Abstract
Yersinia pestis, the causative agent of plague, has been well studied at the molecular and genetic levels, but little is known about the role that host genes play in combating this highly lethal pathogen. We challenged several inbred strains of mice with Y. pestis and found that BALB/cJ mice are highly resistant compared to susceptible strains such as C57BL/6J. This resistance was observed only in BALB/cJ mice and not in other BALB/c substrains. Compared to C57BL/6J mice, the BALB/cJ strain exhibited reduced bacterial burden in the spleen and liver early after infection as well as lower levels of serum interleukin-6. These differences were evident 24 h postinfection and became more pronounced with time. Although a significant influx of neutrophils in the spleen and liver was exhibited in both strains, occlusive fibrinous thrombi resulting in necrosis of the surrounding tissue was observed only in C57BL/6J mice. In an effort to identify the gene(s) responsible for resistance, we measured total splenic bacteria in 95 F2 mice 48 h postinfection and performed quantitative trait locus mapping using 58 microsatellite markers spaced throughout the genome. This analysis revealed a single nonrecessive plague resistance locus, designated prl1 (plague resistance locus 1), which coincides with the major histocompatibility complex of chromosome 17. A second screen of 95 backcrossed mice verified that this locus confers resistance to Y. pestis early in infection. Finally, eighth generation backcrossed mice harboring prl1 were found to maintain resistance in the susceptible C57BL/6J background. These results identify a novel genetic locus in BALB/cJ mice that confers resistance to Y. pestis.
Yersinia pestis is a gram-negative bacterial pathogen and the causative agent of pneumonic and bubonic plague. Bubonic plague results from infection of lymphoid tissue and is fatal in at least 50% of untreated cases. In contrast, pneumonic plague occurs after the bacteria colonize the lung and is nearly 100% lethal (40). Although fewer than 20 cases of plague are reported each year in the United States, approximately 3,000 cases of this infectious disease are reported annually worldwide (6). Due to recurrent outbreaks of plague in India (26) and East Africa, the isolation of a multidrug-resistant Y. pestis strain (12), and the potential of Y. pestis to be used as a bioterrorism agent (20), plague is now considered a reemerging infectious disease. Either a live attenuated strain or a heat-killed virulent strain has been used in the past as a moderately effective vaccine; however, both strains require repeated booster vaccinations to maintain efficacy and are not capable of preventing pneumonic plague (29). Although a phase II clinical trial is under way, a commercially available vaccine to treat Y. pestis infection is not currently available in the United States. This leaves postexposure antibiotic therapy as the only viable option for treatment.
Y. enterocolitica and Y. pseudotuberculosis are two other species of Yersinia, in addition to Y. pestis, that are pathogenic for humans. A significant body of research has focused on virulence factors of these three pathogenic species, all of which possess a conserved 70-kb virulence plasmid (named pCD1 in Y. pestis and pYV in Y. enterocolitica and Y. pseudotuberculosis). This plasmid encodes virulence factors, which include Yersinia outer membrane proteins that are delivered into host cells via a type III secretion system and act to disengage natural host defenses. Many of the Yersinia outer membrane proteins are absolutely essential for virulence in mouse models (4, 42). Y. pestis contains two additional plasmids necessary for virulence, pPCP1 and pMT1 (5).
In contrast to the well-studied virulence factors of Y. pestis, the overall host response to this pathogen is poorly understood. Moreover, the majority of previous studies of Yersinia host defense have focused on Y. enterocolitica as a model organism for Yersinia in general. However, Y. enterocolitica is a poor model for plague as Y. pestis is not an enteric pathogen and causes more acute disease. It has been shown that clearance of Y. enterocolitica infection requires increased production of the proinflammatory cytokines gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), IL-12, and IL-18 (1, 2, 19, 21). However, the precise role of these pleiotropic cytokines during a Y. pestis infection is not well characterized. One study has shown that in vivo administration of TNF-α and IFN-γ can prevent death after exposure to high doses of Y. pestis (33).
An alternative approach to study the host response to a pathogen is to use inbred mouse strains that vary in their resistance to the infection. Through quantitative trait locus (QTL) mapping, specific genes that are responsible for resistance and susceptibility can be identified. This approach has been used for infectious diseases such as tuberculosis (31, 36), leishmaniasis (35), and malaria (7-9). With respect to Yersinia, early work with Y. enterocolitica has shown that C57BL/6 mice are resistant to an intravenous challenge while BALB/c mice are susceptible (16, 17). Through the examination of nine hybrid inbred mouse strains, this phenotypic difference was tentatively assigned to the Es-1 locus on chromosome 8 (16) although no QTL mapping was performed. A subsequent study, however, found no difference in resistance between C57BL/6 and BALB/c mice to Y. enterocolitica infection by the oral route (18). While strain differences in resistance to Y. pestis have not been extensively examined, Congleton et al. (3) have recently identified a 129 substrain as the first inbred mouse line that is resistant to a pgm mutant Y. pestis strain.
In an effort to directly study host genetic factors that are important in resistance to Y. pestis infection, we infected several inbred mouse lines with the pgm mutant strain KIM5 and found that BALB/cJ mice are highly resistant compared to other strains including other BALB/c substrains. Significant differences in bacterial burdens occur within 1 to 2 days of infection and are accompanied by visual differences in immunopathology of several organs. In order to determine the gene(s) responsible for resistance, we performed an F2 screen and found a single locus on chromosome 17 that is associated with resistance to plague. After backcrossing the locus onto the C57BL/6J background, we have confirmed that this region confers most of the resistance of BALB/cJ mice to Y. pestis.
MATERIALS AND METHODS
Mice.
C57BL/6J, BALB/cJ, BALB/cByJ, and CB6F1/J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). BALB/cAnNHsd mice were purchased from Harlan Sprague Dawley, Inc. (Indianapolis, IN). F2 (CB6F1/J × CB6F1/J) and backcrossed (C57BL/6J × CB6F1/J) mice were bred in the Animal Research Facility at the University of Illinois at Urbana-Champaign. All experiments were conducted with female mice and approved by the University of Illinois at Urbana-Champaign Institutional Animal Care and Use Committee.
Bacteria.
The bacterial strain used in this study is the Y. pestis pgm mutant strain KIM5 (41). Bacteria were grown overnight in heart infusion (HI) broth at 23°C. After overnight growth, the bacteria were diluted 1:10 in HI broth and grown to mid-log phase (optical density at 600 nm of ∼0.4). Bacteria were collected by centrifugation, washed twice, and serially diluted in phosphate-buffered saline (PBS) to the appropriate concentration. Mice were intravenously infected via caudal tail vein with a 200-μl inoculum. Doses were verified by plating the inoculum onto HI agar plates. For survival experiments, animals were monitored at least twice daily and culled in a timely fashion upon reaching a moribund state. The Y. enterocolitica strain WA was grown as stated above.
Enumeration of bacterial titer.
Infected mice were sacrificed at the indicated time point by CO2 asphyxiation. Spleen and/or liver were removed aseptically and weighed (where appropriate). Organs were homogenized and serially diluted in PBS. Bacterial burden was determined by plating the dilutions on HI agar plates and incubating them at 23°C for 2 to 3 days.
Cytometric bead assay.
Preinfection serum was obtained from all mice 1 week prior to infection with Y. pestis via tail bleeding. At 24 h postinfection with 105 CFU bacteria, new serum was obtained. Levels of IL-1β, TNF-α, IL-6, IFN-γ, IL-12 (p70), and IL-10 were measured using a Bio-Plex cytokine bead array (Bio-Rad). The limit of detection for this assay was 20 pg/ml.
Histopathology.
Mice were infected with 1 × 104 CFU of Y. pestis, along with control animals injected with an equal volume of PBS. At the 24- and 48-h time points, infected mice were sacrificed along with the control animals at 24 h. The spleen, liver, and lungs were removed from each animal and fixed in 10% buffered formalin for 24 h. Organs were embedded in paraffin wax, sectioned for slides, stained with hematoxylin and eosin, and examined.
Genetic screening and statistical analysis of F2 and backcrossed mice.
Prior to infection, ear biopsies from the mice were obtained, and DNA was extracted. For the F2 screen, 58 polymorphic microsatellite markers covering all 20 chromosomes were selected (http://www.jax.org). The gaps in coverage ranged from 15 to 30 centimorgans (cM). Genotyping was performed through standard PCR and run on 3% agarose gels. QTL mapping was performed with Map Manager QTX (27) (http://www.mapmanager.org) using an additive analysis. Genome-wide significance was determined through the permutation test of the Map Manager QTX software. A total of 10,000 permutations at 1-cM intervals determined that a likelihood ratio statistic (LRS) score of 11.2 or higher was necessary to reach a P of <0.05.
An additional screen of chromosome 17 was performed with 95 F1 backcrossed (CB6F1/J × C57BL/6J) mice using 48-h total splenic CFU as the phenotype. The first 45 cM of chromosome 17 was genotyped using seven individual microsatellite markers (D17Mit164, D17Mit133, D17Mit198, D17Mit24, D17Mit66, D17Mit139, and D17Mit39). Interval mapping of both the F2 and F1 backcross screens was performed with QTL Cartographer (http://statgen.ncsu.edu/qtlcart). The data were analyzed in 1-cM steps and presented as logarithmic odds (LOD) scores.
RESULTS
BALB/cJ mice are resistant to Y. pestis.
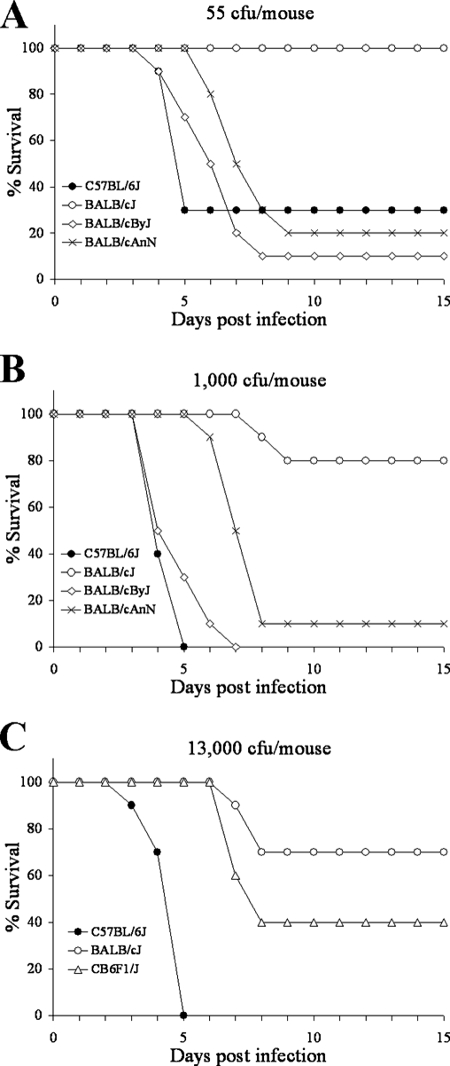
We surveyed various mouse strains for resistance to Y. pestis and found that BALB/cJ mice are highly resistant to this pathogen. It has previously been shown that both BALB/cByJ (38) and BALB/cAnNHsd (24) mice are susceptible to Y. pestis KIM5, with a 50% lethal dose (LD50) near 40 CFU. In order to verify that resistance is unique to the BALB/cJ line, we tested these three substrains, along with susceptible C57BL/6J controls, for resistance to Y. pestis and found that only the BALB/cJ mice are resistant to this pathogen. This is illustrated in Fig. 1A and B, where groups of 10 C57BL/6J, BALB/cJ, BALB/cByJ, and BALB/cAnNHsd mice were infected with 55 or 1,000 CFU and monitored for survival for 15 days. At 55 CFU, less than half of the C57BL/6J, BALB/cByJ, and BALB/cAnNHsd mice survived, verifying that the LD50 of these mice is near 40 to 50 CFU of bacteria. In contrast, all 10 BALB/cJ mice survived this dose. At 1,000 CFU, all C57BL/6J and BALB/cByJ mice and 9 of 10 BALB/cAnNHsd mice succumbed to infection. In contrast, 8 of 10 BALB/cJ animals survived this dose. Interestingly, there appears to be an approximate 3-day delay in death in the BALB/cAnNHsd mice compared to the other susceptible groups. We next wanted to determine the resistance of BALB/cJ mice to a higher infectious dose. Therefore, groups of 10 C57BL/6J and BALB/cJ mice were infected with 13,000 CFU (Fig. 1C). All 10 C57BL/6J but only 3 of 10 BALB/cJ mice succumbed to infection by days 5 and 8, respectively, with no additional deaths taking place thereafter. Based on this and other data, the LD50 for BALB/cJ mice is at least 250-fold higher than the LD50 for the susceptible strains. These results show that, in contrast to C57BL/6J mice and two other BALB/c substrains, BALB/cJ mice are highly resistant to Y. pestis infection.
FIG. 1.
BALB/cJ mice, unlike other BALB/c substrains, are uniquely resistant to Y. pestis. (A and B) Groups of 10 C57BL/6J, BALB/cJ, BALB/cByJ, and BALB/cAnNHsd mice were infected with 55 (A) or 1,000 (B) CFU of Y. pestis KIM5 and monitored for survival for 15 days. (C) Groups of 10 C57BL/6J, BALB/cJ, and CB6F1/J mice were infected with 13,000 CFU of Y. pestis and monitored for survival for 15 days.
We also used this experiment to examine the phenotype of CB6F1/J (BALB/cJ × C57BL/6J) mice. These mice displayed intermediate resistance to Y. pestis with respect to the two parental strains (Fig. 1C). Thus, CB6F1/J mice appear to maintain a relatively high level of resistance compared to the parental C57BL/6J strain. These results indicate that the resistance phenotype of BALB/cJ mice is not recessive.
BALBc/J mice exhibit lower bacterial burden early after infection.
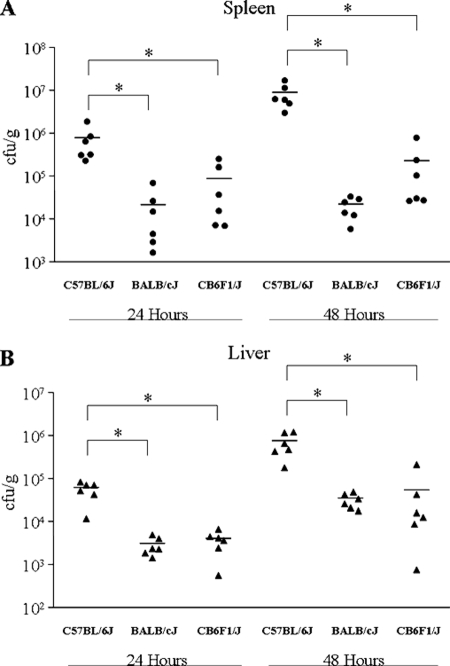
We next examined the early stages of infection by comparing bacterial burden in the spleen and liver of these three strains. Groups of 12 C57BL/6J, BALB/cJ, and CB6F1/J mice were infected with ∼2,000 CFU of Y. pestis, and six mice from each group were sacrificed at 24 and 48 h after infection. The bacterial loads in the spleen and liver of the sacrificed mice were determined by plating serial dilutions of homogenized organs onto HI agar plates. At 24 h postinfection, the spleen of C57BL/6J mice displayed 35- and 9-fold more bacteria than the BALB/cJ and CB6F1/J groups, respectively (Fig. 2A). At 48 h postinfection, the spleen of C57BL/6J mice displayed 410- and 40-fold more bacteria than the BALB/cJ and CB6F1/J groups, respectively. Although less dramatic, these trends of lower bacterial burden in the BALB/cJ and CB6F1/J mice were also observed in the liver (Fig. 2B). For example, at 24 h postinfection, the C57BL/6J mice displayed 19- and 15-fold more bacteria in the liver than the BALB/cJ and CB6F1/J groups, respectively. At 48 h postinfection, the difference remained, with C57BL/6J mice exhibiting 22- and 14-fold higher bacterial titers in the liver than the BALB/cJ and CB6F1/J groups, respectively. Thus, early bacterial burdens closely mirror the survival phenotypes observed, with susceptible C57BL/6J mice exhibiting higher burdens than resistant BALBc/J mice. Overall, the CB6F1/J mice appear to display an intermediate phenotype with respect to the parental strains, although the bacterial burden was not significantly different than the BALB/cJ titers. Since the reduced bacterial load is maintained in CB6F1/J mice, we consider the resistance trait to be nonrecessive.
FIG. 2.
BALB/cJ mice display lower bacterial burden early after infection than the susceptible C57BL/6J strain. Groups of 12 C57BL/6J, BALB/cJ, and CB6F1/J mice were infected with 2,000 CFU of Y. pestis KIM5. At 24 and 48 h postinfection, six mice from each group were sacrificed, and bacterial titers in the spleen (A) and liver (B) of the animals were determined. Horizontal lines represent the mean for each group. Asterisks indicate significant differences between groups as determined by a Student's t test where P is <0.05.
In the C57BL/6J mice, bacterial growth continued in both the spleen and liver from day 1 to day 2 postinfection with 11- and 13-fold increases, respectively. In the BALB/cJ and CB6F1/J mice bacterial growth between days 1 and 2 was almost completely blunted in the spleen but continued in the liver with 11-fold and 13-fold increases, respectively. The overall phenotype of these mice indicates that resistance to Y. pestis in the BALB/cJ line is mediated by early innate immune defenses that control bacterial replication.
BALB/cJ and C57BL/6J mice exhibit differences in immunopathology and cytokine production following Y. pestis infection.
To determine if there were early differences in the disease pathology of the two mouse strains, we chose to examine histological sections of several organs taken from infected animals. Groups of five mice were infected with Y. pestis or injected with PBS as a control and sacrificed 1 or 2 days after infection (Table 1). All infected mice developed suppurative splenitis with multifocal abscesses. Large concentrations of necrotic neutrophils were observed in these abscesses. However, C57BL/6J mice exhibited a higher degree of lymphoid atrophy with a greater loss of defined splenic architecture than BALB/cJ animals. Additionally, the development of vascular fibrinous thrombosis was evident only in the spleens of the C57BL/6J mice, and all animals developed this condition within 2 days.
TABLE 1.
Frequency of observed pathology of Y. pestis-infected mice
| Pathology | Frequency (no. of mice positive/no. of mice scored) in mice sacrificed at:a
|
|||
|---|---|---|---|---|
| C57BL/6J
|
BALB/cJ
|
|||
| 24 h | 48 h | 24 h | 48 h | |
| Suppurative splenitis | 5/5 | 5/5 | 5/5 | 5/5 |
| With multifocal abscesses | 5/5 | 5/5 | 4/5 | 5/5 |
| With vascular fibrinous thrombosis | 1/5 | 5/5 | 0/5 | 0/5 |
| With lymphoid atrophy or apoptosis | 0/5 | 5/5 | 1/5 | 2/5 |
| Multifocal suppurative hepatitis | 5/5 | 5/5 | 4/5 | 5/5 |
| Multifocal suppurative hepatitis with vascular thrombosis | 0/5 | 2/5 | 0/5 | 0/5 |
| Fibrinous effusion in lungs | 0/5 | 5/5 | 0/5 | 0/5 |
| Fibrinous effusion in lungs with vascular thrombosis | 0/5 | 1/5 | 0/5 | 0/5 |
Five mice of a given strain were infected and sacrificed at the times indicated. At least two tissue sections were examined for each of the spleen, liver, and lungs.
All infected mice developed multifocal suppurative hepatitis of various degrees of severity. Vascular thrombosis was seen in the livers of two of five C57BL/6J mice, causing coagulation necrosis of the surrounding hepatocytes, but was not seen in any of the BALB/cJ animals. Another difference in the pathology between the mouse strains was seen in the lungs. While none of the BALB/cJ mice developed fibrinous effusion, all five C57BL/6J animals developed this condition, including one mouse with vascular thrombosis.
Priming of mice with IFN-γ and TNF-α has been shown to protect against Y. pestis (33). This prompted us to compare serum cytokine levels in our mice 24 h after infection with Y. pestis (Table 2). Of the cytokines measured, only IFN-γ and IL-6 were elevated as a result of Y. pestis infection. Interestingly, the induced levels of serum IL-6 in the C57BL6/J mice were approximately threefold higher than those of the BALBc/J animals (Student's t test, P < 0.001). Of the cytokines measured, this was the only significant difference observed between the two strains.
TABLE 2.
Serum cytokine levels of uninfected and infected mice determined by cytometric bead array
| Cytokine | Mean concn (pg/ml ± SD) in:a
|
|||
|---|---|---|---|---|
| C57BL/6J mice
|
BALB/cJ mice
|
|||
| Uninfected | Infected | Uninfected | Infected | |
| IL-1β | 73 ± 28 | 44 ± 25 | 54 ± 19 | 42 ± 41 |
| TNF-α | 57 ± 7 | 58 ± 15 | 62 ± 19 | 65 ± 24 |
| IL-6 | ND | 1252 ± 461 | ND | 473 ± 386 |
| IFN-γ | ND | 403 ± 198 | ND | 289 ± 84 |
| IL-12 (p70) | ND | 36 ± 19 | ND | 27 ± 12 |
| IL-10 | 23 ± 15 | 34 ± 25 | ND | 27 ± 32 |
Mean cytokine concentrations were determined for 10 mice per group using a cytometric bead assay. ND, not detected. The limit of detection for any given cytokine was 20 pg/ml.
A genetic screen reveals a single locus at the H-2 region on chromosome 17 associated with resistance to Y. pestis.
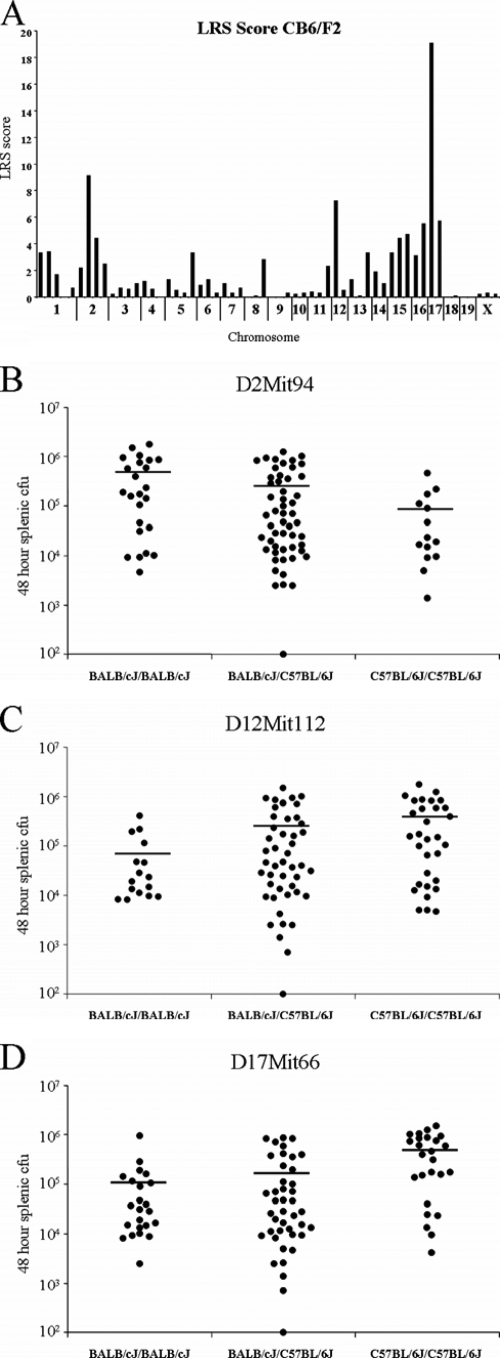
In order to locate the gene(s) responsible for BALB/cJ resistance to Y. pestis, 95 F2 mice were infected with 3,500 bacteria, and total splenic CFU was determined 48 h postinfection. QTL mapping was performed using 58 microsatellite markers spaced throughout the genome at 15- to 30-cM intervals using Map Manager QTX. The screen revealed three major peaks at chromosomes 2, 12, and 17, each of which represents potential resistance loci (Fig. 3A). However, genome-wide significance, as determined by a 10,000 cycle permutation test at 1-cM intervals, revealed a threshold LRS score of 11.2 at the 95% confidence interval. Thus, of the three loci, only the one located on chromosome 17, with an LRS score of 19.1, is significant on a genome-wide scale. We have designated this locus prl1 (plague resistance locus 1).
FIG. 3.
Linkage analysis and interval mapping reveals a single significant QTL on chromosome 17. (A) Ninety-five F2 (CB6/F1J × CB6/F1J) mice were infected with 3,500 CFU of Y. pestis KIM5, and 48 h postinfection total splenic CFU was determined. LRS scores were generated using Map Manager QTX at 58 microsatellite markers across the genome. Based on the Map Manager QTX permutation test (1-cM intervals), the threshold for significance (P < 0.05) is an LRS score greater than 11.2. The CFU counts of all 95 mice based on their genotypes at the two suggestive loci on chromosomes 2 (B) and 12 (C), as well as a single significant locus on chromosome 17 (D), are shown.
In order to assess the heritability of the resistance trait, splenic bacterial burden of the individual mice with respect to the genotype was assessed at each of the loci. The suggestive locus on chromosome 2, D2Mit94, accounts for 9% of the variance according to the analysis; however, the empirical P value is 0.154 according to the permutation test (Fig. 3B). In fact, the trend at this locus indicates that the C57BL/6J allele contributes to resistance. The other suggestive locus, D12Mit112 (Fig. 3C), accounts for 7% of the variance with an empirical P value of 0.373.
The significant locus on chromosome 17 at D17Mit66 (Fig. 3D) accounts for 18% of the variance and has an empirical P value of 0.003, according to the permutation test. Mice homozygous for the BALB/cJ allele, as well as the heterozygous animals, tend to cluster between 104 and 105 CFU, while the mice homozygous for the C57BL/6J allele tend to cluster near 106 CFU. While this is reflective of the F1 results shown in Fig. 2, the discrimination between the three genotypes at D17Mit66 is not as clear as that observed for the parent strains and F1 animals. These results indicate that, while the chromosome 17 locus of BALB/cJ mice makes a major, significant contribution to the phenotype, other minor genes may also play a role in resistance to Y. pestis.
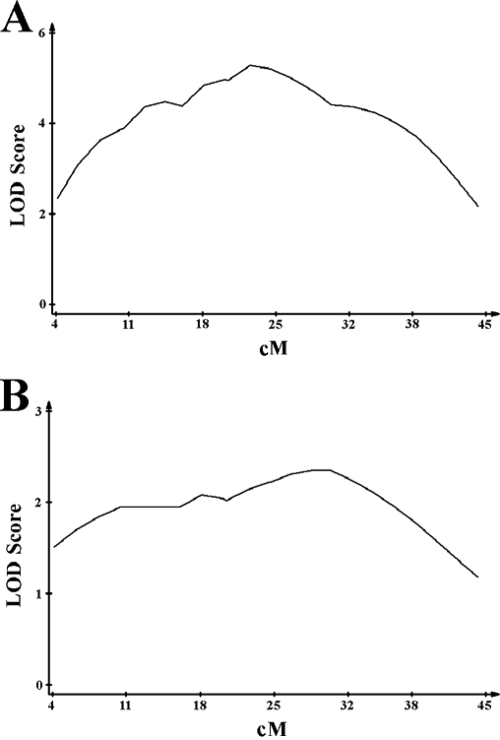
We next analyzed prl1 more closely with seven microsatellite markers on chromosome 17 ranging from 4 to 45 cM. Using QTL Cartographer, we performed interval mapping, in 1-cM intervals, over the first half of the chromosome (Fig. 4A). A clearly defined peak LOD score was observed at approximately 22 cM.
FIG. 4.
Interval mapping at prl1 in F2 and F1 backcrossed mice. The results of the F2 (A) and an F1 backcross (C57BL/6J × CB6/F1J) (B) were subjected to interval mapping using QTL Cartographer across the first 45 cM of chromosome 17 in 1-cM intervals. The estimated peak LOD scores are located at 22 cM for the F2 mice and at 24 and 30 cM for the F1 backcross.
In order to confirm this finding, we performed a second genetic screen on chromosome 17. Since the BALB/cJ resistance locus is at least semidominant, we crossed F1 mice with the susceptible C57BL/6J strain. Examination of these 95 backcrossed mice revealed peak LOD scores at 24 and 30 cM (Fig. 4B). The apparent shift in the peak for this second genetic screen is due to the fact that none of the 95 mice had a crossover between 24 and 30 cM, causing the interval mapping line to skew toward one end. Thus, the LOD peaks at 24 and 30 cM are, in fact, indistinguishable and confirm the peak identified in the original F2 screen. Taken together, these results show that this region of chromosome 17 constitutes a significant Y. pestis resistance locus.
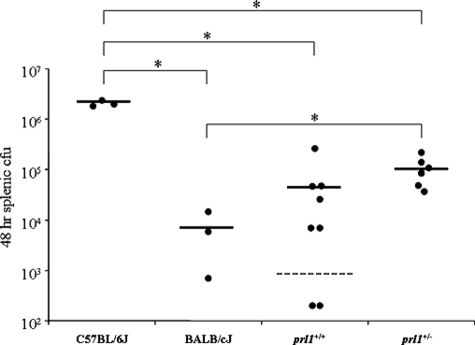
In order to assist with fine-mapping of prl1 and prove that this locus is responsible for the majority of resistance to Y. pestis, we created a more congenic line by backcrossing the first 45 cM of chromosome 17 from BALB/cJ mice to the C57BL/6J background. At generation 8, the mice were intercrossed to generate prl1+/+ and prl1+/− mice. Outside of chromosome 17, these mice theoretically contain less than 0.5% of BALB/cJ DNA. As seen in Fig. 5, mice with either one or two copies of prl1 on the susceptible C57BL/6J background are resistant to Y. pestis and exhibit significantly lower bacterial titers than the C57BL/6J controls. In fact, the bacterial titer of prl1+/+ was not significantly different than that observed in BALB/cJ mice. This was confirmed by an experiment in which three of five prl1+/− mice survived a challenge of 1,000 CFU of Y. pestis compared to zero of five C57BL/6J animals (data not shown). These results clearly indicate that prl1 acts in a semidominant fashion and is responsible for the majority of the resistance observed in BALB/cJ mice.
FIG. 5.
prl1 confers resistance to Y. pestis in a susceptible C57BL/6J background. Eighth generation backcrossed mice were intercrossed, generating prl1+/+ (8 mice) and prl1+/− (6 mice) animals. Mice were infected with 2,000 CFU Y. pestis along with parental controls (3 each). At 48 h postinfection, mice were sacrificed, and total splenic CFU was determined. Horizontal lines represent the average. The dashed line indicates the limit of detection. Asterisks indicate significant differences between groups as determined by a Student's t test where P is <0.05.
DISCUSSION
Y. pestis is a recently evolved, fast-acting, highly lethal pathogen that has been the cause of several deadly plague pandemics over the last 2 millennia. Although the type III secretion system and associated bacterial effector proteins of pathogenic Yersinia species have been extensively studied as virulence factors, comparatively little research has examined the host factors responsible for conferring resistance and/or susceptibility. Upon infection of several inbred mouse lines with Y. pestis, we found that BALB/cJ mice are resistant to this pathogen. We began work on this strain with a phenotypic analysis of infection, followed by a genetic screen to identify QTLs that are associated with resistance.
We along with others have observed that C57BL/6J, BALB/cByJ, and BALB/cAnNHsd mice are all susceptible to Y. pestis KIM5, with an estimated LD50 of less than 50 CFU (24, 38). Unlike the other BALB/c substrains, BALB/cJ mice uniformly survive at this dose. Even at 13,000 CFU, 70% of BALB/cJ mice survive, suggesting that the LD50 is at least 250-fold higher than the estimated LD50 of susceptible mouse lines including C57BL/6J and other BALB/c substrains. The difference in resistance to Y. pestis that we have observed is likely due to the genetic divergence of the BALB/cJ substrain from the other two BALB/c lines since their separation over 70 years ago (23). In fact, phenotypic differences between BALB/c substrains similar to those observed in this study have been well documented for other diseases including Graves’ hyperthyroidism (37), experimental allergic encephalomyelitis (39), and Taenia crassiceps cysticercosis (10).
Early studies have revealed that a variety of mouse strains, including BALB/cBy, are susceptible to an intravenous infection of Y. enterocolitica, with reported LD50 values ranging from 200 to 600 CFU (17). Thus, we were interested in ascertaining the generality of our results to another Yersinia species. To this end, we infected 10 BALB/cJ mice intravenously with 2,000 CFU of Y. enterocolitica WA and, in three independent experiments, observed that all of the mice succumbed within 9 days (data not shown). These results are in sharp contrast to what we have reported here for Y. pestis. Thus, it is likely that despite sharing many of the same virulence traits, host defenses that are effective for resistance to Y. enterocolitica and Y. pestis differ. Indeed, Y. enterocolitica is naturally acquired orally, and a recent study found that mouse strain-specific differences in host susceptibility can be greatly affected by the route of infection (18).
An analysis of serum cytokine levels revealed that, among those examined, only IFN-γ and IL-6 increased appreciably 24 h postinfection and that only the levels of IL-6 were significantly different between the two mouse strains (Table 2). It is highly possible that this difference is due to greater numbers of bacteria in the C57BL/6J animals. The general lack of induction of inflammatory cytokines by Y. pestis probably reflects the fact that at 37°C these gram-negative bacteria shift the synthesis of their lipopolysaccharide to a tetra-acylated form that is no longer capable of stimulating Toll-like receptor 4 (32). This appears to be an important virulence mechanism as a genetically engineered Y. pestis strain that synthesizes the hexa-acylated stimulatory form of lipopolysaccharide is no longer virulent in mice. Additionally, earlier studies have shown that priming with both TNF-α and IFN-γ prior to Y. pestis infection fully protects mice from death (33). Our results indicate that differential induction of TNF-α and/or IFN-γ is not responsible for the increased resistance of BALB/cJ mice to Y. pestis.
Within 24 h postinfection significantly higher bacterial burdens in the spleen and liver of C57BL/6J mice were already evident and became even more pronounced at 48 h. These results indicate that enhanced resistance of BALB/cJ mice to Y. pestis is likely due to innate immune defenses that provide protection early in the infection. A recent study which tracked a green fluorescent protein-labeled strain of Y. pestis in infected mice suggests that the bacteria reside within splenic macrophages and are controlled extracellularly by neutrophil activity (25). Both strains of mice in our study displayed suppurative splenitis and hepatitis early in infection, consistent with the pathologies observed by others (22). However, of the two strains, only the susceptible C57BL/6J mice exhibited pronounced vascular thrombosis and fibrinous effusion in the lungs, which is likely associated with bacterial escape and ultimately contributes to the death of the animal.
The resistance of F1 mice to high-dose Y. pestis infection showed that BALB/cJ resistance is not recessive. This provided the basis for an F2 genome scan which revealed a single significant resistance locus, designated prl1, that maps to the major histocompatibility complex (MHC) region on chromosome 17. Although this region is usually associated with antigen-presenting H-2 genes of adaptive immunity, it also contains many genes associated with innate immune defense, including complement components, certain TNF family members, and stress-related proteins (15). Resistance of mice to a variety of pathogens including Plasmodium berghei (14), T. crassiceps (10), Chlamydia pneumoniae (30), and Streptococcus pyogenes (13) has been mapped to the MHC region. Similar to what we have found for Y. pestis, an early innate immune defense appears to be responsible for the observed resistance to S. pyogenes.
Although very little is known about genetic differences between BALB/c substrains, a dramatic difference is known to occur within the MHC region at Qa-2, a locus that encodes “nonclassical” class I antigens. In this regard, unequal crossing over of Q8 and Q9 genes has occurred in BALB/cJ mice, leading to a Qa-2low phenotype (28). Moreover, the BALB/cBy substrain is Qa-2null due to an additional deletion of genomic DNA that has occurred between the Q6 and Q7 genes (28, 34). Using BALB/c substrains and F1 backcrosses, one group of investigators has correlated Qa-2 expression with resistance to T. crassiceps (11). Whether these genetic differences in the Qa-2 region play a role in the resistance of BALB/cJ mice to Y. pestis is presently unknown.
We have begun fine-mapping of prl1 to identify the candidate gene(s) responsible for resistance to Y. pestis. The identification of such a host gene should allow for a better understanding of natural clearance of plague in both rodents and humans.
Acknowledgments
We thank the staff of the Animal Core Facility at the University of Illinois for their excellent assistance in animal husbandry.
This work was supported by NIH grants AI056148 and AI057153 (to J.L.X.) and AI056148 (to R.I.T.).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 23 June 2008.
REFERENCES
- 1.Autenrieth, I. B., and J. Heesemann. 1992. In vivo neutralization of tumor necrosis factor-alpha and interferon-gamma abrogates resistance to Yersinia enterocolitica infection in mice. Med. Microbiol. Immunol. 181333-338. [DOI] [PubMed] [Google Scholar]
- 2.Bohn, E., and I. B. Autenrieth. 1996. IL-12 is essential for resistance against Yersinia enterocolitica by triggering IFN-gamma production in NK cells and CD4+ T cells. J. Immunol. 1561458-1468. [PubMed] [Google Scholar]
- 3.Congleton, Y. H., C. R. Wulff, E. J. Kerschen, and S. C. Straley. 2006. Mice naturally resistant to Yersinia pestis Δpgm strains commonly used in pathogenicity studies. Infect. Immun. 746501-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelis, G. R. 2002. The Yersinia Ysc-Yop “type III” weaponry. Nat. Rev. Mol. Cell Biol. 3742-752. [DOI] [PubMed] [Google Scholar]
- 5.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 621315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis, D. T., K. L. Gage, N. Gratz, J. D. Poland, and E. Tikhomirov. 1999. Plague manual: epidemiology, distribution, surveillance and control. World Health Organization, Geneva, Switzerland.
- 7.Foote, S. J., R. A. Burt, T. M. Baldwin, A. Presente, A. W. Roberts, Y. L. Laural, A. M. Lew, and V. M. Marshall. 1997. Mouse loci for malaria-induced mortality and the control of parasitaemia. Nat. Genet. 17380-381. [DOI] [PubMed] [Google Scholar]
- 8.Fortin, A., A. Belouchi, M. F. Tam, L. Cardon, E. Skamene, M. M. Stevenson, and P. Gros. 1997. Genetic control of blood parasitaemia in mouse malaria maps to chromosome 8. Nat. Genet. 17382-383. [DOI] [PubMed] [Google Scholar]
- 9.Fortin, A., L. R. Cardon, M. Tam, E. Skamene, M. M. Stevenson, and P. Gros. 2001. Identification of a new malaria susceptibility locus (Char4) in recombinant congenic strains of mice. Proc. Natl. Acad. Sci. USA 9810793-10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fragoso, G., E. Lamoyi, A. Mellor, C. Lomeli, T. Govezensky, and E. Sciutto. 1996. Genetic control of susceptibility to Taenia crassiceps cysticercosis. Parasitology 112119-124. [DOI] [PubMed] [Google Scholar]
- 11.Fragoso, G., E. Lamoyi, A. Mellor, C. Lomeli, M. Hernandez, and E. Sciutto. 1998. Increased resistance to Taenia crassiceps murine cysticercosis in Qa-2 transgenic mice. Infect. Immun. 66760-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galimand, M., A. Guiyoule, G. Gerbaud, B. Rasoamanana, S. Chanteau, E. Carniel, and P. Courvalin. 1997. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N. Engl. J. Med. 337677-680. [DOI] [PubMed] [Google Scholar]
- 13.Goldmann, O., A. Lengeling, J. Bose, H. Bloecker, R. Geffers, G. S. Chhatwal, and E. Medina. 2005. The role of the MHC on resistance to group a streptococci in mice. J. Immunol. 1753862-3872. [DOI] [PubMed] [Google Scholar]
- 14.Goncalves, L. A., P. Almeida, M. M. Mota, and C. Penha-Goncalves. 2008. Malaria liver stage susceptibility locus identified on mouse chromosome 17 by congenic mapping. PLoS ONE 3e1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruen, J. R., and S. M. Weissman. 1997. Evolving views of the major histocompatibility complex. Blood 904252-4265. [PubMed] [Google Scholar]
- 16.Hancock, G. E., R. W. Schaedler, and T. T. MacDonald. 1988. Multigenic control of resistance to Yersinia enterocolitica in inbred strains of mice. Infect. Immun. 56532-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock, G. E., R. W. Schaedler, and T. T. MacDonald. 1986. Yersinia enterocolitica infection in resistant and susceptible strains of mice. Infect. Immun. 5326-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handley, S. A., P. H. Dube, P. A. Revell, and V. L. Miller. 2004. Characterization of oral Yersinia enterocolitica infection in three different strains of inbred mice. Infect. Immun. 721645-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hein, J., A. Sing, M. S. Di Genaro, and I. B. Autenrieth. 2001. Interleukin-12 and interleukin-18 are indispensable for protective immunity against enteropathogenic Yersinia. Microb. Pathog. 31195-199. [DOI] [PubMed] [Google Scholar]
- 20.Inglesby, T. V., D. T. Dennis, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, J. F. Koerner, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, M. Schoch-Spana, K. Tonat, et al. 2000. Plague as a biological weapon: medical and public health management. JAMA 2832281-2290. [DOI] [PubMed] [Google Scholar]
- 21.Jordan, M., I. G. Otterness, R. Ng, A. Gessner, M. Rollinghoff, and H. U. Beuscher. 1995. Neutralization of endogenous IL-6 suppresses induction of IL-1 receptor antagonist. J. Immunol. 1544081-4090. [PubMed] [Google Scholar]
- 22.Kerschen, E. J., D. A. Cohen, A. M. Kaplan, and S. C. Straley. 2004. The plague virulence protein YopM targets the innate immune response by causing a global depletion of NK cells. Infect. Immun. 724589-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Les, E. P. 1990. A brief history of the two substrains of BALB/c, BALBcJ, and BALBcByJ available from animal resources. JAX Notes no. 443. The Jackson Laboratory. http://jaxmice.jax.org/jaxnotes/archive/443a.html.
- 24.Leung, K. Y., B. S. Reisner, and S. C. Straley. 1990. YopM inhibits platelet aggregation and is necessary for virulence of Yersinia pestis in mice. Infect. Immun. 583262-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukaszewski, R. A., D. J. Kenny, R. Taylor, D. G. Rees, M. G. Hartley, and P. C. Oyston. 2005. Pathogenesis of Yersinia pestis infection in BALB/c mice: effects on host macrophages and neutrophils. Infect. Immun. 737142-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahesh, S., J. Shukla, U. Tuteja, and H. V. Batra. 2005. Molecular detection of Yersinia pestis isolates of Indian origin by using Pla specific monoclonal antibodies. Comp. Immunol. Microbiol. Infect. Dis. 28131-144. [DOI] [PubMed] [Google Scholar]
- 27.Manly, K. F., R. H. Cudmore, Jr., and J. M. Meer. 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12930-932. [DOI] [PubMed] [Google Scholar]
- 28.Mellor, A. L., J. Antoniou, and P. J. Robinson. 1985. Structure and expression of genes encoding murine Qa-2 class I antigens. Proc. Natl. Acad. Sci. USA 825920-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, K. F. 1970. Effectiveness of live or killed plague vaccines in man. Bull. W. H. O. 42653-666. [PMC free article] [PubMed] [Google Scholar]
- 30.Min-Oo, G., L. Lindqvist, A. Vaglenov, C. Wang, P. Fortin, Y. Li, B. Kaltenboeck, and P. Gros. 2008. Genetic control of susceptibility to pulmonary infection with Chlamydia pneumoniae in the mouse. Genes Immun. 9383-388. [DOI] [PubMed] [Google Scholar]
- 31.Mitsos, L. M., L. R. Cardon, A. Fortin, L. Ryan, R. LaCourse, R. J. North, and P. Gros. 2000. Genetic control of susceptibility to infection with Mycobacterium tuberculosis in mice. Genes Immun. 1467-477. [DOI] [PubMed] [Google Scholar]
- 32.Montminy, S. W., N. Khan, S. McGrath, M. J. Walkowicz, F. Sharp, J. E. Conlon, K. Fukase, S. Kusumoto, C. Sweet, K. Miyake, S. Akira, R. J. Cotter, J. D. Goguen, and E. Lien. 2006. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat. Immunol. 71066-1073. [DOI] [PubMed] [Google Scholar]
- 33.Nakajima, R., and R. R. Brubaker. 1993. Association between virulence of Yersinia pestis and suppression of gamma interferon and tumor necrosis factor alpha. Infect. Immun. 6123-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama, K., S. Tokito, C. Pannetier, H. Nakauchi, and G. Gachelin. 1991. MHC gene Q8/9d of the BALB/cJ mouse strain cannot encode a Qa-2,3 class I antigen. Immunogenetics 33225-234. [DOI] [PubMed] [Google Scholar]
- 35.Roberts, L. J., T. M. Baldwin, J. M. Curtis, E. Handman, and S. J. Foote. 1997. Resistance to Leishmania major is linked to the H2 region on chromosome 17 and to chromosome 9. J. Exp. Med. 1851705-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez, F., T. V. Radaeva, B. V. Nikonenko, A. S. Persson, S. Sengul, M. Schalling, E. Schurr, A. S. Apt, and C. Lavebratt. 2003. Multigenic control of disease severity after virulent Mycobacterium tuberculosis infection in mice. Infect. Immun. 71126-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seetharamaiah, G. S., and K. J. Land. 2006. Differential susceptibility of BALB/c and BALB/cBy mice to Graves’ hyperthyroidism. Thyroid 16651-658. [DOI] [PubMed] [Google Scholar]
- 38.Straley, S. C., and M. L. Cibull. 1989. Differential clearance and host-pathogen interactions of YopE− and YopK− YopL− Yersinia pestis in BALB/c mice. Infect. Immun. 571200-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teuscher, C., E. P. Blankenhorn, and W. F. Hickey. 1987. Differential susceptibility to actively induced experimental allergic encephalomyelitis and experimental allergic orchitis among BALB/c substrains. Cell Immunol. 110294-304. [DOI] [PubMed] [Google Scholar]
- 40.Titball, R. W., and E. D. Williamson. 2001. Vaccination against bubonic and pneumonic plague. Vaccine 194175-4184. [DOI] [PubMed] [Google Scholar]
- 41.Une, T., and R. R. Brubaker. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect. Immun. 43895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viboud, G. I., and J. B. Bliska. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 5969-89. [DOI] [PubMed] [Google Scholar]