Abstract
The genomes of several species of mycoplasma have been sequenced. Most of these species rely on the glycolytic pathway for energy production, with the one exception of Ureaplasma, a species that breaks down urea as its principle source of acquiring energy. Several species, including as Mycoplasma arthritidis, are nonglycolytic and can use arginine as their source of energy. Described here are the genome sequence and a transposon library of M. arthritidis. The genome of 820,453 bp is typical in size for a mycoplasma and contains two large families of genes that are predicted to code for phase-variable proteins. The transposon library was constructed using a minitransposon that inserts stably into the mycoplasma genome. Of the 635 predicted coding regions, 218 were disrupted in a library of 1,100 members. Dispensable genes included the gene coding for the MAM superantigen and genes coding for ribosomal proteins S15, S18, and L15.
Mycoplasma arthritidis is a natural pathogen of rats, causing severe polyarthritis. Arthritis in mice can be experimentally induced by injection of M. arthritidis into the tail vein. Disease in mice is more chronic than in rats, with lesions similar to those seen in human rheumatoid arthritis (25). Factors thought to contribute to the virulence of M. arthritidis are MAM, a potent superantigen secreted by the mycoplasma, and the M. arthritidis adhesins (MAAs) that have a role in cytadherence (32).
The genome sequences of several species of mycoplasma are known (see http://cbi.labri.fr/outils/molligen/). Other than Ureaplasma, which utilizes the hydrolysis of urea to generate ATP, all of the mycoplasma species that have sequenced genomes are glycolytic. Some species of mycoplasma such as M. arthritidis are nonglycolytic, and thus it was anticipated that the sequence of the M. arthritidis genome would reveal new aspects of mycoplasma physiology. Nonglycolytic mycoplasmas generally catabolize arginine as a major source of energy, and, indeed, the required genes for arginine catabolism were identified in the genome sequence of M. arthritidis.
Described here is the genome sequence of the virulent M. arthritidis strain 158L3-1 (3, 31). Strain 158L3-1 is a lysogen containing the 16-kb genome of the MAV1 bacteriophage. No additional prophages were noted in the genome. The sequence revealed features offering insight into the mechanisms by which the mycoplasma causes chronic inflammation, including two families of genes that are predicted to code for phase-variable proteins and a predicted gene product related to but distinct from MAM.
We also describe a transposon library of M. arthritidis strain 158, the nonlysogenic parent of 158L3-1. The library was created using minitransposons derived from Tn4001 that inserts into the genome at essentially random sites. The genomic location of the minitransposon was determined for 1,113 library members. Using criteria for gene inactivation previously developed for transposon mutagenesis of Mycoplasma pulmonis, 218 of the predicted protein coding regions, including three ribosomal protein genes, were inactivated.
MATERIALS AND METHODS
Genome sequencing.
M. arthritidis strain 158 is pathogenic in rats and mice and is the parent of the MAV1 lysogen strain 158L3-1 (3, 31). M. arthritidis was cultured in Edward broth or Edward agar as previously described (30). Genomic DNA was isolated by using an Easy-DNA kit (Invitrogen). Prior to cell lysis, washed cells were incubated with Protein Degrader according to manufacturer's directions. The nucleotide sequence of the genome (eight times coverage of the genome) was determined by a subcontractual arrangement with The Institute for Genomic Research (now known as the J. Craig Venter Institute). Although some species of mycoplasma have complex genetic elements that can complicate assembly and closure (2), the determination of the genome sequence of M. arthritidis was straightforward and was accomplished by using a random shotgun strategy.
Genome analysis.
Open reading frames (ORFs) and the potential start and stop sites of genes were identified using the NCBI GLIMMER 3 server located at http://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi (5). Gene products were annotated by analysis using a combination of BLAST and the Clusters of Orthologous Groups database at NCBI. The identification of orthologs in the genomes of Bacillus subtilis, M. pulmonis, and Mycoplasma genitalium was facilitated by additional BLAST analysis at http://genolist.pasteur.fr/SubtiList/, http://genolist.pasteur.fr/MypuList/, and http://cbi.labri.fr/outils/molligen/. Final annotations of each ORF to determine its likelihood as a coding region and its most likely 5′ start took into consideration potential start codons (ATG and GTG start codons were favored over TTG starts), alignments to orthologs, and potential Shine-Dalgarno sequences. Proteins with predicted signal peptides or transmembrane regions were detected by using the InterProScan Web service at http://www.ebi.ac.uk/Tools/webservices/services/interproscan. Potential promoters in genes containing upstream poly(T) or poly(A) tracts were identified using BPROM at http://www.softberry.com/berry.phtml. Amino acid sequence identity between protein pairs was calculated using the default setting of the SIM application at http://us.expasy.org/. Clustal W analysis of protein families was performed using MacVector (Accelerys), version 7.2.
Library construction.
Plasmid pTF85 carrying the minitransposon Tn4001TF1 was constructed by inserting the tetM gene into the BamHI site of pMT85, a plasmid containing a mini Tn4001 but without a tetracycline resistance determinant (36). The tetM gene was obtained by amplifying a 2.3-kb region of transposon Tn916 using the primer pair GTAATGGATCCCTCTCTTTGATAAAAAATTG and CTTATTGGATCCGAAACCATATTTATATAACAAC. Plasmid pTF20 carrying the minitransposon Tn4001TF2 was constructed by amplifying a 3.2-kb region comprising tetM and its upstream promoter and downstream terminator sequences from Tn916 using the primers GGTTGTGGATCCTTGTGGGTACTTTTAGGGC and CTTATTGAATTCGAAACCATATTTATATAACAAC. This fragment was ligated into the BamHI/EcoRI sites of pMT85. The gentamicin resistance determinant of pMT85 was then removed by digestion with BglII and XbaI, the vector was blunt-ended with T4 DNA polymerase, and an SalI linker, CCGGTCGACCGG (NEB), was inserted into the blunted BglII/XbaI site.
Mycoplasma strain 158 was transformed using the polyethylene glycol-mediated method as described previously (29). Plasmids pTF85 and pTF20 do not replicate in mycoplasmas, and transformants are obtained only when the minitransposon transposes into the mycoplasma genome. Transformants were selected on agar supplemented with 5 μg of tetracycline/ml. Individual transformant colonies were picked and grown at 37°C in 1 ml of broth containing tetracycline. Each culture was frozen at −80°C in broth supplemented with 15% glycerol. Most (90%) of the library consists of transformants containing Tn4001TF1, our first-generation minitransposon for M. arthritidis. Later, we switched to using Tn4001TF2 because higher transformation frequencies were obtained.
Mapping of transposon insertion sites.
The genomic location of the transposon was determined for each transformant by DNA sequence analysis of the junction between the transposon and the adjacent genomic DNA. The junction was amplified by inverse PCR. The sequence of the PCR product was compared to the complete genome sequence for identification of the transposon insertion site. The inverse PCR conditions and primers were similar to those used to map the transposon location in transformants of M. pulmonis (24) except that genomic DNA was digested with Sau3AI instead of NlaIII, and the primers for inverse PCR were CCTTCGGAAAAAGAGTTGGTAGC and TCCTGCGTTATCCCCTGATTC. The inverse PCR product was purified from an agarose gel, and the nucleotide sequence was determined using the primer TTTGAGTGAGCTGATACCGCTCGC.
Confirmation of gene dispensability.
To confirm that an ORF was dispensable, at least one transformant with the gene disrupted was analyzed by direct PCR to verify that the transposon integration site was correct and to determine whether an intact copy of the gene might be present in the template preparation. To confirm the genomic location of the transposon, the primer described above to sequence the inverse PCR products was paired with a gene-specific primer that annealed to sequences in the adjacent genomic DNA. A negative control consisted of amplification of a wild-type mycoplasma DNA preparation that lacked the transposon. The possible presence of an intact copy of the gene was assessed by using two gene-specific primers that flanked the site of transposon integration. The lack of a PCR product would indicate that no intact gene was present in the transformant. Template DNA from the wild-type parent strain was used as a positive control. If the PCR data confirmed the location of the transposon and indicated that no intact copy of the gene was present in the transformant, it was concluded that the gene was indeed mutable. If the transposon location was confirmed but a PCR product corresponding to an intact copy of the gene was also obtained, the transformant was subcloned. Individual filter clones were analyzed by direct PCR as described above to determine whether the transposon still resided in the gene and whether an intact copy of the gene was present in the template. Again, if in any subclone the transposon disrupted the gene and no intact copy of the gene was identified by PCR, it was concluded that the gene was mutable.
Nucleotide sequence accession number.
The M. arthritidis genome sequence was deposited in the GenBank database under accession number CP001047.
RESULTS
Genome sequence.
M. arthritidis strain 158L3-1 has a circular genome of 820,453 bp. The G+C content of 30.7 mol% is low for most bacteria but higher than that of many mycoplasma genomes. A total of 635 predicted coding regions and 36 predicted RNA genes occupy over 90% of the genome. The absence of genes in M. arthritidis coding for hexokinase and phosphofuctokinase is consistent with an organism that does not undergo glycolysis. As is the case for some other mycoplasmas, M. arthritidis lacks many genes coding for enzymes involved with nucleotide and amino acid biosynthesis as well as the GroEL and GroES chaperones. Missing were genes coding for either subunit of ribonucleotide reductase, an essential enzyme for B. subtilis grown in minimal medium (16). These genes are also absent in some other species of mycoplasma and are present but nonessential in M. pulmonis (12). The absence of ribonucleotide reductase suggests that deoxynucleoside triphosphates must be acquired from the host. The gene coding for signal peptidase II but not signal peptidase I was identified. M. arthritidis like other mycoplasmas has a high number of genes (>40) that are predicted to code for lipoproteins, necessitating the need for signal peptidase II activity.
As predicted for an organism that catabolizes arginine, genes coding for arginine deiminase, ornithine carbamoyltransferase, and carbamate kinase were identified. The genes coding for the latter two enzymes are apparently organized as an operon. M. arthritidis may be able to catabolize compounds in addition to arginine for energy production (20). Genes coding for the enzymes required to convert glycerol to pyruvate and then to lactate were identified. Other predicted enzymes include alcohol dehydrogenase and acetate kinase, suggesting that acetylaldehyde and acetyl phosphate can be substrates for generating ATP. The predicted gene products are inconsistent with a report of oxidation of fatty acids in M. arthritidis (27).
Other than the bacteriophage MAV1 genome, no mobile elements were evident in the genome of 158L3-1. The MAV1 genome is integrated at nucleotide positions 706579 through 722218. A 230-bp fragment that is homologous to the left end of the MAV1 genome was identified at nucleotide positions 38110 to 38340. The location of this MAV1 DNA fragment may be of interest because it is 228 bp upstream of the 5′ end of the mam gene (Marth_orf036), suggesting that mam may have originally been of phage origin. Compared to the MAV1 genome sequence that was reported previously (30), the MAV1 sequence of 158L3-1 has one nucleotide addition, one deletion, and two substitutions. These differences result in a single amino acid change in the predicted MAV1 DNA integrase and a lengthening of the MAV1 HtpN protein by 23 amino acids at its amino terminus. Other than the MAV1 integrase, the only other potential site-specific DNA recombinase encoded by the genome is the Marth_orf195 gene product. This predicted protein has 62 amino acids that share amino acid similarity with a portion of the carboxy half of the IS1630 transposase from Mycoplasma penetrans. Marth_orf195 may be a truncated gene coding for a nonfunctional recombinase. M. arthritidis, therefore, appears to lack the intricate site-specific DNA inversion systems found in some species of mycoplasma (14, 15, 17, 22).
A barrier to genetic transformation of M. arthritidis is the MarI restriction enzyme, an isoschizomer of AluI (29). Strain 158L3-1 DNA is resistant to cleavage by AluI and contains AGCT sequences modified by methylation of the C nucleotide. In addition to MarI, the MAV1 genome is predicted to code for a restriction and modification system of unknown specificity (30). No obvious gene that would encode the MarI endonuclease was identified in the genome sequence, but a candidate for the gene encoding the MarI DNA methyltransferase is Marth_orf138. The Marth_orf138 product has features characteristic of cytosine-specific DNA methyltransferases and has 75% amino acid sequence identity to the HhaI methyltransferase. Marth_orf138 was likely acquired as a result of horizontal gene transfer because the top hit by BLASTn analysis of the nucleotide collection (nonredundant nucleotide) database was the HhaI DNA methyltransferase gene of Haemophilus haemolyticus (71% nucleotide identity; E value of 6e-123). It is apparent that M. arthritidis lacks the complex type I restriction systems found in some mycoplasmas such as M. pulmonis (11). Marth_orf681 is related to genes coding for the HsdM subunit of type I restriction enzymes, but no type I activity in M. arthritidis is predicted because absent are genes coding for the other subunits required for enzymatic activity, HsdS and HsdR.
Phase variation and gene families.
One mechanism by which many mycoplasmas evade the host's adaptive immune responses is through the phase-variable production of critical surface proteins (6). In some cases, phase variation is achieved by slipped-strand mispairing (SSM) at a run of homonucleotides located upstream of the gene's coding region (9). Gain or loss of nucleotides in this region acts as an on/off switch for promoter activity by changing the spacing between the promoter's −10 and −35 regions. To identify sequences that might be associated with phase variation, we examined the genome of 158L3-1 to identify homonucleotide repeats of at least 10 bp in length. Twenty-nine homonucleotide repeat regions were identified. Twenty-five of these repeats were located in the predicted promoter region of genes that likely code for phase-variable surface proteins (Table 1). Many of the predicted phase-variable genes are members of one of two families (the massive surface protein, or Msp, and major surface antigen, or MSA, families). One of the Msp family members (Marth_orf471) was not initially identified as having a homonucleotide repeat because the length of the repeat was only 7 bp. However, the structure of the promoter region of this gene was essentially the same as for other family members. Therefore, a total of 26 gene products are predicted to be subject to phase variation. In nearly all cases, the homonucleotide repeat on the gene's coding strand consisted of poly(T). This is in contrast to the vlp genes of Mycoplasma hyorhinis, which have poly(A) in the promoter region of the coding strand (35).
TABLE 1.
Predicted phase-variable genes containing homonucleotide repeats in the putative promoter
| Gene | Product (mass [kDa]) | Translation start position (nt [direction of transcription])a | Repeatb
|
Promoter length (bp)d | ||
|---|---|---|---|---|---|---|
| Homonucleotide | No. of repeats | Position (bp)c | ||||
| Marth_orf057 | MspA (266) | 58223 (−) | Poly(T) | 12 | 143-131 | 129 |
| Marth_orf133 | Hypothetical protein (116) | 127760 (−) | Poly(T) | 15 | 104-87 | 82 |
| Marth_orf150 | MspI (272) | 146094 (−) | Poly(T) | 12 | 142-130 | 128 |
| Marth_orf220 | MSA family lipoprotein (30) | 211858 (−) | Poly(T) | 14 | 114-100 | 98 |
| Marth_orf221 | MSA family lipoprotein (28) | 213028 (−) | Poly(T) | 14 | 114-100 | 98 |
| Marth_orf223 | MSA family lipoprotein (19) | 214124 (−) | Poly(T) | 14 | 115-101 | 99 |
| Marth_orf224 | MSA family lipoprotein (28) | 215283 (−) | Poly(T) | 13 | 118-105 | 103 |
| Marth_orf226 | Lipoprotein (29) | 216472 (−) | Poly(T) | 14 | 112-98 | 96 |
| Marth_orf227 | Lipoprotein (30) | 218158 (−) | Poly(T) | 15 | 121-108 | 106 |
| Marth_orf229 | Lipoprotein (34) | 219322 (−) | Poly(T) | 10 | 116-106 | 104 |
| Marth_orf316 | MSA family lipoprotein (14) | 283591 (−) | Poly(T) | 16 | 141-125 | 123 |
| Marth_orf321 | Membrane protein (34) | 286893 (+) | Poly(T) | 15 | 78-63 | 59 |
| Marth_orf358 | MspB (220) | 320793 (+) | Poly(T) | 15 | 145-130 | 128 |
| Marth_orf459 | MSA family protein (15) | 398310 (+) | Poly(T) | 13 | 133-120 | 118 |
| Marth_orf462 | MSA family lipoprotein (21) | 399209 (+) | Poly(T) | 13 | 118-105 | 103 |
| Marth_orf469 | MspC (310) | 400579 (+) | Poly(T) | 14 | 145-131 | 129 |
| Marth_orf471 | MspJ (38) | 409106 (+) | Poly(T) | 7 | 137-130 | 128 |
| Marth_orf481 | MspD (294) | 419732 (−) | Poly(T) | 15 | 140-125 | 123 |
| Marth_orf492 | MspE (337) | 423935 (+) | Poly(T) | 15 | 144-129 | 127 |
| Marth_orf497 | MspF (338) | 433902 (+) | Poly(T) | 12 | 137-125 | 123 |
| Marth_orf508 | Hypothetical protein (84) | 456060 (−) | Poly(A) | 12 | 78-66 | 58 |
| Marth_orf566 | MSA family lipoprotein (28) | 508346 (+) | Poly(T) | 13 | 104-91 | 89 |
| Marth_orf647 | MspG (308) | 581582 (+) | Poly(T) | 12 | 142-130 | 128 |
| Marth_orf653 | MspH (277) | 590327 (+) | Poly(T) | 14 | 145-131 | 129 |
| Marth_orf727 | MspK (157) | 669401 (+) | Poly(T) | 12 | 140-128 | 122 |
| Marth_orf793 | MAA2 MSA lipoprotein (58) | 736230 (−) | Poly(T) | 12 | 118-106 | 104 |
Direction of transcription is indicated as follows: +, upstream; −, downstream. nt, nucleotide.
Sequence upstream of the gene on the coding strand.
Distance from repeat to translation start site.
Distance from the start of the −10 region (consensus TATAAT) of the putative promoter to translation start site.
Four of the identified homonucleotide repeats may have no bearing on gene expression. A 14-homonucleotide repeat (nucleotides 775605 to 775618) was located downstream of each of two converging ORFs. The poly(A) repeat located just 8 bp before the translation start site of Marth_orf832 is probably located downstream of the gene's promoter and may not disrupt transcription initiation through SSM. Because M. arthritidis is thought to require arginine for growth, it would be surprising if the length of the poly(A) sequence located 69 bp upstream of the translation start site of the arcA gene (Marth_orf873) affected gene expression. Indeed, the predicted −10 region of the arcA promoter is located far upstream, 220 bp from the translation start site. The arcA promoter, including the homonucleotide repeats, has been combined with a lacZ reporter, inserted into the M. arthritidis chromosome, and tested for its ability to drive transcription (10). LacZ activity was detected, and no indication of phase variation was noted, suggesting that this homonucleotide repeat has no effect on transcription in its native context. Marth_orf783, another gene with an upstream homonucleotide repeat, is predicted to encode a cytoplasmic protein. Although exceptions exist, phase-variable proteins are most often localized to the cell surface. Marth_orf783 may be expressed constitutively despite the poly(A) repeat that precedes the translation start site by 75 bp because the predicted −10 region of the promoter is located further upstream, 139 bp from the translation start site.
Eleven of the 26 predicted phase-variable proteins belong to the Msp family. Each of the MspA through MspK proteins is considered to be phase variable; the proteins are encoded by ORFs Marth_orf057, Marth_orf358, Marth_orf469, Marth_orf481, Marth_orf492, Marth_orf497, Marth_orf647, Marth_orf653, Marth_orf150, Marth_orf471, and Marth_orf727 (Table 1), respectively. An additional Msp protein (MspL, encoded by Marth_orf0275) may not be phase variable because the promoter lacks homonucleotide repeats. Most of the Msp proteins have a predicted mass of 200 or 300 kDa. The proteins apparently have a single transmembrane domain and should be exposed on the outside surface of the mycoplasma except for a short cytoplasmic domain at the amino terminus. There are no Msp orthologs identified in other bacteria.
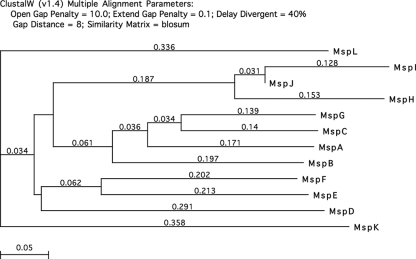
A Clustal W analysis of the Msp proteins is illustrated in Fig. 1. The most divergent proteins are MspK and MspL, but these proteins are clearly in the Msp family. For example, MspK and MspI share 40.3% amino acid sequence identity in a 1,005-residue region, and MspL and MspF share 43.7% amino acid identity in a 955-residue region. The shortest protein is MspJ (337 amino acids), which shares 95.7% amino acid sequence identity with MspI in a 308-residue region. A 5-kb region that includes the mspJ gene (Marth_orf471) and the 3′ half of the mspD gene (Marth_orf00481) is shown in Fig. 2. Four different single nucleotide changes in this region of the 158L3-1 genome (deletion of T at nucleotide position 410113, insertion of A at position 410668, substitution of C for T at 411146, and deletion of A at 411505) would merge ORFs Marth_orf471, Marth_orf472, and additional downstream sequences into a single gene that would code for a 922-amino-acid protein that shares 91.8% sequence identity with MspI. It is probable that Marth_orf471 and Marth_orf472 are remnants of a single ancestral msp gene.
FIG. 1.
Clustal W analysis of the Msp protein family. The scale indicates the expected number of substitutions per site.
FIG. 2.
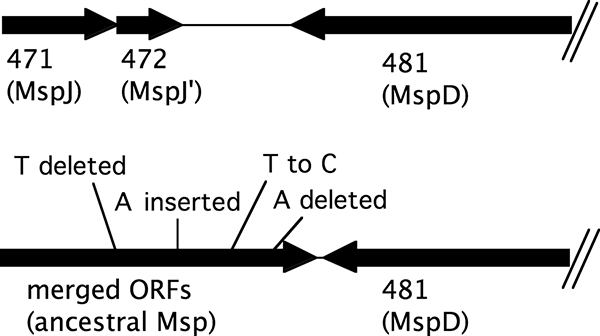
Schematic of a 5-kb region (nucleotides 409106 to 414105) of the genome sequence illustrating the nucleotide substitutions that would merge the ORFs of strain 158L3-1 (top) into an ancestral msp gene (bottom). Numbers refer to Marth_orf designations. Not all of Marth_orf00481 is shown.
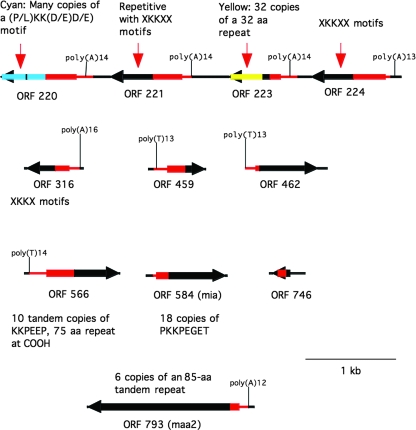
Another set of genes identified in the genome sequence of 158L3-1 is designated the MSA family (Fig. 3). Most of the genes in this family share strong nucleotide similarity in their 5′ untranslated regions and in the regions coding for the protein's signal peptide sequence. Nucleotide similarity extends into the promoter region of most of the msa genes. These genes (Marth_orf220, -221, -223, -224, -316, -459, -462, -566, and -793) are predicted to be phase-variably expressed based on the presence of homonucleotide repeats in the promoter region (Table 1). Marth_orf793 codes for the MAA2 protective antigen and has been shown previously to be phase-variably expressed due to changes in the poly(T) region of the promoter (34). Marth_orf584 is in the MSA family but has a different promoter structure lacking homonucleotide repeats. Marth_orf584 codes for MIA (major immunodominant antigen), the major surface antigen recognized by serum from infected animals and previously referred to as M. arthritidis ORF 619 (26). Marth_orf746 is the only MSA family member that lacks nucleotide similarity to other family members in the 5′ untranslated region. Instead, Marth_orf746 is similar to Marth_orf220, -221, -223, -224, -316, -459, -566, and -584 in a central region of the coding sequence. Most MSA family members have repetitive domains (Fig. 3). The maa2 and mia genes have tandem repeat regions in their coding regions that have been shown previously to undergo size variation (gain or loss of repeat units) as a result of SSM (26, 34).
FIG. 3.
Schematic illustrating features of MSA family members including the locations and lengths of poly(A) or poly(T) tracts in promoter regions. Other than black regions, which are nonhomologous, regions of like color are homologous.
Potential virulence factors.
The MAM superantigen (Marth_orf036) has been implicated as having a major role in pathogenesis. The gene product has a signal peptide sequence that is absent in the mature, secreted protein of 213 amino acids (4). In addition to its superantigen activity, MAM also has nuclease activity (8). Marth_orf729 is predicted to code for a protein that shares 49.4% amino acid identity with MAM. The Marth_orf729 gene product may be an integral membrane protein because it lacks an obvious signal peptide sequence but has a predicted transmembrane domain near the amino terminus. There is significant sequence divergence between the Marth_orf729 gene product and the region of MAM (amino acids 11 to 38 of the mature protein) associated with lymphocyte proliferation activity (4). Perhaps the Marth_orf729 protein is a nuclease that lacks superantigen activity.
The MAA proteins are thought to contribute to adherence to host tissue, and MAA2 may not be the only MSA family member (Fig. 3) with a role in adherence. Some proteins within the MSA family such as MAA2 and MIA are strongly recognized by the immune system, and the phase-variable production of many of the MSA proteins likely contributes to immune avoidance (26, 33). Other than putative adhesins and MAM, little is known about factors that might contribute to pathogenesis. The M. arthritidis genome codes for peptide methionine sulfoxide reductase (Marth_orf807), required for full virulence in M. genitalium (7), and a potential hemolysin (Marth_orf698).
M. arthritidis transposon library.
Unlike previous descriptions of mycoplasmal transposon libraries (12, 13), the M. arthritidis library was created with minitransposons that could initially insert into the genome but then would be unable to transpose to secondary sites. The minitransposon Tn4001TF1 was used to generate the first 1,000 mutants in the library, and minitransposon Tn4001TF2 was used for the remaining mutants. With rare exceptions, the genomic location of the transposon was different for each member of the library. A total of 1,113 different genomic sites for transposon insertion were mapped. We estimate that the library contains null mutations in 226 different genes, using the criteria that null mutations are produced when the transposon truncates at least 10% from the 5′ end or 15% from the 3′ end of an ORF. These criteria are based in part on results from transposon mutagenesis of M. pulmonis (12). An exception, however, was made for predicted lipoprotein gene products. A lipoprotein gene was considered inactivated even if less than 10% of the 5′ end was truncated, provided that the fatty acid addition site would be missing from the remainder of the gene product. Our final table of mutated genes (see Table S1 in the supplemental material) has 218 entries, not 226. For eight genes, analysis of the transposon's genomic location by direct PCR failed to confirm the inverse PCR sequencing data, or an intact copy of the gene was detected by direct PCR in each of the subclones of the transformant. Similar findings of genes that were disrupted in the primary transformant but not in subclones thereof have been reported for transposon mutagenesis of M. genitalium (13) and M. pulmonis (12).
Of the 218 genes disrupted in M. arthritidis (see Table S1 in the supplemental material), 36 have essential homologs in M. genitalium, 26 have essential homologs in M pulmonis, and 18 have homologs that are essential in both species (Table 2). Only a few of the dispensable M. arthritidis genes that are essential in one or both of the other species of mycoplasma have potential paralogs within the M. arthritidis genome that might render the genes expendable. Perhaps most interesting are the six dispensable M. arthritidis genes that have essential homologs in B. subtilis. Three of these are the ribosomal protein S15, S18, and L15 genes. The truncated protein in the disrupted S15, S18, and L15 genes would be 78%, 61%, and 58%, respectively, of the full-length wild-type protein. Other genes important for protein synthesis that were disrupted in the library included those coding for methionyl-tRNA formyltransferase and polypeptide deformylase. Although these genes are essential in M. genitalium and M. pulmonis, formylation is not essential for initiation of protein synthesis in all bacteria, and Mycoplasma hyopneumoniae lacks these genes altogether (18, 23, 28). Other genes of interest that were disrupted in the library included those coding for potential virulence factors such as the MAM superantigen, the Marth_orf698 hemolysin, and several members of the MSA family (Marth_orf220, -221, -224, -459, -746, and -793).
TABLE 2.
Disposable M. arthritidis genes that are essential in M. pulmonis or M. genitalium
| Locus taga | Product | Gene symbol | Ortholog
|
Essential gene in:d
|
M. arthritidis paraloga | Essential gene in B. subtilisd | ||
|---|---|---|---|---|---|---|---|---|
| M. pulmonisb | M. genitaliumc | M. pulmonis | M. genitalium | |||||
| 012 | RNA pseudouridylate synthase family | rluC | 1170 | 209 | No | Yes | ||
| 019f | Oligopeptide ABC transporter/Valyl-tRNA synthetase | oppF | 2860 | 080 | No | Yes | 399 | |
| 104 | Phosphonate ABC transporter | 2250 | 289 | Yes | No | |||
| 125 | M. hominis P60-related lipoprotein | 0070 | NA | Yes | NA | |||
| 168 | Conserved hypothetical protein | NAe | 432 | NA | Yes | |||
| 169 | GTP-binding protein, putative translation factor | ychF | 1650 | 024 | Yes | No | ||
| 243 | RNase HIII | rnhC | 3000 | 199 | Yes | Yes | ||
| 258 | ATP synthase F1 alpha subunit | atpA | 7000 | 401 | No | Yes | ||
| 269 | Inorganic pyrophosphatase | ppa | 4700 | 351 | Yes | Yes | ||
| 277 | Transketolase | tktA | 5110 | 066 | Yes | No | Yes | |
| 366 | Putative esterase or lipase, membrane protein | 3130 | NA | Yes | NA | |||
| 369 | Ribosomal protein S15 | rpsO | 3280 | 424 | Yes | Yes | Yes | |
| 377 | Hypoxanthine phosphoribosyltransferase | hpt | 5510 | 458 | Yes | Yes | Yes | |
| 388 | Holliday junction DNA helicase motor protein | ruvA | 6570 | 358 | No | Yes | ||
| 389 | Holliday junction DNA helicase | ruvB | 6580 | 359 | No | Yes | ||
| 422 | Ribosomal protein L15 | rplO | 5690 | 169 | Yes | Yes | Yes | |
| 498 | ATP synthase subunit B | atpD | 6990 | 399 | Yes | Yes | ||
| 499 | ATP synthase subunit A | atpA | 7000 | 401 | No | Yes | ||
| 520 | COF family HAD hydrolase protein, conserved | 2440 | 265 | Yes | Yes | |||
| 562 | Predicted helicase | 1220 | 140 | No | Yes | |||
| 611 | Oligoendopeptidase | pepF | 3210 | 183 | No | No | ||
| 626 | Proline iminopeptidase | NA | 020 | NA | Yes | |||
| 628 | Spermidine/putrescine ABC transporter | potD | 4220 | 045 | Yes | Yes | 634 | |
| 634 | Spermidine/putrescine ABC transporter | potD | 4220 | 045 | Yes | Yes | 628 | |
| 635 | GTP-binding protein | era | 3800 | 387 | Yes | Yes | ||
| 637 | Bacteriocin exporter and processing endoprotease | 3760 | 390 | No | Yes | |||
| 656 | Formamidopyrmidine-DNA glycosylase MutM | fpg | 3100 | 262.1 | Yes | No | ||
| 668 | Putative Na+ driven multidrug efflux pump | norM | 2930 | NA | Yes | NA | ||
| 687 | Conserved hypothetical protein | 2260 | 103 | No | Yes | |||
| 694 | Ribosomal protein S18 | rpsR | 6090 | 092 | No | Yes | Yes | |
| 698 | Putative hemolysin | hlyC | 5140 | 146 | Yes | Yes | ||
| 699 | Polypeptide deformylase | def | 6890 | 106 | Yes | Yes | ||
| 708 | Oligopeptide ABC transporter, permease | oppB | 2830 | 077 | Yes | Yes | 402 | |
| 709 | Conserved protein | 1760 | NA | Yes | NA | |||
| 755 | tRNA modification protein | trmE | 0130 | 008 | Yes | Yes | ||
| 789 | Glucose inhibited division protein A | gidA | 2530 | 379 | Yes | Yes | ||
| 815 | Uracil-DNA glycosylase | ung | 6150 | 097 | No | Yes | ||
| 816 | Methionyl-tRNA formyltransferase | fmt | 0450 | 365 | Yes | Yes | Yes | |
| 821 | Sugar ABC transporter ATP-binding protein | 6190 | 119 | No | Yes | |||
| 822 | Sugar ABC transporter permease protein | 6180 | 120 | No | Yes | |||
| 830 | DHH family phosphoesterase | 6920 | 190 | Yes | Yes | 830 | ||
| 831 | DHH family phosphoesterase | 0220 | 190 | No | Yes | 831 | ||
| 852 | Exodeoxyribonuclease V, alpha subunit | recD | 7820 | NA | Yes | NA | ||
| 865 | Phosphate transport system regulatory protein | phoU | NA | 409 | NA | Yes | ||
The number represents the M. arthritidis locus tag of the form Marth_orf#.
M. pulmonis locus tag, MYPU_#.
M. genitalium locus tag, MG#.
Essential genes in M. pulmonis, M. genitalium, and B. subtilis are as reported, respectively, by French et al. (12), Glass et al. (13), and Kobayashi et al. (16).
NA, not available.
Marth_019 appears to be a fusion of oppF and the valyl tRNA synthetase gene valS. All transposon disruptions were in the oppF portion of the gene.
None of the genes involved in catabolism of arginine or in the conversion of glycerol to pyruvate, the triose arm of the Embden-Meyerhof-Parnas pathway, were disrupted in the library. These pathways are most likely required to meet the mycoplasma's needs for ATP. The genome has four genes (tktA, rpiB, xfp, and rpe) involved in the pentose phosphate pathway, at least three of which are dispensable. Other genes involved with carbohydrate metabolism, a predicted alcohol dehydrogenase (Marth_orf738) and lactate dehydrogenase (Marth_orf109), are also dispensable.
DISCUSSION
Signal peptidase I activity should be present in M. arthritidis because some proteins such as the secreted MAM superantigen have a typical signal peptide sequence that is absent in the mature protein (4). Species of mycoplasma such as M. pulmonis, Mycoplasma synoviae, Mycoplasma gallisepticum, and M. hyopneumoniae have an annotated signal peptidase I gene that is missing in many other species including Mycoplasma pneumoniae, M. genitalium, Mycoplasma agalactiae, Ureaplasma parvum, and now M. arthritidis (see http://cbi.labri.fr/outils/molligen/home.php). Signal peptidase I activity in species that lack an obvious signal peptidase I gene is probably catalyzed by a novel enzyme.
Although phase-variable proteins have been described for several species of mycoplasma, the number of such proteins predicted from the genome sequence of M. arthritidis is high compared to M. pulmonis, M. genitalium, and M. pneumoniae but probably not as high as some other species such as M. gallisepticum (19, 21). The total length of the coding regions for the genes listed in Table 1 is more than 90 kb, accounting for 11% of the genome. Homonucleotide tracts located within the promoter regions of genes that are phase-variably expressed were first described in M. hyorhinis (35). The frequency of phase variation is high, about 10−3 per CFU, and results from SSM events that vary the length of the repeats and thus the spacing between the −10 and −35 sites. With 26 genes demonstrating phase variation at a frequency of 10−3, about 1 CFU in 40 will undergo a phase variation event each generation. Several of the phase-variable genes also have tandem repeats in the coding region that would be subject to SSM, resulting in variation in the size of the protein, as has been shown previously for the MIA protein of M. arthritidis (Marth_orf00584) (26). Cultures of M. arthritidis consist of complex cell populations producing a varied repertoire of proteins. Not all transformants in the transposon library will be isogenic, and care must be taken in the assessment of mutant phenotypes.
Based on the number of genes that are essential for growth in M. genitalium, about one-half of the M. arthritidis genome should consist of essential genes. The nonessential half of the genome, around 400 kb, would represent the effective target size for transposon integration. Within the nonessential regions of the genome, the density of insertion sites would be about one site per 360 bp (400 kb of genome/1,100 transposon insertion sites). Genome coverage of the library is thus inadequate to conclude that any gene not hit by the transposon is essential.
We had anticipated that unconfirmable mutants would be rare in the M. arthritidis library. In minitransposons, the transposase gene is not inside the transposon but is located elsewhere on the vector backbone. Consequently, Tn4001TF1 and Tn4001TF2 should not be capable of transposition into secondary sites. Indeed, no evidence of secondary transposition of the minitransposons was detected. Previous studies of transposon mutagenesis in mycoplasma have reported unconfirmable mutants—genes that were disrupted by the transposon in primary transformants but could not be subsequently verified (12, 13). These reports did not use a minitransposon. As Tn4001 and its derivatives transpose by an excision-insertion mechanism, we had attributed unconfirmable mutants to active transposition within the genome. In the current study using minitransposons, the location of the transposon could not be confirmed for only 32 of 338 (9%) primary transformants. For our M. pulmonis library constructed with the non-minitransposon Tn4001T, confirmation of the location of the transposon was not obtained for 97 of 464 (21%) primary transformants. Thus, the use of a minitransposon did not eliminate the occurrence of unconfirmable transposon insertions but did reduce them by 50%.
Considering the essential roles of ribosomal proteins S15, S18, and L15 in most bacteria, the dispensability of these proteins in M. arthritidis is surprising. Although we have knocked out the S18 gene in M. pulmonis, the S15, S18, and L15 genes are essential in M. genitalium. It appears that the structure of the ribosome may differ, perhaps in subtle ways, between species of mycoplasma. The ribosomes of other bacteria may exhibit different properties as well because six of the 30S ribosomal proteins including S15 are nonessential in Escherichia coli (1).
Supplementary Material
Acknowledgments
This work was supported by NIH grant AR44252.
Editor: A. Camilli
Footnotes
Published ahead of print on 23 June 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Bubunenko, M., T. Baker, and D. L. Court. 2007. Essentiality and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J. Bacteriol. 1892844-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. C. Rocha, and A. Blanchard. 2001. The complete genome of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 292145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapper, B., A.-H. T. Tu, W. L. Simmons, and K. Dybvig. 2004. Bacteriophage MAV1 is not associated with virulence of Mycoplasma arthritidis. Infect. Immun. 727322-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, B. C., K. L. Knudston, A. Oliphant, A. D. Sawitzke, A. Pole, M. Manohar, L. S. Benson, E. Ahmed, and C. L. Atkin. 1996. The sequence of the Mycoplasma arthritidis superantigen, MAM: identification of functional domains and comparison with microbial superantigens and plant lectin mitogens. J. Exp. Med. 1831105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 274636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denison, A. M., B. Clapper, and K. Dybvig. 2005. Avoidance of the host immune system through phase variation in Mycoplasma pulmonis. Infect. Immun. 732033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhandayuthapani, S., M. W. Blaylock, C. M. Bebear, W. G. Rasmussen, and J. B. Baseman. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 1835645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diedershagen, M., S. Overbeck, S. Arlt, B. Plumakers, M. Lintges, and L. Rink. 2007. Mycoplasma arthritidis-derived superantigen (MAM) displays DNase activity. FEMS Immunol. Med. Microbiol. 49266-271. [DOI] [PubMed] [Google Scholar]
- 9.Dybvig, K. 1993. DNA rearrangements and phenotypic switching in prokaryotes. Mol. Microbiol. 10465-471. [DOI] [PubMed] [Google Scholar]
- 10.Dybvig, K., C. T. French, and L. L. Voelker. 2000. Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis. J. Bacteriol. 1824343-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dybvig, K., R. Sitaraman, and C. T. French. 1998. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc. Natl. Acad. Sci. USA 9513923-13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French, C. T., P. Lao, A. E. Loraine, B. T. Matthews, H. Yu, and K. Dybvig. 2008. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol. Microbiol. doi: 10.1111/j. 1365-2958.2008.06262.x. [DOI] [PMC free article] [PubMed]
- 13.Glass, J. I., N. Assas-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchinson III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glew, M. D., M. Marenda, R. Rosengarten, and C. Citti. 2002. Surface diversity in Mycoplasma agalactiae is driven by site-specific DNA inversions within the vpma multigene locus. J. Bacteriol. 1845987-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horino, A., Y. Sasaki, T. Sasaki, and T. Kenri. 2003. Multiple promoter inversions generate surface antigenic variation in Mycoplasma penetrans. J. Bacteriol. 185231-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouillie, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. M. L. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 1004678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lysnyansky, I., Y. Ron, and D. Yogev. 2001. Juxtaposition of an active promoter to vsp genes via site-specific DNA inversions generates antigenic variation in Mycoplasma bovis. J. Bacteriol. 1835698-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton, D. T., C. Creuzenet, and D. Mangroo. 1999. Formylation is not essential for initiation of protein synthesis in all eubacteria. J. Biol. Chem. 27422143-22146. [DOI] [PubMed] [Google Scholar]
- 19.Papazisi, L., T. S. Gorton, G. Kutish, P. F. Markham, G. F. Browning, D. K. Nguyen, S. Swartzell, A. Madan, G. Mahairas, and S. J. Geary. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain Rlow. Microbiology 1492307-2316. [DOI] [PubMed] [Google Scholar]
- 20.Pollack, J. D. 1992. Carbohydrate metabolism and energy conservation, p. 181-200. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, DC.
- 21.Rocha, E. P. C., and A. Blanchard. 2002. Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 302031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sitaraman, R., A. M. Denison, and K. Dybvig. 2002. A unique, bifunctional site-specific DNA recombinase from Mycoplasma pulmonis. Mol. Microbiol. 461033-1040. [DOI] [PubMed] [Google Scholar]
- 23.Steiner-Mosonyi, M., C. Creuzenet, R. A. B. Keates, B. R. Strub, and D. Mangroo. 2004. The Pseudomonas aeruginosa initiation factor IF-2 is responsible for formylation-independent protein initiation in P. aeruginosa. J. Biol. Chem. 27952262-52269. [DOI] [PubMed] [Google Scholar]
- 24.Teachman, A. M., C. T. French, H. Yu, W. L. Simmons, and K. Dybvig. 2002. Gene transfer in Mycoplasma pulmonis. J. Bacteriol. 184947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu, A.-H., J. R. Lindsey, T. R. Schoeb, A. Elgavish, H. Yu, and K. Dybvig. 2002. Role of bacteriophage MAV1 as a mycoplasmal virulence factor for the development of arthritis in mice and rats. J. Infect. Dis. 186432-435. [DOI] [PubMed] [Google Scholar]
- 26.Tu, A.-H. T., B. Clapper, T. R. Schoeb, A. Elgavish, J. Zhang, L. Liu, H. Yu, and K. Dybvig. 2005. Association of a major protein antigen of Mycoplasma arthritidis with virulence. Infect. Immun. 73245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VanDemark, P. J., and P. F. Smith. 1965. Nature of butyrate oxidation by Mycoplasma hominis. J. Bacteriol. 89373-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasconcelos, A. T. R., H. B. Ferreira, C. V. Bizarro, S. L. Bonatto, M. O. Carvalho, P. M. Pinto, D. F. Almeida, L. G. P. Almeida, R. Almeida, L. Alves-Filho, E. N. Assunc, V. A. C. Azevedo, M. R. Bogo, M. M. Brigido, M. Brocchi, H. A. Burity, A. A. Camargo, S. S. Camargo, M. S. Carepo, D. M. Carraro, J. C. de Mattos Cascardo, L. A. Castro, G. Cavalcanti, G. Chemale, R. G. Collevatti, C. W. Cunha, B. Dallagiovanna, B. P. Dambrós, O. A. Dellagostin, C. Falcão, F. Fantinatti-Garboggini, M. S. S. Felipe, L. Fiorentin, G. R. Franco, N. S. A. Freitas, D. Frías, T. B. Grangeiro, E. C. Grisard, C. T. Guimarães, M. Hungria, S. N. Jardim, M. A. Krieger, J. P. Laurino, L. F. A. Lima, M. I. Lopes, E. L. S. Loreto, H. M. F. Madeira, G. P. Manfio, A. Q. Maranhão, C. T. Martinkovics, S. R. B. Medeiros, M. A. M. Moreira, M. Neiva, C. E. Ramalho-Neto, M. F. Nicolás, S. C. Oliveira, R. F. C. Paixão, F. O. Pedrosa, S. D. J. Pena, M. Pereira, L. Pereira-Ferrari, I. Piffer, L. S. Pinto, D. P. Potrich, A. C. M. Salim, F. R. Santos, R. Schmitt, M. P. C. Schneider, A. Schrank, I. S. Schrank, A. F. Schuck, H. N. Seuanez, D. W. Silva, R. Silva, S. C. Silva, C. M. A. Soares, K. R. L. Souza, R. C. Souza, C. C. Staats, M. B. R. Steffens, S. M. R. Teixeira, T. P. Urmenyi, M. H. Vainstein, L. W. Zuccherato, A. J. G. Simpson, and A. Zaha. 2005. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 1875568-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voelker, L. L., and K. Dybvig. 1996. Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J. Bacteriol. 1786078-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voelker, L. L., and K. Dybvig. 1999. Sequence analysis of the Mycoplasma arthritidis bacteriophage MAV1 genome identifies the putative virulence factor. Gene 233101-107. [DOI] [PubMed] [Google Scholar]
- 31.Voelker, L. L., K. E. Weaver, L. J. Ehle, and L. R. Washburn. 1995. Association of lysogenic bacteriophage MAV1 with virulence of Mycoplasma arthritidis. Infect. Immun. 634016-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Washburn, L. R., and B. C. Cole. 2002. Mycoplasma arthritidis pathogenicity: membranes, MAM and MAV1, p. 473-490. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum, New York, NY.
- 33.Washburn, L. R., and E. J. Weaver. 1997. Protection of rats against Myco-plasma arthritidis-induced arthritis by active and passive immunizations with two surface proteins. Clin. Diagn. Lab. Immunol. 4321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Washburn, L. R., K. E. Weaver, E. J. Weaver, W. Donelan, and S. Al-Sheboul. 1998. Molecular characterization of Mycoplasma arthritidis variable surface protein MAA2. Infect. Immun. 662576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yogev, D., R. Rosengarten, R. Watson-McKown, and K. S. Wise. 1991. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 104069-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmerman, C.-U., and R. Herrmann. 2005. Synthesis of a small, cysteine-rich, 29 amino acids long peptide in Mycoplasma pneumoniae. FEMS Microbiol. Lett. 253315-321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.