suggests that this microorganism uses a siderophore-mediated transport mechanism. Several strains of C. jejuni are able to utilize ferric enterobactin as an iron source (17, 35). A number of C. jejuni gene sequences have been identified, which encode proteins with significant sequence similarity to proteins in the Campylobacter coli siderophore transport system (19, 33). Most of the corresponding gene sequences have an upstream Fur box. Fur acts as a negative corepressor during transcription of iron transport genes (33). Binding of Fur to the promoter of some of these genes in C. jejuni has been shown directly using mobility shift assays (20, 37).
Conditions of iron starvation in C. jejuni lead to the overexpression of a 76-kDa outer membrane protein, now called the Campylobacter ferric receptor (CfrA). CfrA has considerable sequence similarity with BfrA, a iron-siderophore transporter in Bordetella bronchiseptica (17, 45). In growth plate studies, cfrA deletion mutants are incapable of utilizing enterobactin as an iron source (33). Deletion mutants of cfrA are also unable to establish a successful infection in chick colonization assays (33). Consequently, CfrA has been reannotated as the ferric enterobactin transporter in C. jejuni.
Several iron-siderophore transporters from Escherichia coli outer membranes have been studied from a structural perspective (16, 40, 47). Crystal structures of FhuA, FecA, FepA, and Cir have been determined to high resolution. The proteins all possess a β-barrel topology with a plug domain that is implicated in siderophore binding and transport (6, 47). The plug domain is situated in the middle of the β-barrel and prevents the direct transport of the iron-loaded siderophore from the extracellular environment into the periplasm. Despite the high-resolution structural information, the mechanism of transport by iron-siderophore transporters across the membrane remains to be established. One limiting factor has been the lack of efficient protocols for reconstituting the iron-siderophore transporters and their associated proteins into model membrane systems that are amenable to biochemical and biophysical studies.
Given the prevalence and potentially severe consequences of C. jejuni infection, the outer membrane putative siderophore receptor/transporter CfrA is a potential therapeutic target. As a first step toward better understanding the role played by CfrA in iron transport in C. jejuni, we cloned the cfrA gene from chromosomal DNA and present here a protocol for the expression, purification, and reconstitution of CfrA into model membranes. The membrane-reconstituted CfrA adopts an overall β-sheet secondary structure that is accessible to aqueous solvent and yet highly resistant to thermal denaturation. The structural and biophysical properties of CfrA are consistent with those of other iron-siderophore transporters. Sequence alignments and a homology model of CfrA also show that CfrA contains many of the conserved structural motifs or sequences found in other iron-siderophore transporters. Surprisingly, regions of the protein that are predicted to play a role in ferric enterobactin binding exhibit variations in sequence relative to other iron-siderophore transporters, including the ferric enterobactin transporter, FepA. In fact, key aromatic residues known to play an important role in ferric enterobactin binding/transport in FepA are absent in CfrA. The homology model and sequence comparisons suggest that FepA and CfrA use different mechanisms to interact with ferric enterobactin. The altered sequences of the binding sites suggest that it may be possible to exploit structural differences between CfrA and FepA to develop CfrA-specific therapeutics.
MATERIALS AND METHODS
Construction of the cfrA plasmid.
The cfrA gene was amplified from C. jejuni chromosomal DNA (NCTC 11168) using PCR with the primers listed in Table 1. PCRs were performed in an Eppendorf Mastercycler Gradient (Eppendorf) with 30 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 2.5 min. PCR products were ligated into the pET20b vector (Novagen). The ligation mixture was transformed into chemically competent E. coli XL1-Blue without further manipulation. Resulting transformants were screened for plasmids of appropriate size. Inserts were sequenced to ensure gene integrity. The final plasmid used in these studies was designated pCN10 (CfrA N-terminal His10 tagged).
TABLE 1.
Primer sequences designed for construction of CC6, CN6, CC8, and CN10 CfrA constructsa
| Primer | Sequence (5′-3′)b |
|---|---|
| CfrA-5-Bam-10-His | GCGCGGATCCGCACCACCACCACCACCACCACCACCACCACGACGACGACGACAAGCAAAATGTAGAACTAGATAGC |
| CfrA-3-Xho-10H-NS | GCGCCTCGAGGTGGTGGTGGTGCTTGTCGTCGTCGTCAAAGTTACCATTGATAG |
| CfrA-5-Bam-8-His | GCGCGGATCCGCACCACCACCACCACCACCACCACGACGACGACGACAAGCAAAATGTAGAACTAGATAGC |
| CfrA-3-Xho-8H-NS | GCGCCTCGAGGTGGTGCTTGTCGTCGTCGTCAAAGTTACCATTGATAG |
| CfrA-5-Bam-His | GCGCGGATCCGCACCACCACCACCACCACAAAATGTAGAACTAGATAGC |
| CfrA-3-Xho-H-NS | GCGCCTCGAGAAAGTTACCATTGATAG |
| CfrA-C-Bam-5 | GCGCGGATCCGCAAAATGTAGAACTAGATAGC |
| CfrA-N-Xho-3NS | GCGCCTCGAGTTAAAAGTTACCATTGATAG |
Abbreviations: CC6, CfrA with a C-terminal His6 tag; CN6, CfrA with an N-terminal His6 tag; CC8, CfrA with a C-terminal His6 tag; CN10, CfrA with an N-terminal His10 tag.
Enzyme cleavage sites for cloning purposes are underlined.
Protein expression.
Chemically competent E. coli strain C41 (DE3) was transformed with pCN10 and grown on Luria-Bertani (LB) plates (29). After overnight incubation, single colonies were subcultured into 50 ml of LB medium supplemented with 100 μg of ampicillin (Shelton Scientific)/ml (LB-Amp100) and incubated further for 18 h at 37°C. Cultures were then diluted 1:300 into 16 liters of fresh LB-Amp100, followed by incubation at 37°C. Protein expression was induced at an optical density at 600 nm of 0.6 by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside; MBI Fermentas) to a final concentration of 0.2 mM. After 3 h at 26°C, the cultures were pelleted, and cells were resuspended in 100 ml of lysis buffer (300 mM NaCl, 50 mM NaH2PO4 [pH 7.0]). Cells were frozen until use.
Protein purification.
The resuspended cells were incubated for 1 h at 4°C in lysis buffer containing 2 mM Tris(2-carboxyethyl)phosphine (TCEP), 1 μg of leupeptin/ml, 1 μg of aprotinin/ml, 1 μg of soybean trypsin inhibitor/ml, and 1 mM benzamidine (all from Calbiochem); 1 mM phenylmethylsulfonyl fluoride (Sigma); 100 μg of lysozyme (Sigma)/ml; 10 mM β-mercaptoethanol (Sigma); and 30 μg of DNase (Roche)/ml and then lysed by three passes through an Emulsiflex C3 (Avestin, Inc.) at 18,000 lb/in2. The cell extract was centrifuged at 7,000 × g for 20 min at 4°C to remove the nonlysed cells, and the membrane protein fraction was isolated by centrifugation at 135,000 × g. Membrane fraction protein concentration was determined by BCA protein assay (Pierce).
Membrane proteins were solubilized by incubating 1 g of the membrane protein for 1 h at 4°C in lysis buffer containing 1% (wt/vol) n-dodecyl-β-d-maltoside (DDM; Anatrace), 2 mM TCEP, and 10 mM β-mercaptoethanol. After centrifugation at 100,000 × g, the solubilized protein in the supernatant was incubated for 1 h at 4°C with Superflow cobalt-based resin (Clontech). The resin was transferred to a glass column, and nonspecifically bound protein was removed with three buffer washes (three column volumes each) with increasing imidazole concentrations (0, 10, and 50 mM imidazole) (Sigma) in lysis buffer supplemented with 0.075% (wt/vol) DDM. Protein was eluted from the resin in 150 mM imidazole, 300 mM NaCl, 50 mM NaH2PO4, and 0.075% (wt/vol) DDM at pH 7.0. Protein fractions were concentrated using a Centricon 30,000-Da cutoff concentrator (Millipore) and then further purified on a Superdex-200 gel filtration column (GE Healthcare) equilibrated with lysis buffer supplemented with 2 mM TCEP, 10 mM β-mercaptoethanol, and 0.07 5% (wt/vol) DDM.
The purified CfrA was reconstituted into a lipid membrane by diluting the solubilized protein 1:20 into a solution containing 0.13 μM soybean asolectin (Sigma) and 1% (wt/vol) sodium cholate (Sigma) in lysis buffer. After the sample was mixed for 5 min, the dilute solution was placed in dialysis tubing (12,000- to 14,000-Da cutoff; Fisher), and the samples were dialyzed five times against 2 liters of lysis buffer at 4°C. The reconstituted vesicles were pelleted at 100,000 × g, resuspended in 5 mM Tris-HCl-0.02% (wt/vol) azide at pH 7.5 (Sigma), and stored at −80°C.
Sucrose gradients.
Step sucrose gradients were prepared by layering 2 ml each of 40% (wt/vol), 20% (wt/vol), 10% (wt/vol) sucrose and 3 ml of 0% (wt/vol) sucrose in a plastic centrifuge tube. A portion (150 μg) of protein was pipetted directly into the 0% (wt/vol) sucrose. The sample was centrifuged for 20 h at 100,000 × g. Fractions were assayed for both protein (BCA assay; Pierce) and lipid (phospholipid C; Wako Chemicals) to determine the lipid/protein ratios.
FTIR spectroscopy.
Fourier transform infrared (FTIR) spectra were recorded on either an FTS 40 or an FTS 7000 spectrometer (Varian), both equipped with a deuterated triglycine sulfate detector. For the analysis of protein secondary structure, 200 μg of the protein was exchanged into 2 mM 2H2O phosphate buffer (pH 7.0) by repeated centrifugation and resuspension cycles. After 72 h at 4°C, the samples were pelleted and redissolved in 35 μl of 2 mM 2H2O phosphate buffer (pH 7.0) and subjected to five freeze-thaw cycles. Each sample was then sandwiched between two CaF2 windows separated by a 12-μm Teflon spacer and placed in a thermostatically controlled FTIR transmission cell (Harrick Scientific). Spectra were recorded at a resolution of 2.0 cm−1, averaging 4,000 scans. For thermal denaturation, 256 scan spectra were recorded at 2°C temperature intervals between 35 and 95°C. The temperature was varied with a computer-controlled circulating water bath, with 20 min equilibrations between temperatures.
The extent of hydrogen-deuterium exchange after 72 h in 2H2O was assessed by recording FTIR spectra using the attenuated total reflectance technique. The reconstituted samples were deposited from 2 mM 2H2O (pH 7.0) buffer on a germanium internal reflection optical element, and the sample was immersed in 100 mM NaCl and 5 mM NaH2PO4-2H2O buffer (see Results). All spectra were analyzed using Grams/AI (v.7.01; Galactic) and plotted using Graphpad Prism (Graphpad Software, Inc.).
Homology model.
The homology model was constructed by using Swiss-Model (39). The plug and β-barrel domains were modeled independently using Cir and FepA, respectively, as templates. Collectively, the two domains represent a full-length model of CfrA. The majority (95%) of the ϕ and φ angles of the model are found in favorable regions of the Ramachandran plot, with proline and glycine making up most of the remaining 5%.
RESULTS
Expression, purification, and membrane reconstitution of CfrA.
The cfrA gene was cloned from the chromosomal DNA of C. jejuni (NCTC 11168) and constructs containing either a C-terminal His6 or an N-terminal His6 tag were placed into the pET20b vector under the control of the T7 promoter. The IPTG-induced expression of both proteins was monitored in E. coli strains C41 and C43, which are less sensitive to the expression of toxic proteins and recommended for the expression of membrane proteins (29). In 3-h expression trials, the C41 strain produced higher levels of both the C-terminal and the N-terminal His6-tagged proteins than the C43 strain (data not shown). All further expression studies were performed using the C41 strain.
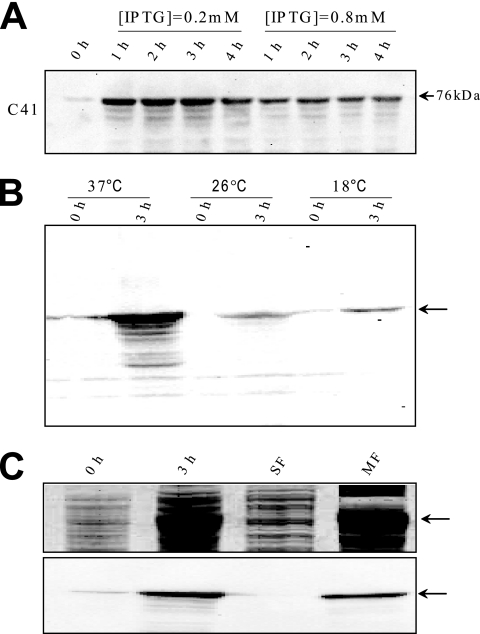
The effect of inducer (IPTG) concentration, over the range of 0.2 to 0.8 mM, on the expression of CfrA was examined. Western blots probed using an anti-His antibody show that expression levels do not increase at concentrations greater than 0.2 mM IPTG (Fig. 1A). In fact, a slight reduction in protein quantity is observed with increasing IPTG concentration, possibly due to a negative effect on cell viability. Also, the overexpression of CfrA is accompanied by the production of degradation products during the 4-h induction trials. To reduce degradation, both the N- and C-terminal His6-tagged CfrAs were expressed at lower induction temperatures of 26 and 18°C (Fig. 1B) (3). Although the amount of degradation may be reduced at lower temperatures, the quantity of expressed protein diminishes substantially. To balance the need for both quality and quantity, the remaining induction experiments and purification trials were performed with an induction temperature of 26°C. Note that the expressed CfrA is found associated with the pelleted membrane fraction (Fig. 1C).
FIG. 1.
Expression of CfrA in E. coli strain C41. Transformed C41 cells were grown at 37°C, and the expression of the N-terminal His6 CfrA examined under various induction conditions. (A) Representative Western blot probed with an anti-His antibody showing the expression of the N-terminal His6 CfrA at 0, 1, 2, 3, and 4 h after induction of expression in the presence of 0.2 and 0.8 mM IPTG. Expression was performed at 37°C. (B) Western blot probed with an anti-His antibody comparing expression of His6 CfrA at 37, 26, and 18°C at time zero and 3 h after induction. (C) SDS-PAGE gel with Coomassie blue stain (upper panel) and a Western blot probed with an anti-His antibody (lower panel) showing expression at time zero and 3 h postinduction, as well as the cellular localization of CfrA. Lane 1, T = 0 h prior to induction; lane 2, T = 3 h postinduction; lane 3, SF (soluble fraction); and lane 4, MF (membrane fraction).
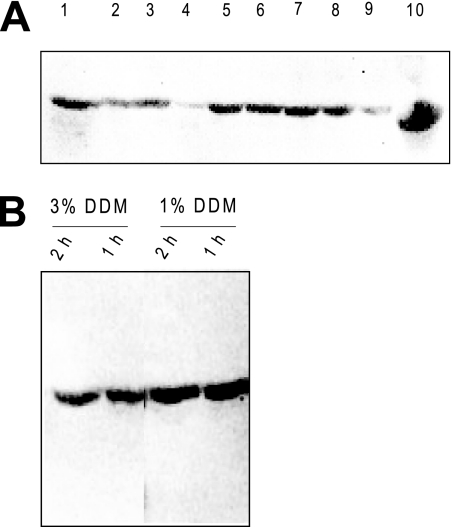
To purify CfrA, we first examined various detergents for their ability to solubilize the protein from its membrane environment. Each detergent solubilization trial was performed at a concentration 1% (wt/vol) above the critical micellar concentration (CMC) of the utilized detergent (23). Of the 17 detergents tested, 9 were able to solubilize appreciable amounts of CfrA from the membrane fraction. Unfortunately, a consistent maximum yield approaching only ∼50% of the total CfrA was obtained with all nine detergents (Fig. 2A). Increasing the detergent concentration and/or the solubilization time did not improve the quantity of solubilized protein (Fig. 2B). One possible explanation for the incomplete solubilization is that the nonsolubilized protein is found in inclusion bodies, which pellet with the membrane fraction. Consistent with this possibility, higher levels of solubilized protein were obtained using the denaturing agent guanidinium hydrochloride as a “solubilizing” agent (data not shown). Biochemical procedures commonly used to isolate proteins from inclusion bodies also led to improved isolation of CfrA (data not shown). No further attempts were thus made to increase the yield of solubilized protein.
FIG. 2.
Solubilization of the N-terminal His6 CfrA. Membrane fractions, containing the expressed N-terminal His6 CfrA, were incubated with the noted detergent for 1 h at 4°C and then centrifuged at 100,000 × g. The supernatants, which contain the solubilized protein, were then analyzed by SDS-PAGE. (A) Western blot with anti-His antibody of soluble fractions from detergents successful in solubilizing CfrA from the membrane. Lane 1, sodium dodecanoyl sarcosinate; lane 2, anzergent 3-14; lane 3, fos mea-10; lane 4, fos-choline; lane 5, cymal 5; lane 6, dodecyl maltoside; lane 7, decyl maltoside; lane 8, nonyl glucoside; lane 9, octyl glucoside; lane 10, control showing sample prior to solubilization. (B) Western blot with anti-His antibody comparing the amount of CfrA solubilized with 1- and 2-h solubilization times at both 1 and 3% DDM.
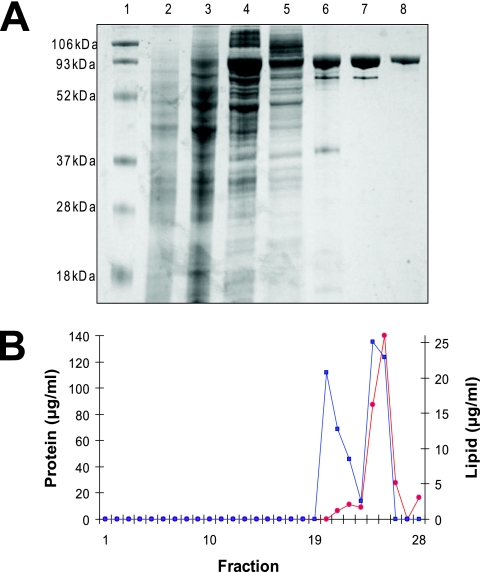
The C-terminal and N-terminal His6 constructs were purified using a cobalt affinity column, followed by size-exclusion chromatography on a Superdex 200 column. Both the N- and C-terminal His6-tagged proteins showed poor binding to the affinity column, possibly due to inaccessibility of the histidine tags. A C-terminal His8-tagged construct was produced, but the improvement in binding to the affinity column was marginal. We finally constructed an N-terminal His10-tagged version of CfrA with an enterokinase cleavage site. The extended tag greatly improved CfrA affinity for the cobalt column. The expression and solubilization of the N-terminal His10-tagged version of CfrA were consistent with the results obtained with the His6-tagged version of the protein discussed above. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) confirms that the affinity column, followed by size-exclusion chromatography is sufficient to purify a His10-tagged protein with a molecular mass of 84 kDa, which is close to that expected for CfrA (76 kDa). Mass spectrometry confirmed the identity of the purified CfrA (472 residues were sequenced from a total of 696 amino acids; Proteomics Resource Centre of the Ottawa Institute of Systems Biology).
Note that a second band is observed in lanes 6 and 7 on the SDS-PAGE gel running at a molecular mass of 73 kDa (Fig. 3). β-Barrel membrane proteins tend to be highly stable structures that are resistant to SDS denaturation (5, 24). A second band running at a lower molecular size on SDS-PAGE gels has been observed with other β-barrel membrane proteins and has been attributed to protein that is not completely denatured by SDS (24). Mass spectrometry confirms that the lower-molecular-weight protein has the expected sequence for CfrA (131 residues were sequenced from a total of 696 amino acids; Proteomics Resource Centre of the Ottawa Institute of Systems Biology). More importantly, boiling the sample prior to SDS-PAGE eliminates the lower-molecular-weight component (Fig. 3, lane 8), confirming that it is due to partially folded CfrA (5).
FIG. 3.
Expression, purification, and reconstitution of N-terminal His10-tagged CfrA. (A) SDS-PAGE with Coomassie blue staining showing protein purity at each step of the purification of CfrA. Lane 1, protein standards; lane 2, whole-cell lysis prior to induction; lane 3, whole-cell lysis 3 h postinduction at 26°C; lane 4, membrane fraction; lane 5, detergent-solubilized protein; lane 6, affinity-purified CfrA; lane 7, affinity-purified CfrA after size-exclusion chromatography; lane 8, CfrA reconstituted into asolectin membranes. Note that only the sample in lane 8 was boiled prior to loading. (B) Sucrose density gradients were used to assess the reconstitution success of CfrA into asolectin lipids. A total of 150 μg of reconstituted CfrA was subjected to sucrose density gradients. Fractions of 400 μl were collected by pipetting, beginning with the least dense (fraction 1, 0% sucrose) to the most dense (fraction 28, 40% sucrose). Individual fractions were assayed for protein (red) and lipid (blue) concentration as described in Materials and Methods.
CfrA was reconstituted into lipid membranes by first diluting the DDM-solubilized protein above the CMC for DDM with a buffered solution containing cholate-solubilized soybean asolectin lipids and then dialyzing away both detergents with several buffer changes. For these initial studies, we chose to reconstitute CfrA into asolectin. We chose this complex lipid mixture because we have had consistently excellent results reconstituting both prokaryotic and eukaryotic membrane proteins into this lipid environment. The dialysis protocol was sufficient to both decrease the two detergents to undetectable levels (data not shown; (13) and form lipid vesicles with incorporated CfrA. The reconstituted CfrA routinely contains two distinct populations of vesicles that vary in terms of the amount of incorporated protein (Fig. 3B). The lipid/protein ratios of the two vesicles populations were estimated by an enzymatic assay (see Materials and Methods) to be 220:1 (mol/mol) and 20:1 (mol/mol), although the assay is specific for phosphatidylcholine and thus underestimates the total lipid content. Phosphatidylcholine represents only 14 to 24% of the total asolectin lipid (Sigma-Addrich). The actual lipid/protein ratios of the two vesicle population were thus 900:1 to 1,600:1 and 80:1 to 150:1, mol/mol, respectively. The less-dense, higher-lipid/protein-ratio vesicles are lost (remain in the supernatant) upon centrifugation and resuspension in 2H2O (see below).
Structural and biophysical characterization of CfrA.
Iron-siderophore transporters adopt a characteristic β-barrel structure with an N-terminal plug domain located within the lumen of the β-barrel. Unlike α-helical transmembrane domain proteins, iron-siderophore transporters exhibit high thermal stability and yet are relatively accessible to aqueous solvent. We used FTIR spectroscopy to ascertain whether affinity purified and reconstituted CfrA exhibits the structural and biophysical properties characteristic of other iron-siderophore transporters. For comparison purposes, we examined similar properties of an α-helical transmembrane domain protein, the nicotinic acetylcholine receptor (nAChR).
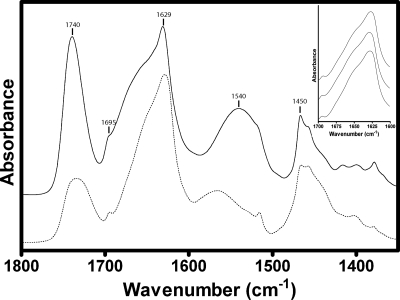
FTIR spectra of the membrane-reconstituted CfrA recorded dried from 1H2O buffer and in 2H2O buffer are presented in Fig. 4. The spectra exhibit intense lipid vibrations centered near 1,740 cm−1 and 1,450 cm−1, as well as intense protein vibrations between 1,700 cm−1 and 1,600 cm−1 and centered near 1,545 cm−1 (1H2O) or 1,450 cm−1 (2H2O). The relative intensities of the lipid vibrations are much higher in spectra recorded in 1H2O than in 2H2O. This is because the centrifugation and resuspension cycles used to exchange the membranes into the denser 2H2O buffer isolates the lower lipid/protein ratio and thus higher-density vesicles. We estimated the lipid/protein molar ratio of the vesicles isolated in 2H2O using FTIR to be roughly 200:1 (13). This value is close to the lipid/protein ratio of the higher density CfrA vesicles isolated using the sucrose gradients (80 to 150:1), as judged by our enzymatic phospholipid C assay.
FIG. 4.
FTIR spectra of CfrA recorded in 1H2O (solid line) and 2H2O (dashed line) buffer. The 1H2O spectrum was recorded after evaporating the bulk solvent under nitrogen. The spectrum of CfrA in 2H2O was recorded after prior exposure of CfrA to 2H2O buffer for 72 h at 4°C. The absorbance of 2H2O has been subtracted. Lipid vibrations are labeled at 1,740 and 1,450 cm−1. Protein amide I vibrations due to peptides in β-sheet secondary structures are labeled at 1,695 and 1,629 cm−1. The protein amide II vibration near 1,540 cm−1 undergoes a downshift in frequency upon peptide 1H/2H exchange to overlap with the lipid vibrations near 1,450 cm−1. Spectra are each an average of 4,000 scans. Spectra were baseline corrected between 1,775 and 1,350 cm−1 and normalized for presentation according to the intensities of the amide I band. The inset shows three representative amide I band contours, each recorded from separate expressions or purifications of CfrA.
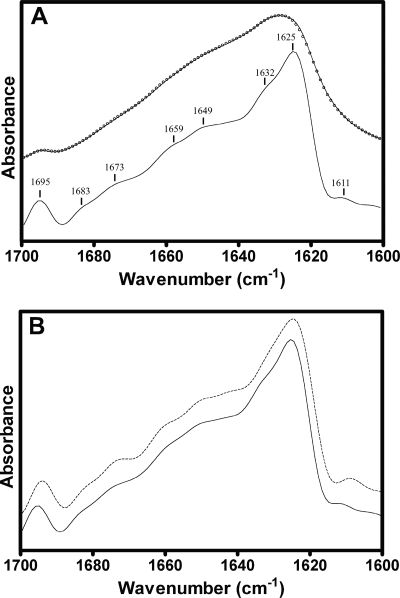
The secondary structure of CfrA was estimated by analyzing the protein amide I band (1,600 to 1,700 cm−1), which is due to the C=O stretching vibrations of the polypeptide backbone coupled to both N-H in-plane bending and C-N stretching (43). The band undergoes little change in shape upon exchange of peptide hydrogen for deuterium, suggesting that a relatively small proportion of the peptides in CfrA form random/loop structures (43). In 2H2O, the band is broad and asymmetric, with a maximum near 1,629 cm−1 and a weaker shoulder near 1,695 cm−1; both features are highly characteristic of predominantly β-sheet proteins, in general, and iron-siderophore transporters, in particular (30, 43). The reproducibility of the amide I band shape is displayed in the inset of Fig. 4 for three independent reconstitutions. In contrast, α-helical proteins give rise to symmetric amide I band shapes, with a maximum located near 1,650 cm−1 (43).
Resolution enhancement reveals the component bands that contribute to the broad amide I contour. Peaks identified from the deconvolved spectrum were used as input to curve fit the amide I contour and thus estimate the secondary structure of CfrA, as shown in Fig. 5 (26). The validity of the curve fit was assessed by comparing the resolution enhanced curve fit spectrum with the experimental resolution-enhanced data (Fig. 5B). Both the predicted β-sheet and the α-helical contents of CfrA, ca. 44 and 8%, respectively (Table 2), are remarkably close to the known β-sheet and α-helical contents of similar iron-siderophore transporters FhuA, FecA, and FepA (ca. 48 and 6%, respectively) (21, 47). The remaining amide I component bands have been attributed to unordered structures and 310 helices, according to the band assignments of Jackson and Mantsch (21) and Goormaghtigh et al. (18), although the assignment of weak amide I component bands to specific types of secondary structures is often problematic (27).
FIG. 5.
Resolution-enhanced and curve fit of the amide I band in the FTIR spectra of CfrA. (A) FTIR spectrum of CfrA in 2H2O (upper solid line) superimposed with the curve fit analysis (circles). Peaks identified from the resolution-enhanced spectrum (lower solid trace) were used as component bands for curve fitting and thus estimation of the secondary structure content. Spectra were baseline corrected between 1,775 and 1,325 cm−1. Resolution-enhanced spectral deconvolution parameters of γ of 8 and smoothing of 75% were used. (B) FTIR spectra of the amide I band of the resolution-enhanced experimental spectrum (solid line) and the resolution-enhanced curve fit spectrum (dashed line).
TABLE 2.
Secondary structure of CfrA
| Band frequency (cm−1)a | Band assignmentb | % Secondary structurec |
|---|---|---|
| 1,695 | β-Sheet | 1 |
| 1,683 | β-Turn | 6 |
| 1,673 | β-Sheet | 6 |
| 1,659 | 310-Helix | 24 |
| 1,649 | α-Helix | 9 |
| 1,641 | Unordered | 17 |
| 1,633 | β-Sheet | 12 |
| 1,624 | β-Sheet | 25 |
Integral membrane proteins tend to be resistant to peptide hydrogen deuterium exchange. In many cases, the number of unexchanged peptide hydrogens remaining after prolonged exposure to 2H2O correlates with the proportion of residues that resides within the hydrophobic environment of the lipid bilayer (25). The amide II band, which is due to in-plane N-1H bending coupled to C-N stretching, undergoes a substantial downshift in frequency from 1,545 to 1,450 cm−1 upon exchange of peptide N-1H for N-2H. The relative intensity at 1,545 cm−1 thus provides a measure of the extent of peptide hydrogen-to-deuterium exchange.
The aqueous solvent accessibility of CfrA was assessed by comparing the extent of peptide 1H/2H exchange in CfrA after precisely 72 h of exposure to 2H2O to that of the α-helical transmembrane domain protein, nAChR.
The extent of peptide 1H/2H exchange in CfrA after 72 h of exposure to 2H2O at 4°C was measured by comparing the residual amide II band area to the amide II band areas in spectra recorded dried from 1H2O (100% N-1H) and in 2H2O after boiling in 2H2O for 10 min (i.e., 100% N-2H). Spectra were initially compared at pH 7.0 (data not shown). Subsequently, spectra were recorded from the same samples at pH 2.0 by exchanging the buffer in the FTIR sample accessory. The pH 2.0 spectra are presented in Fig. 6, because the overlapping vibrations of carboxylic acid side chains (Asp and Glu) shift from near 1,580 cm−1 (deprotonated, at pH 7.0) to near 1,720 cm−1 (protonated, pH 2.0), thus providing an unobstructed view of the residual amide II band (26). This allows for a more accurate visual assessment of the residual amide II band intensity. The spectral analysis suggests that 75% ± 0.8% (four independent experiments using three different membrane preps) of the peptide N-1H have exchanged for N-2H in CfrA after 3 days of exposure to 2H2O at 4°C (Fig. 6A). Note that the precision of our measurement from spectrum to spectrum is high, but the accuracy of the number is sensitive to the accuracy of the 100% N-1H value, which was calculated from a CfrA sample dried from H2O.
FIG. 6.
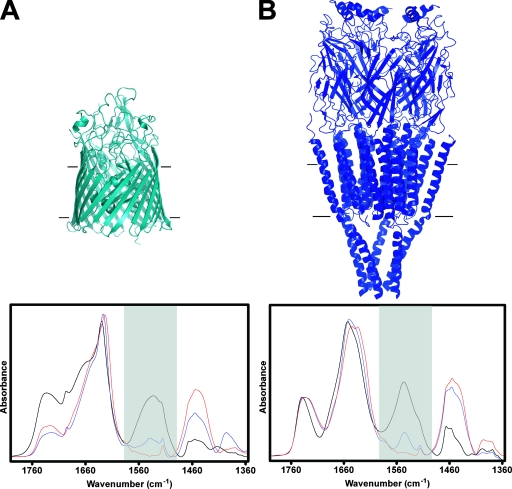
Comparison of the extent of peptide hydrogen-deuterium exchange for CfrA (A) and nAChR (B), both after exposure to 2H2O buffer (pH 7.0) for 72 h at 4°C. In each case, the extent of exchange was estimated by comparing the residual amide II intensity (shaded area, peptide N-1H vibrations) in spectra recorded after 72 h of exposure to 2H2O (blue line) to the intensities in spectra recorded from samples in 1H2O (black line, 100% N-1H) and in 2H2O after exposure to conditions that lead to complete 1H/2H exchange (red line). Conditions for complete 1H/2H were different for the respective proteins: the CfrA sample was boiled for 10 min, whereas the nAChR was heated to 70°C for 10 min. The appropriate buffer spectrum has been subtracted from each spectrum. A spectrum of each sample was initially recorded at pH 7.0 (data not shown). The buffer was then switched to an identical buffer at pH 2.0 to shift the vibrations of carboxyl side chains (Asp and Glu) and thus provide an unobstructed view of the amide II band intensity. For comparison purposes, ribbon diagrams of FepA (A) and nAChR (B) show the relative proportions of each protein that are thought to reside within the lipid bilayer.
For comparison, we also recorded spectra under identical exchange conditions from another integral membrane protein, the nAChR (structure shown in Fig. 6B). A similar percentage (75 to 80%) of peptide hydrogens in the nAChR, which has an exclusively α-helical transmembrane domain, undergo peptide hydrogen-deuterium exchange after 72 h of exposure to 2H2O (4). The number of peptide hydrogens that are resistant to the 1H/2H exchange in the nAChR, however, correlates with the number of peptide hydrogens that are found in the transmembrane domain. In contrast to the nAChR, the percentage of peptide hydrogens in CfrA that exchange for deuterium after 72 h of exposure to 2H2O is much smaller than the proposed number of peptide hydrogens in known iron-siderophore transporter structures that are located within the transmembrane domain (70% of the residues are typically found in the β-strands of the barrel, with the plug domain and N-terminal extension comprising 18 and 12% of the amino acids, respectively) (47). This comparison between CfrA and the nAChR highlights the fact that CfrA is relatively exposed to aqueous solution. A high solvent exposure has been noted for other β-barrel transmembrane structures, including iron-siderophore transporters (11). The hydrogen-deuterium exchange data are thus also consistent with CfrA adopting a transmembrane β-barrel structure.
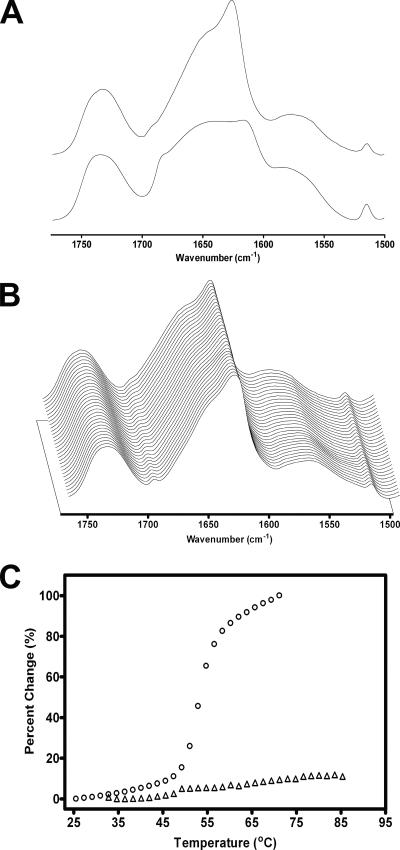
Finally, we examined the thermal denaturation of CfrA by recording FTIR spectra as a function of temperature in Fig. 7. Denaturation of CfrA upon boiling in 2H2O for 1 h leads to a substantially altered amide I band shape with new peak maxima near 1,614 and 1,685 cm−1 (Fig. 7A, bottom trace) (30). In contrast, FTIR spectra recorded from CfrA at temperatures ranging from 35 to 95°C show no detectable changes in amide I band shape, indicating that CfrA remains in its native conformation over the entire temperature range of the experiment (Fig. 7B). The high thermal stability of CfrA is emphasized when one compares its thermal denaturation profile to that of the nAChR, which denatures at roughly 55°C (Fig. 7C) (14). The high thermal stability CfrA is consistent with the high thermal stabilities reported for other β-barrel proteins (42). The secondary structure, hydrogen exchange characteristics, and thermal stability of CfrA are all consistent with both a β-barrel structure and the proposed role of CfrA as a predominantly β-barrel outer membrane iron-siderophore transporter.
FIG. 7.
Thermal denaturation of CfrA. (A) Comparison of a spectrum of CfrA recorded at 95°C (upper trace) to a spectrum recorded after CfrA was boiled for 1 h and thus denatured (lower line). (B) Three-dimensional stack plot showing FTIR spectra of CfrA recorded in 2H2O at pH 7.0 with increasing temperatures from 35°C (front spectra) to 95°C (back spectra). (C) Changes in intensity at 1,681 cm−1 plotted as a function of temperature for CfrA (▵) compared to the intensity changes observed in similar spectra recorded from the nAChR (○) (14).
Structural model of CfrA.
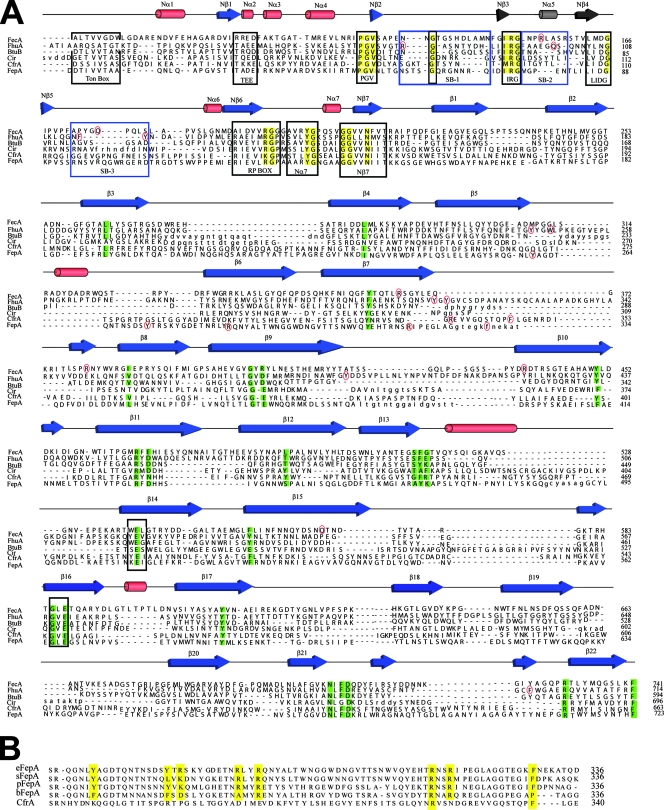
The FTIR data provide a structural rationale for performing sequence alignments with other iron-siderophore transporters and for constructing a homology model (39). The model was first attempted using Cir, as a template, since this TonB-dependent transporter has the highest sequence identity with CfrA (29% overall sequence identity, E value of 4e−65). Cir is expressed in many strains of E. coli, where it binds and transports ferric ion complexed to linear catecholates, such as dihydroxybenzoyl serine. The crystal structure of Cir, however, exhibits poor electron density in some of the extracellular loops connecting the transmembrane β-strands (7). This, coupled with the fact that the sequence of CfrA is longer than that of Cir (CfrA has 696 residues versus 663 for Cir), made it difficult to model the β-barrel. Alternatively, a structure was generated by modeling the plug and β-barrel domains separately, using Cir as a template for the plug (45% sequence identity, E value of 1e−23) and FepA as a template for the β-barrel (24% sequence identity, E value of 6e−19). Cir and FepA were chosen since they exhibit the highest sequence identities for the independent domains modeled. FepA is also an E. coli TonB-dependent iron transporter but transports iron complexed to the endogenous catecholate siderophore, enterobactin (8). The three-dimensional models of both the β-barrel and the plug domains are shown in Fig. 9A. A flattened schematic of the β-barrel showing conserved residues is compared to a similar structure of FepA in Fig. 9B.
FIG. 9.
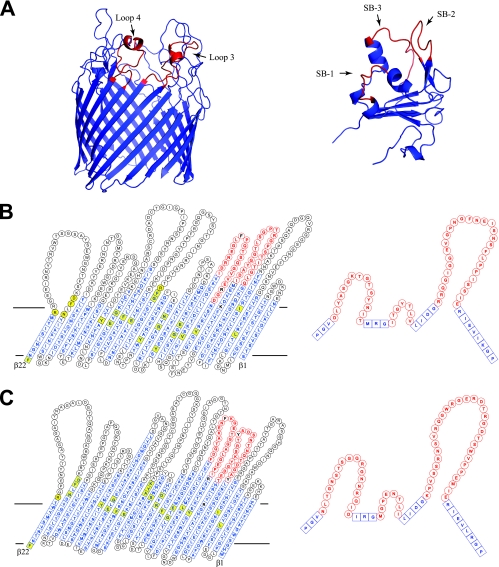
Topology of iron-siderophore transporters. (A) Ribbon representation of the β-barrel and plug domains of the CfrA model. Barrel loops predicted to participate in ligand binding are indicated in red. Substrate-binding loops of the plug domain are also shown in red. (B and C) Comparison of unrolled flat representation of the barrel domain (left panel) and the plug domain (right panel) for CfrA (B) and FepA (C). Blue boxes in the barrel domain indicate residues located in the β-strands of the barrel domain. Blue boxes in the plug domain indicate conserved motifs within the plug domain. Black circles indicate residues located in the loop region of the proteins. Residues predicted to be involved in ligand binding are indicated in black text. Residues that interact with the plug domain are also shown in yellow. The predicted position of the membrane bilayer for the barrel domain is outlined with horizontal lines.
The model predicts, as expected given the chosen templates, that CfrA adopts a 22-strand antiparallel β-barrel with the N-terminal of the protein folded into the lumen of the β-barrel forming the plug domain. The strands of the β-barrel range in length from 7 to 23 amino acids. The β-barrel exhibits long extracellular loops and shorter periplasmic turns connecting the β-strands. The plug, which blocks passage of large molecules from the extracellular to the periplasmic solutions, contains a β-sheet core with several loop regions (SB-1, SB-2, and SB-3) that extend toward the extracellular surface of the β-barrel. These loops likely play a role in siderophore binding and transport (see below).
The existence of several structural motifs, which are highly conserved in other iron-siderophore transporters, provides confidence in the modeled structure of CfrA, particularly in the plug domain (11). Nine conserved structural motifs of the plug, identified by Chimento et al., are found in CfrA (black boxes in Fig. 8), including the Ton box and the PGV, IRG, LIDG, and RP motifs (11). The Ton box interacts with the energy transducer, TonB, and thus plays a role in the conformational switch associated with iron transport. The PGV, IRG, LIDG, and RP motifs define the location of the three loops of the plug domain (SB-1, SB-2, and SB-3), which play a role in substrate binding. These binding loops are held in place by a charge cluster involving two conserved arginine residues in the IRG and RP motifs. These arginines form salt bridges with two glutamate residues located in strands 14 and 16 of the β-barrel, as discussed below (11). The location of these and other motifs allow us to define with precision the location of the different secondary structures in the plug domain sequence of CfrA, as well as the potential residues in SB-1, SB-2, and SB-3 of the plug that likely interact with the putative substrate(s) of CfrA.
FIG. 8.
Alignments of CfrA model with other iron-siderophore transporters. (A) The aligned sequences of CfrA, Cir, and FepA were manually added to the structure-based sequence alignment of crystallized iron-siderophore transporters performed by Buchanan and coworkers (47). Secondary structural elements are depicted as red cones (helices) and blue arrows (β-strands). Conserved residues, conserved motifs, and substrate-binding loops within the plug domain are indicated in yellow, black boxes, and blue boxes, respectively. Conserved residues from the barrel that interact with the plug domain are also shown in yellow. Residues that participate in binding of ferrichrome, diferric dicitrate, and enterobactin, as discussed in the text, are indicated by red circles. (B) Sequence alignments of CfrA with enterobactin transporters from difference bacterial species adapted from (32). Bacterial species: e, Escherichia coli; s, Salmonella enterica serovar Typhimurium; p, Pseudomonas aeruginosa; b, Bacillus pertussis.
Although the lower sequence conservation in the β-barrel domain makes the precise definition of some of the β-strands in the β-barrel equivocal, several conserved residues are present. In particular, two acidic residues observed at conserved locations of β-strands 14 and 16 of the barrel likely form a charge cluster with the conserved arginine residues located in the IRG and RP motifs of the plug domain (see above). These conserved glutamic acid residues, shaded blue in Fig. 8 and 9, allow us to accurately place the sequence in register in these two β-strands. Other conserved residues in the β-barrel have also been shown to form interactions with the plug domain and thus provide further confidence in the sequence alignment of CfrA with the other iron-siderophore transporters (gray in Fig. 8 and 9) (11). Unfortunately, these conserved residues are found mostly in the C-terminal half of the β-barrel.
The identified conserved residues and structural motifs in our homology model provide confidence in our modeled structure and allow us to compare the sequence of CfrA to those of other iron-siderophore transporters, particularly in regions thought to play an important role in siderophore binding and/or transport. Current data indicate that CfrA acts as a transporter of ferric enterobactin (33). FepA is the endogenous E. coli enterobactin transporter. We thus expected CfrA to exhibit a high sequence similarity to FepA in regions thought to play a role in enterobactin binding and/or transport. As shown in the sequence alignments of Fig. 8B and the homology model of Fig. 9, however, there are considerable sequence variations, suggesting that the mechanisms used by CfrA and FepA to interact with ferric enterobactin may differ substantially (see Discussion).
DISCUSSION
Expression, purification, reconstitution, and structural characterization of CfrA.
We report here the successful full-length cloning and expression of an outer membrane protein implicated in iron-siderophore transport in the enteric pathogen, C. jejuni. The protein was expressed in the E. coli strain, C41, with typical yields of ∼7 mg of purified CfrA per 16 liters of culture, of which 4 to 5 mg was typically reconstituted into lipid bilayer vesicles. Although these yields are less than anticipated, they are not entirely unexpected for the expression of a C. jejuni membrane protein in E. coli. For example, Mamelli et al. were able to express the C. jejuni membrane protein, CadF, in E. coli in large quantities, but these authors were only able to do so with a small truncated version of the protein (24). The truncated CadF was also targeted to inclusion bodies. We were unable to completely solubilize CfrA from the membrane fraction (which includes inclusion bodies), suggesting that some of the expressed protein may be targeted to inclusion bodies. Further, high-level expression of CfrA led to degradation of the expressed protein. E. coli may not be efficient at expressing C. jejuni membrane proteins. Despite the encountered problems with the expression of CfrA, we were still able to obtain intact protein in milligram quantities, which is sufficient for structural characterization.
We also present an efficient protocol for the reconstitution of the expressed and purified CfrA into lipid membranes. We utilized the relatively mild detergent, DDM, to solubilize and purify CfrA from the bacterial cell membranes, since it is an excellent detergent for maintaining membrane protein structure or function in the solubilized state. Unfortunately, DDM has a very low CMC and is thus difficult to remove by dialysis to form reconstituted protein-lipid vesicles. Consequently, protein incorporated vesicles are often formed in the presence of exogenous lipids using hydrophobic BioBeads to remove the DDM. However, we found this approach leads to loss of protein and incomplete detergent removal (data not shown). We therefore diluted our DDM-solubilized CfrA with sufficient cholate-solubilized exogenous lipid to bring the concentration of DDM below its CMC. We then dialyzed away both detergents and efficiently formed protein-lipid complexes. We found this simple dilution-dialysis procedure removed both DDM and cholate from the solution to undetectable levels. Sucrose gradients confirm that the CfrA is associated with lipids, although two populations of vesicles with differing lipid/protein ratios were typically formed.
The ability to efficiently reconstitute CfrA into lipid bilayers opens the door for more detailed biochemical and biophysical studies of CfrA structure and function. Reconstituting CfrA in the presence of other components of the TonB system may allow us to probe in detail the mechanisms of siderophore-mediated iron transport.
FTIR spectra of the purified and reconstituted CfrA are highly characteristic of a predominantly β-sheet protein. We estimated the secondary structure content of CfrA to be ∼44% β-sheet and ∼10% α-helix; both are surprisingly close to the secondary structure of the iron transporter FecA estimated from the crystal structure (47). More importantly, the FTIR spectra are CfrA are similar to those recorded from another iron-siderophore transporter, FhuA demonstrating the similarity in structure (30). CfrA also exhibits a high thermal stability and is resistant to SDS denaturation and yet is relatively accessible to solvent and thus undergoes relatively extensive peptide hydrogen-deuterium exchange. These findings are all consistent with CfrA adopting a transmembrane β-barrel, since similar biophysical properties have been observed for other transmembrane β-barrel proteins (5, 16). The data accumulated in the present study provide the first structural evidence that CfrA adopts a topology consistent with an iron-siderophore transporter.
Homology model of CfrA.
Our homology model of CfrA created using Cir and FepA as templates for the plug and β-barrels, respectively, provides additional structural evidence that CfrA is a putative iron-siderophore transporter. The model exhibits many of the conserved sequences and/or structural motifs found in other iron-siderophore transporters, including the Ton box, PGV, IRG, RP, and LIDG motifs of the plug domain. Numerous conserved residues in the β-barrel domain that, in other iron-siderophore transporters, interact with conserved residues in the plug domain are also found at equivalent sites in CfrA. Of particular interest are two glutamic acid residues (E484 and E547) of β-strands 14 and 16, which form a charge cluster with arginines in the IRG and RP boxes of the plug domain.
Selectivity of siderophore binding.
C. jejuni grows in culture using exogenously supplied ferric enterobactin as an iron source (33). CfrA deletion mutants of C. jejuni, however, are not capable of transporting ferric enterobactin, suggesting that CfrA is an enterobactin transporter (33). Based on these data, we expected our homology model and sequence comparisons to reveal binding site motifs consistent with those found in the E. coli enterobactin transporter, FepA.
We first compared our homology model and the sequence of CfrA with the structures and sequences of FhuA and FecA, since structures of both of these iron-siderophore transporters have been determined in the presence of bound siderophores. The structures provide direct insight into the nature of siderophore-transporter interactions (22, 47). FhuA transports the cyclic hexapeptide, ferrichrome, which coordinates iron using hydroxamate functional groups. Ferrichrome interacts with FhuA predominantly via aromatic residues located in the third binding loop of the plug domain, SB-3 (F115 and Y116), and loops 3 (Y244 and W246), 4 (Y313 and Y315), 5 (Y391), and 11 (F702) of the β-barrel. R81 and Q100 of the plug SB-1 and SB-2 binding loops, respectively, interact directly with the hydroxamate functional groups that chelate the bound ferric iron (22). Significantly, both the structural model of CfrA and the sequence comparisons reveal an absence of aromatic residues at comparable sites to FhuA, even taking into account possible errors in the registers of the individual β-strands. The arginine and glutamine residues of SB-1 and SB-2 in FhuA are also missing, although a lysine residue is located close by in SB-1 of CfrA. The homology model predicts that iron-bound ferrichrome is not a high-affinity substrate for CfrA, in agreement with the inability of C. jejuni NCTC 11168 to acquire iron from ferrichrome (A. Stintzi, unpublished data).
Structures of FecA have been resolved with both diferric dicitrate and dicitrate bound to the plug and β-barrel domains (47). In contrast to FhuA, the binding site of FecA exhibits a large number of positively charged arginines that interact with the highly negative citrate molecule. These include R155 of SB-1, R365 of β-strand 7, R380 of loop 4, and R438 of β-strand 10. Two other residues involved in binding, Q176 and Q570, are found in SB-3 and β-strand 15, respectively (47). Even allowing for substantial flexibility in the registers of the β-strands, there are few positively charged residues in CfrA that could play a comparable role in substrate binding. As expected, the sequence alignment and modeled structure thus predict that ferric citrate is also not a substrate for CfrA.
Finally, we compared the modeled structure and sequence of CfrA with that of FepA. Although bound ferric enterobactin was not resolved in the published structure of FepA, thus precluding direct structural analysis of FepA-enterobactin interactions, there is considerable biochemical data available concerning FepA-enterobactin interactions (8, 10, 32). The catecholate siderophore, enterobactin, has a net negative charge, suggesting that aromatic and basic residues play a role in siderophore binding (36). These postulates are supported by mutagenesis studies with FepA, which show that the Y260A and F329A play a role in enterobactin binding affinity and transport efficiency, although Y260 appears to be essential to both processes (10, 32). Combinatorial mutations of two aromatic residues, Y272A and F329A, or two arginine residues, R286A and R316A, suggest the involvement of all four residues in binding/transport. As shown in Fig. 8B, Klebba and coworkers have demonstrated the across-species conservation of both aromatic and positively charged residues in a region of FepA that contains most of the residues known to play an important role in ferric enterobactin binding and transport (32).
Surprisingly, the sequence of CfrA does not match up as well with that of FepA in the regions of the protein that are predicted to play a role in substrate binding, as might be expected for an enterobactin transporter. CfrA does not exhibit a tyrosine residue at a position comparable to Y260 of FepA, even allowing for considerable errors in the aligned register of the two sequences. This variation is significant because Y260 is the only residue in FepA identified thus far that upon mutation alone (i.e., not in combination with another residue) leads to a substantial reduction in binding affinity and/or transport (2). CfrA also lacks a tyrosine or other comparable aromatic residue at a position consistent with Y272 in FepA. CfrA does exhibit a positively charged lysine at a position that matches R286 in FepA. A shift in the sequence will bring F338 of CfrA in register with the F329 in FepA (Fig. 8B) and thus R336 of CfrA to within two residues of R316 of FepA, but this shift in register would then shift Y328 of CfrA out of register with its conserved counterpart in FepA.
The striking lack of sequence conservation between CfrA and FepA in a region known to be central to enterobactin binding in FepA is illustrated clearly in Fig. 8B. Also, the three binding loops of the plug domain, shown to interact directly with bound siderophores in FhuA and FecA, exhibit little sequence similarity between CfrA and FepA in terms of either aromatic or charged residues (blue boxes in Fig. 8A). The structural and sequence comparisons show that the putative enterobactin binding site of CfrA exhibits substantial sequence variations relative to comparable regions of the enterobactin binding site of FepA.
An intriguing possibility is that CfrA lacks key residues involved in ferric enterobactin binding and thus has a lower binding affinity for the siderophore. Lower affinity enterobactin binding might correlate with a broader siderophore binding selectivity. Unlike E. coli, the genome of C. jejuni appears to code for only one TonB-dependent siderophore transporter since Cj0178 was recently shown to mediate iron acquisition from ferric-lactoferrin (28). Given that CfrA is potentially the lone siderophore transporter in C. jejuni, it may have evolved to bind and transport a broader range of siderophores with lower affinity in order to ensure survival of C. jejuni in hostile environments.
Given that C. jejuni is a common food-borne pathogen that leads to considerable health problems, this microorganism is of considerable interest as a therapeutic target. Deletion of the cfrA gene greatly diminishes the virulence of C. jejuni in chick colonization assays (33). Inhibitors of CfrA function should thus be effective at treating C. jejuni infection. The sequence and structural comparisons strongly suggest that CfrA from C. jejuni and FepA from E. coli bind enterobactin using different mechanisms or binding interactions. Given that the two proteins exhibit structural differences in their enterobactin binding sites, it may be possible to exploit the structural differences between CfrA and FepA to develop CfrA-specific therapeutics. Such an approach might allow one to target CfrA containing C. jejuni in the intestine, while leaving the endogenous micro flora unharmed.
In summary, we have cloned, expressed, purified, membrane reconstituted, and structurally characterized a 76-kDa protein that has been identified as the potential iron-siderophore transporter in C. jejuni, CfrA. We show that CfrA is membrane associated and can be expressed in E. coli and purified or reconstituted into a membrane environment in milligram quantities sufficient for structural characterization. The protein exhibits a predominantly β-sheet secondary structure, is highly thermally stable, and has peptide hydrogens that are relatively accessible to aqueous solvent, a finding consistent with other iron-siderophore transporter. Structure-based sequence alignments and a homology model show that CfrA exhibits all of the conserved structural motifs found in other iron-siderophore transporters, particularly in the proposed plug domain. Our analysis provides the first structural evidence that CfrA is the putative iron-siderophore transporter in C. jejuni. There are substantial sequence variations in the putative siderophore binding site of CfrA relative to other iron-siderophore transporters, including the enterobactin transporter FepA. The data suggest that CfrA and FepA use different mechanism to interact with the siderophore enterobactin.
Acknowledgments
We thank Alain Stintzi for helpful comments and for providing the C. jejuni chromosomal DNA (NCTC 11168) and Lopamudra Dey for the purified nAChR.
Footnotes
Published ahead of print on 13 June 2008.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27215-237. [DOI] [PubMed] [Google Scholar]
- 2.Annamalai, R., B. Jin, Z. Cao, S. M. Newton, and P. E. Klebba. 2004. Recognition of ferric catecholates by FepA. J. Bacteriol. 1863578-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auer, M., M. J. Kim, M. J. Lemieux, A. Villa, J. Song, X. D. Li, and D. N. Wang. 2001. High-yield expression and functional analysis of Escherichia coli glycerol-3-phosphate transporter. Biochemistry 406628-6635. [DOI] [PubMed] [Google Scholar]
- 4.Baenziger, J. E., and N. Methot. 1995. Fourier transform infrared and hydrogen/deuterium exchange reveal an exchange-resistant core of alpha-helical peptide hydrogens in the nicotinic acetylcholine receptor. J. Biol. Chem. 27029129-29137. [DOI] [PubMed] [Google Scholar]
- 5.Bonhivers, M., M. Desmadril, G. S. Moeck, P. Boulanger, A. Colomer-Pallas, and L. Letellier. 2001. Stability studies of FhuA, a two-domain outer membrane protein from Escherichia coli. Biochemistry 402606-2613. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan, S. K. 1999. Beta-barrel proteins from bacterial outer membranes: structure, function and refolding. Curr. Opin. Struct. Biol. 9455-461. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan, S. K., P. Lukacik, S. Grizot, R. Ghirlando, M. M. Ali, T. J. Barnard, K. S. Jakes, P. K. Kienker, and L. Esser. 2007. Structure of colicin I receptor bound to the R-domain of colicin Ia: implications for protein import. EMBO J. 262594-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 656-63. [DOI] [PubMed] [Google Scholar]
- 9.Butzler, J. P. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10868-876. [DOI] [PubMed] [Google Scholar]
- 10.Cao, Z., Z. Qi, C. Sprencel, S. M. Newton, and P. E. Klebba. 2000. Aromatic components of two ferric enterobactin binding sites in Escherichia coli FepA. Mol. Microbiol. 371306-1317. [DOI] [PubMed] [Google Scholar]
- 11.Chimento, D. P., R. J. Kadner, and M. C. Wiener. 2005. Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins 59240-251. [DOI] [PubMed] [Google Scholar]
- 12.Clarke, T. E., L. W. Tari, and H. J. Vogel. 2001. Structural biology of bacterial iron uptake systems. Curr. Top. Med. Chem. 17-30. [DOI] [PubMed] [Google Scholar]
- 13.daCosta, C. J., and J. E. Baenziger. 2003. A rapid method for assessing lipid:protein and detergent:protein ratios in membrane-protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 5977-83. [DOI] [PubMed] [Google Scholar]
- 14.daCosta, C. J., D. E. Kaiser, and J. E. Baenziger. 2005. Role of glycosylation and membrane environment in nicotinic acetylcholine receptor stability. Biophys. J. 881755-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson, A. D., and J. Deisenhofer. 2002. TonB-dependent receptors-structural perspectives. Biochim. Biophys. Acta 1565318-332. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 2822215-2220. [DOI] [PubMed] [Google Scholar]
- 17.Field, L. H., V. L. Headley, S. M. Payne, and L. J. Berry. 1986. Influence of iron on growth, morphology, outer membrane protein composition, and synthesis of siderophores in Campylobacter jejuni. Infect. Immun. 54126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goormaghtigh, E., J. M. Ruysschaert, and V. Raussens. 2006. Evaluation of the information content in infrared spectra for protein secondary structure determination. Biophys. J. 902946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerry, P., J. Perez-Casal, R. Yao, A. McVeigh, and T. J. Trust. 1997. A genetic locus involved in iron utilization unique to some Campylobacter strains. J. Bacteriol. 1793997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes, K., F. Mulholland, B. M. Pearson, C. Pin, J. McNicholl-Kennedy, J. M. Ketley, and J. M. Wells. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151243-257. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, M., and H. H. Mantsch. 1995. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 3095-120. [DOI] [PubMed] [Google Scholar]
- 22.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95771-778. [DOI] [PubMed] [Google Scholar]
- 23.Loll, P. J. 2003. Membrane protein structural biology: the high throughput challenge. J. Struct. Biol. 142144-153. [DOI] [PubMed] [Google Scholar]
- 24.Mamelli, L., J. M. Pages, M. E. Konkel, and J. M. Bolla. 2006. Expression and purification of native and truncated forms of CadF, an outer membrane protein of Campylobacter. Int. J. Biol. Macromol. 39135-140. [DOI] [PubMed] [Google Scholar]
- 25.Methot, N., and J. E. Baenziger. 1998. Secondary structure of the exchange-resistant core from the nicotinic acetylcholine receptor probed directly by infrared spectroscopy and hydrogen/deuterium exchange. Biochemistry 3714815-14822. [DOI] [PubMed] [Google Scholar]
- 26.Methot, N., M. P. McCarthy, and J. E. Baenziger. 1994. Secondary structure of the nicotinic acetylcholine receptor: implications for structural models of a ligand-gated ion channel. Biochemistry 337709-7717. [DOI] [PubMed] [Google Scholar]
- 27.Methot, N., B. D. Ritchie, M. P. Blanton, and J. E. Baenziger. 2001. Structure of the pore-forming transmembrane domain of a ligand-gated ion channel. J. Biol. Chem. 27623726-23732. [DOI] [PubMed] [Google Scholar]
- 28.Miller, C. E., J. D. Rock, K. A. Ridley, P. H. Williams, and J. M. Ketley. 2008. Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni. J. Bacteriol. 1901900-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260289-298. [DOI] [PubMed] [Google Scholar]
- 30.Moeck, G. S., P. Tawa, H. Xiang, A. A. Ismail, J. L. Turnbull, and J. W. Coulton. 1996. Ligand-induced conformational change in the ferrichrome-iron receptor of Escherichia coli K-12. Mol. Microbiol. 22459-471. [DOI] [PubMed] [Google Scholar]
- 31.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton, S. M., J. S. Allen, Z. Cao, Z. Qi, X. Jiang, C. Sprencel, J. D. Igo, S. B. Foster, M. A. Payne, and P. E. Klebba. 1997. Double mutagenesis of a positive charge cluster in the ligand-binding site of the ferric enterobactin receptor, FepA. Proc. Natl. Acad. Sci. USA 944560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 1864714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, S. F. 2002. The physiology of Campylobacter species and its relevance to their role as food-borne pathogens. Int. J. Food Microbiol. 74177-188. [DOI] [PubMed] [Google Scholar]
- 35.Pickett, C. L., T. Auffenberg, E. C. Pesci, V. L. Sheen, and S. S. Jusuf. 1992. Iron acquisition and hemolysin production by Campylobacter jejuni. Infect. Immun. 603872-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raymond, K. N., E. A. Dertz, and S. S. Kim. 2003. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 1003584-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ridley, K. A., J. D. Rock, Y. Li, and J. M. Ketley. 2006. Heme utilization in Campylobacter jejuni. J. Bacteriol. 1887862-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schalk, I. J., W. W. Yue, and S. K. Buchanan. 2004. Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol. Microbiol. 5414-22. [DOI] [PubMed] [Google Scholar]
- 39.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 313381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, B. S., B. Kobe, R. Kurumbail, S. K. Buchanan, L. Venkatramani, D. Van Der Helm, and J. Deisenhofer. 1998. Crystallization and preliminary X-ray analysis of ferric enterobactin receptor FepA, an integral membrane protein from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 54697-699. [DOI] [PubMed] [Google Scholar]
- 41.Snelling, W. J., M. Matsuda, J. E. Moore, and J. S. Dooley. 2005. Campylobacter jejuni. Lett. Appl. Microbiol. 41297-302. [DOI] [PubMed] [Google Scholar]
- 42.Sukumaran, S., C. Zscherp, and W. Mantele. 2004. Investigation of the thermal stability of porin from Paracoccus denitrificans by site-directed mutagenesis and Fourier transform infrared spectroscopy. Biopolymers 7482-86. [DOI] [PubMed] [Google Scholar]
- 43.Tamm, L. K., and S. A. Tatulian. 1997. Infrared spectroscopy of proteins and peptides in lipid bilayers. Q. Rev. Biophys. 30365-429. [DOI] [PubMed] [Google Scholar]
- 44.van Vliet, A. H., and J. M. Ketley. 2001. Pathogenesis of enteric Campylobacter infection. Symp. Ser. Soc. Appl. Microbiol. 3045S-56S. [DOI] [PubMed] [Google Scholar]
- 45.van Vliet, A. H., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 1805291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wooldridge, K. G., and P. H. Williams. 1993. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 12325-348. [DOI] [PubMed] [Google Scholar]
- 47.Yue, W. W., S. Grizot, and S. K. Buchanan. 2003. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J. Mol. Biol. 332353-368. [DOI] [PubMed] [Google Scholar]