Abstract
Phagocytosis of Candida albicans by either primary bone marrow-derived mouse macrophages or RAW 264.7 cells upregulated transcription of PRA1, which encodes a cell wall/membrane-associated antigen previously described as a fibrinogen binding protein. However, a pra1 null mutant was still able to bind fibrinogen, showing that Pra1p is not uniquely required for fibrinogen binding. As well, Pra1 tagged with green fluorescent protein did not colocalize with AlexaFluor 546-labeled human fibrinogen, and while PRA1 expression was inhibited when Candida was grown in fetal bovine serum-containing medium, Candida binding to fibrinogen was activated by these conditions. Therefore, it appears that Pra1p can play at most a minor role in fibrinogen binding to C. albicans. PRA1 gene expression is induced in vitro by alkaline pH, and therefore its activation in phagosomes suggested that phagosome maturation was suppressed by the presence of Candida cells. LysoTracker red-labeled organelles failed to fuse with phagosomes containing live Candida, while phagosomes containing dead Candida underwent a normal phagosome-to-phagolysosome maturation. Immunofluorescence staining with the early/recycling endosomal marker transferrin receptor (CD71) suggested that live Candida may escape macrophage destruction through the inhibition of phagolysosomal maturation.
Candida albicans is an opportunistic pathogen that can cause both superficial mucosal infections and more-serious disseminated bloodstream infections, the latter typically in immunocompromised patients or in patients with severe injuries or underlying diseases, such as diabetes (62). The roles of the different immune system components in the detection, defense, and elimination of this fungal pathogen are under extensive study (19, 57, 68). The morphological switching ability of this yeast and its cell surface-associated antigens also play important roles in both the pathogen's recognition by the host immune cells and its capacity to escape from destruction (7, 36, 58, 64, 66-68). For example, the yeast form of C. albicans is recognized by the Toll-like receptor 4 and the dectin-1 receptor, which stimulate release of proinflammatory cytokines, thus favoring Candida elimination (27, 60). However, both yeast and hyphal cells are also recognized by Toll-like receptor 2, which mediates the release of anti-inflammatory cytokines, thus favoring Candida survival (8, 59). Other receptors, such as DC-SIGN, MR, Mac-1, and Gal3, are also involved in C. albicans binding (reviewed in references 19, 64, and 66). The membrane/cell wall composition of C. albicans cells that represent the target for many receptors varies depending on environmental cues such as temperature, pH, and the presence of serum (12, 73). The differential expression of proteins includes in particular glycosyltransferases that modify the glycan components of the cell surface, resulting in differential recognition by the immune system (7, 58). Overall, Candida albicans surface proteins can bind many host proteins, including fibrinogen (10), complement fragments (34), plasminogen (13), and extracellular matrix proteins such as collagen, fibronectin, and laminin (28).
Because of the complexity of the host immune response, it has proven useful to investigate the behavior of pathogens that interact with isolated elements of the innate immune system. Many studies have looked at the consequences of Candida albicans cells growing in the presence of cultured macrophages. Such cultured cells, while far from providing a picture of the complete immune response, allow a focus on specific elements of the process. For example, the recognition that the yeast-to-hypha transition was an important component of the ability of the pathogen to escape phagocytosis was emphasized through studies using the J774A cell line (44). Subsequent work using this mouse macrophage line identified transcriptional consequences of the host-pathogen interaction on the pathogen (46). In addition, we have previously shown that, although RAW 264.7 mouse macrophages can also phagocytose and kill Candida, specific C. albicans mutant strains can more efficiently escape the macrophage although they are less virulent in the whole mouse tail vein injection test of pathogenicity (48). As an alternative to macrophage lines, primary cells can be used to investigate pathogen killing; these cells may be less standardized than established cell lines but are potentially more representative of the in vivo cellular status (53) and can be derived from hosts with interesting genetic modifications to immune system molecules (52).
We are interested in C. albicans genes that are modulated during the pathogen's interaction with mouse macrophages, especially those genes that encode proteins that localize to the cell membrane/wall. Among such genes, we noted PRA1 to be highly upregulated. Pra1p is a cell wall-associated protein found in both the yeast and hyphal forms of C. albicans: Pra1p becomes highly glycosylated in hyphal-form cells (12) and elicits a strong immune response in infected patients (77). Pra1p was also identified as mp58 (58-kDa mannoprotein) or Fbp1 (fibrinogen binding protein) (45, 78). An attractive model for a fibrinogen-binding role of Pra1p would be to link Candida cells to the Mac-1 receptor through fibrinogen (21, 22, 74), so the expression of this protein during phagocytosis could facilitate Candida adsorption to and escape from macrophages. However, because the addition of fibrinogen can inhibit lymphocyte adsorption to Candida, there is evidence against this model (21). As well, it was recently suggested that a soluble 250-kDa complex containing Pra1p was a direct ligand of the Mac-1 receptor, eliminating the need for a bridging role of fibrinogen (72).
After pathogen internalization and the formation of the phagosome, this organelle undergoes a maturation process: it fuses sequentially with sorting endosomes (pH 6.1) (often referred to as early endosomes), late endosomes (pH 5.5 to 6.0, with active proteases), and finally lysosomes (pH 4.5 to 5.5, with mature proteases) (16, 31, 63). As a result of these fusion events, the phagosomal lumen becomes a highly acidic and oxidizing environment, endowed with a variety of hydrolytic enzymes that can effectively digest its contents (32, 37). This maturation process ultimately leads to the degradation of the engulfed components and the formation of hydrolyzed fragments of the pathogen available for antigen presentation. Because normal phagocytosis should lead to acidification of the phagosome environment, it is surprising that PRA1 is upregulated in the macrophage because in vitro the expression of PRA1 and other genes, such as PHR1, is triggered by alkaline pH (15). This modulation is abolished in a rim101 mutant, suggesting that PRA1 expression is under the control of the Rim101p transcription factor. Newman et al. (61) had previously shown that, in human macrophages, live but not heat-killed Candida could inhibit lysosomal fusion to phagosomes. In this study, we investigated the expression of the PRA1 gene in the context of phagocytosis, including lysosomal fusion to the Candida phagosome, and the colocalization with fibrinogen by use of a green fluorescent protein (GFP)-tagged Pra1p-expressing strain.
MATERIALS AND METHODS
Reagents.
Dulbecco's modified Eagle's medium, Iscove modification of Dulbecco's medium (IMDM), Dulbecco's phosphate-buffered saline (D-PBS), AlexaFluor 546 (AF546) human fibrinogen conjugate, Trizol, Micro-FastTrack 2.0 kit, Superscript III, LysoTracker red DND-99, Hoechst 33342, and Prolong gold were purchased from Invitrogen (Rockville, MD). Fetal bovine serum (FBS), minimum essential medium with Earle's balanced salt solution (MEM-EBSS), and HEPES were purchased from Hy-Clone (Logan, UT). FBS was heat inactivated at 56°C for 30 min. Unless otherwise cited, all other reagents were purchased from Sigma (St. Louis, MO).
Strains and cell lines.
The C. albicans strains used in this study are listed in Table 1. Unless otherwise specified, the strains were grown overnight at 30°C in yeast extract-peptone-dextrose medium (YPD). The RAW 264.7 mouse macrophage line, kindly provided by A. Descoteaux (IAF, Laval, Canada), was maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated FBS (D-10). Bone marrow-derived macrophages (BMDM) were prepared from bone marrow cells isolated from 10-week-old BALB/c mice and differentiated in macrophages in MEM-EBSS supplemented with 10% heat-inactivated FBS, 10% L cell-conditioned medium as a source of CSF-1 (14), 2 mM l-glutamine, 15 mM HEPES, 100 IU/ml penicillin, and 100 μg/ml streptomycin (MEM-10), as described before (23, 76). The medium was changed every 2 to 3 days until macrophage differentiation and confluence were reached 10 to 14 days later. Proper differentiation of bone marrow cells into macrophages was assessed by flow cytometry analysis using rat anti-mouse fluorescein isothiocyanate-labeled F4/80 (BioLegend, San Diego, CA) and phycoerythrin (PE)-labeled CD11b (Cedarlane Laboratories Ltd., Hornby, ON, Canada) or appropriate rat isotypic control antibodies (PE-labeled rat immunoglobulin G2b or fluorescein isothiocyanate-labeled rat immunoglobulin G2b; Cedarlane Laboratories Ltd.). For assays with macrophages, C. albicans cells were washed twice in PBS, counted with a hemacytometer, and resuspended at the required concentration in warmed medium immediately before the interaction. RAW 264.7 cells were seeded the day before at 2.2 × 105 cells/cm2, and BM cells were seeded 10 to 14 days before at 3.3 × 104 cells/cm2 in differentiation medium. At time zero, culture medium was replaced with fresh medium of a Candida suspension to give a final multiplicity of infection (MOI) of 1 for live Candida cells (at macrophage confluence, 6.6 × 105 C. albicans cells/cm2 for RAW 264.7 cells and 2.2 × 105 C. albicans cells/cm2 for BMDM) or 5 for fixed Candida cells. The cells were incubated at 37°C with 5% CO2. For transcription profile analysis, control cultures consisting of Candida only were initiated at the same time under the same culture conditions. Fixed Candida cells were prepared the day before from an overnight culture in YPD (yeast form) or from an additional 1.5-h incubation under the same medium/culture conditions (37°C, 5% CO2 in D-10 or MEM-10 at 108 cells/30 ml). After a quick wash in D-PBS, cells were fixed for 30 min at room temperature in 4% paraformaldehyde in PBS containing Complete protease inhibitors (Roche Diagnostics, Laval, Canada). Fixed cells were then washed three times in PBS and stored at 4°C in PBS containing Complete protease inhibitors until use.
TABLE 1.
Candida strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SC5314 | Wild type | 20 |
| CAI4 | ura3::imm434/ura3::imm434 | 20 |
| CAI4-GFP | ura3::imm434/ura3::imm434 pAM5.6 | 2 |
| RM1000 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG | 56 |
| CAM33 | RM1000 pra1::HIS1/PRA1 | This study |
| CAM35 | RM1000 pra1::HIS1/pra1::URA3 | This study |
| CAM38 | CAI4 PRA1::PRA1-GFP::URA3 | This study |
Candida RNA isolation and cDNA labeling.
One to four 150-mm dishes were harvested by a quick wash in D-PBS, followed by the addition of 10 ml/plate (Candida only) or 15 ml/plate (Candida with macrophages) of Trizol reagent. Cells were collected and centrifuged at 12,000 × g. Pellets contained intact Candida cells, and the pellet containing Candida with macrophages was washed two more times with Trizol to remove contaminating macrophage DNA and RNA. Intact Candida cell pellets were quickly frozen at −80°C. Total RNA was isolated using a hot phenol extraction protocol repeated three times (40). Total RNA was further purified using an RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Quantification was assessed by absorbance reading (Nanodrop; Thermo Fisher Scientific, Montreal, Canada). The quality of mRNA was assessed using an RNA 6000 Nano Lab-on-a-Chip kit and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Eight to 20 μg of total RNA (equal amounts for each pair) was reverse transcribed in cDNA and purified as described before (55) by using Superscript III. Unincorporated dyes were removed using a CyScribe GFX purification kit (GE Healthcare Bio-Sciences Inc.). Procedures used for hybridization, washing, and scanning were done as described before (55), with the exception that a slide booster hybridization station (Advalytix; Olympus America Inc., Concord, MA) was used for microarray hybridization for 16 h at 42°C.
Microarray analysis.
We measured the fluorescence ratios between C. albicans cultures incubated with macrophages for 60, 120, or 240 min against that of control cultures incubated in the absence of macrophages. Each time point was assayed four times using independently produced cultures. To account for the possibility of dye bias, each series of four hybridization experiments included two Cy3/Cy5 and two Cy5/Cy3 comparisons. We eliminated from our analysis 60 probes that showed significant cross-hybridization with total RNA isolated from mouse macrophages. Normalization and statistical analysis were conducted with GeneSprings GX (Agilent Technologies, Santa Clara, CA). To select significantly modulated transcripts at each time point, we used volcano plots that combine a Welch t test (P values of <0.05) with a 1.5-fold-change cutoff.
Northern blot analysis.
mRNA was isolated using a Micro-FastTrack 2.0 kit according to the manufacturer's instructions. Northern blot analysis was performed as described previously (2), using 1 μg of mRNA per lane. Probes were made using a Rediprime II labeling kit (GE Healthcare Life Sciences, Baie D'Urfe, Canada) according to the manufacturer's instructions. Detection of specific RNAs was performed as previously described (1). The 500-bp PRA1 probe was obtained from a BstEII/ClaI fragment of pAM13 (PRA1 gene cloned in BamHI/PstI restriction sites of pVec). The other probes were PCR amplified from SC5314 genomic DNA: the 300-bp PHR1 probe used oligonucleotides 5′ GCTAACCGTCCACGTTTGTTC 3′ and 5′ TGGTGGCAAATTAGTTGCAGC 3′, and the 203-bp TEF2 control probe used oligonucleotides 5′ GTCCATGGTACAAGGGTTGG 3′ and 5′ ACCGGCTTTGATGATACCAG 3′.
Deletion of PRA1.
Oligonucleotides used in this study are listed in Table 2. As shown in Fig. 1A, the entire PRA1 open reading frame (orf19.3111) was deleted in two steps by homologous recombination using a PCR-based cassette method, as described previously (17, 29). Briefly, 100-bp oligonucleotides oAM98 and oAM99, consisting of 80 nucleotides of the 5′ (oAM98) or 3′ (oAM99) untranslated region (UTR) of PRA1 and 22 nucleotides of BlueScript plasmid, were used to amplify a URA3 or a HIS1 cassette from pBS-cURA3 (a kind gift of A. P. Brown) or pBS-CaHIS1 (a kind gift from C. Bachewich). The PCR fragments were purified on a QIAquick PCR purification column (Qiagen, Valencia, CA) and transformed sequentially into RM1000 (ura/ura his1/his1) by use of a modified rapid lithium acetate transformation protocol (11) with an overnight incubation at 30°C with the DNA, followed by a 15-min heat shock at 44°C (79). Transformants were plated on the appropriate selection medium and screened by PCR (Fig. 1B).
TABLE 2.
Oligonucleotides used in this study
| Name | Description | No. of nucleotides | Sequencea | Source or reference |
|---|---|---|---|---|
| oAM86 | PRA1 5′ UTR plus PstI | 35 | AAAACTGCAGAAGATGAGCTCGACTCTTCATCATC | This study |
| oAM87 | PRA1 3′ UTR | 23 | GTGACAAGTCGACCCAATCGGAC | This study |
| oAM92 | PRA1 end, GFP start forward | 53 | CATGCAGATGGTGAAGTCCACTGTATGAGTAAAGGAGAAGAACTTTTCACTGG | This study |
| oAM93 | PRA1 end, GFP start reverse | 53 | CCAGTGAAAAGTTCTTCTCCTTTACTCATACAGTGGACTTCACCATCTGCATG | This study |
| oAM94 | GFP end, PRA1 3′ UTR, forward | 61 | CACATGGCATGGATGAACTATACAAATAATTGTTAAGTTCAGGCATTAAACAATTTTTAAG | This study |
| oAM95 | GFP end, PRA1 3′ UTR, reverse | 61 | CTTAAAAATTGTTTAATGCCTGAACTTAACAATTATTTGTATAGTTCATCCATGCCATGTG | This study |
| oAM98 | PRA1 5′ UTR PCR cassette | 100 | CAACAATATCTCGTTGGAAAAGACCTTTGTTTGGTTAATCATTTTTTTTATTCACATCTATAATCACAAACTTTCTCTcgtaatacgactcactataggg | This study |
| oAM99 | PRA1 3′ UTR PCR cassette | 100 | TGCAATTAATCTTATTAATTCAAGCTATAAAAGATATCCATGAAACACCTTAAAAATTGTTTAATGCCTGAACTTAACaagggaacaaaagctgggtacc | This study |
| H1 | HIS1 forward | 20 | GGTACCTGGAGGATGAGGAG | C. Bachewich |
| H2 | HIS1 reverse | 25 | AATATTTATGAGAAACTATCACTTC | C. Bachewich |
| U1 | URA3 forward | 22 | TTGAAGGATTAAAACAGGGAGC | 17 |
| U2 | URA3 reverse | 24 | ATACCTTTTACCTTCAATATCTGG | 17 |
Underlining indicates the PstI site, boldface type indicates the ATG codon from the GFP sequence, and lowercase lettering indicates the URA or HIS cassette common sequence.
FIG. 1.
Disruption of PRA1 (orf19.3111). (A) PCR-based cassette method for disruption of PRA1 in two steps. The thick black bar represents genomic DNA at the PRA1 locus, and the white rectangles represent the PRA1 gene coding sequence. The PCR cassettes used for the disruption are composed of a selectable marker (gray ovoid rectangle) flanked by two 80-nucleotide segments from the PRA1 5′ UTR or 3′ UTR locus (small white rectangles) for the homologous recombination of the cassettes. The first allele of PRA1 was replaced by the HIS1 marker, and the second allele was replaced by the URA3 marker. Small arrows represent the orientations and approximate positions of oligonucleotides (Table 2) used for PCR analysis and confirmation of the disruption. (B) Confirmation of disruption by PCR. The parent strain RM1000, the first-allele-disrupted strain CAM33, and the second-allele-disrupted strain CAM35 were analyzed by PCR using the oligonucleotides described for panel A. A 1-kb DNA ladder (Invitrogen, Carlsbad, CA) was used for size reference. PCR with oligonucleotides oAM86 and oAM87 produced a 2.8-kb DNA fragment for the PRA1 wild-type allele, as seen for strains RM1000 and CAM33 (lanes 3 and 8), and a 3.2-kb fragment when PRA1 was replaced by the HIS1 marker, as seen for strains CAM33 and CAM35 (lanes 8 and 13), or the URA3 marker, as seen for strain CAM35 (lane 13). The proper integration of the markers was confirmed using external oligonucleotides of the PRA1 locus (oAM86 or oAM87) together with internal oligonucleotides for the markers HIS1 (H2 or H1, respectively) (lanes 1,2, 6, 7, 11, and 12) and URA3 (U2 or U1, respectively) (lanes 4, 5, 9, 10, 14, and 15).
End point dilution survival assay.
The end point dilution survival assay was performed as described previously (48). Briefly, Candida cells were counted and serially fourfold diluted in macrophage medium in 96-well microplates with or without mouse macrophages. After a 24-h incubation at 37°C, 5% CO2, Candida colonies from wells where colonies could be visualized were counted and compared to the numbers of colonies from wells of the same dilution containing macrophages.
Fibrinogen binding.
SC5314 or CAM35.1 Candida cells were incubated on acid-washed coverslips for 2 h at 37°C, 5% CO2 in IMDM supplemented with 10% FBS (I-10) with or without human fibrinogen at a final concentration of 250 μg/ml. They were then incubated on ice for 10 min, followed by the addition of AF546-conjugated human fibrinogen at a final concentration of 25 μg/ml. After an additional 10-min incubation on ice, cells were washed, fixed, and mounted as described in “Immunofluorescence (CD71 labeling)” below.
Microscopy.
Phase contrast and epifluorescence pictures were taken using a Leica DMIRE2 inverted microscope (Leica Microsystems Canada) equipped with a Hamamatsu cooled charge-coupled-device camera at ×200, ×400, ×630, or ×1,000 magnification, using the appropriate filters. Openlab software (Improvision, MA) was used for image acquisition. Macrophage nuclei were stained using Hoechst 33342 at a final concentration of 1 μg/ml.
PRA1-GFP construct.
A C-terminal GFP-tagged Pra1 protein was constructed using a PCR fusion strategy. Chimeric oligonucleotides forward oAM92 (24 bases upstream of the PRA1 stop codon and 29 bases from the first ATG coding sequence of GFP) and reverse oAM95 (35 bases from the PRA1 stop codon toward the 3′ UTR and 26 bases from the GFP 3′-end coding sequence) were used to amplify GFP from plasmid pGFP26 (51). Oligonucleotides oAM86 (1 kb upstream of the PRA1 coding sequence, containing a PstI restriction site for subcloning) and oAM93 (reverse of oAM92) were used on SC5314 genomic DNA to amplify the PRA1 gene without its stop codon and its 5′ UTR, and oligonucleotides oAM94 and oAM87 were used to amplify the 3′ UTR of PRA1 including its stop codon. The three PCR fragments were sequentially reamplified in two steps using the appropriate oligonucleotides to generate the Pra1-GFP C-terminal fusion protein. The resulting 3.5-kb PCR fragment was subcloned into pVec (47) using PstI and BamHI (840 bp downstream of the GFP stop codon) restriction sites to generate pAM19. Several clones were verified by sequencing. Finally, pAM19.5 was ApaI digested and transformed into the CAI4 ura−/− C. albicans strain. Colony PCR using an external oligonucleotide and an internal oligonucleotide was used to verify the appropriate integration of the construct at the PRA1 locus. Clones were also tested for their ability to become GFP positive under inducing conditions (high pH, using IMDM).
Fibrinogen and PRA1 colocalization.
AF546-conjugated human fibrinogen at a final concentration of 25 μg/ml was added to overnight cultures of CAM38.3 (Pra1-GFP-expressing Candida) or CAM35.1 (pra1 null) cells in IMDM or I-10 at 37°C and incubated on ice for 1 h. Cultures were then washed twice in PBS, followed by microscopic observation at ×630 magnification.
Effect of FBS on Pra1-GFP expression.
Overnight cultures of CAM38.3 cells in IMDM at 30°C were diluted 10 times in fresh IMDM with or without 10% FBS. They were incubated for an additional 4 h at the same temperature with agitation. Ten percent FBS was added to the IMDM control culture just prior to microscopic observation at ×630 or ×1,000 magnification.
Time-lapse experiment.
Bone marrow cells were seeded in a Bioptechs petri dish prepared as previously described (48) in BMDM differentiation medium for 10 days. For time-lapse experiments, MEM-EBSS was replaced with Leibovitz's L15 medium, omitting penicillin and streptomycin. BMDM were then preincubated for 30 min at 37°C with 300 nM of LysoTracker red DND-99. Fixed hyphal or live CAI4-GFP cells were then added at an MOI of 1 to give a final LysoTracker red DND-99 concentration of 50 nM. Phase, green, and red images were captured every 10 min using the appropriate filters.
Immunofluorescence (percentage of phagocytosis).
Macrophages were seeded in 24-well plates as described earlier, and culture medium was replaced with warmed medium containing GFP-expressing C. albicans to obtain the desired MOI. At the end of interactions, cells were washed two times in culture medium and stained with an anti-Candida antibody as described previously (42). Epifluorescence was monitored using the appropriate filters, at ×400 magnification. The percentage of phagocytosis was determined by counting the number of macrophages containing at least one Candida cell divided by the total number of macrophages, using Openlab and ImageJ software. The percentage of phagocytosis was calculated from a total of 12 images from three independent experiments for each time point. Each image contained a mean of 120 BMDM or 225 RAW 264.7 cells.
Immunofluorescence (CD71 labeling).
BM cells were seeded on acid-washed coverslips at 3 × 105 cells per well in a six-well plate. Washed Candida cells were then incubated with macrophages at an MOI of 1 for the times indicated in Fig. 8. After a quick wash with PBS, cells were fixed for 5 min with fresh 4% paraformaldehyde in PBS, washed three times in PBS, permeabilized with 0.2% NP-40 in PBS for 10 min, washed, and blocked with StartingBlock (Pierce, Thermo Fisher Scientifique, Montreal, Canada). Coverslips were then incubated with a 1/100 dilution of PE-conjugated anti-mouse CD71 (transferrin receptor) or isotype controls (Cedarlane Laboratories, Burlington, Canada) in blocking buffer for 1 h at 37°C. They were then washed three times in PBS containing 0.05% Tween 20 and once with PBS, dried, and mounted in Prolong gold.
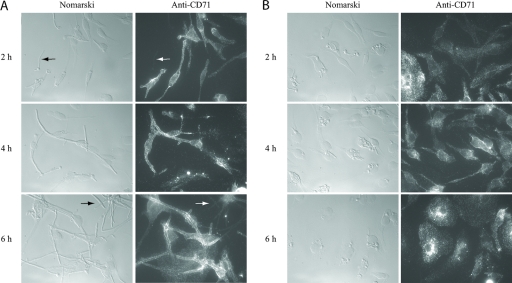
FIG. 8.
Transferrin receptor (CD71) staining of primary BMDM in the presence of Candida cells. SC5314 wild-type cells live (A) or fixed (in hyphal form) (B) were incubated for the indicated times at 37°C, 5% CO2 and stained with PE-labeled anti-CD71 antibodies as described in Materials and Methods. Cells were visualized at ×630 magnification. Arrows show unengulfed Candida cells that were not stained.
Statistics.
Unless otherwise stated, statistical analysis of the data was performed using Student's t test and results were considered significant at P values of <0.05.
Microarray data accession number.
Microarray data have been deposited into the National Center for Biological Information (NCBI)'s Gene Expression Omnibus (GEO) database under accession number GSE11399.
RESULTS
Macrophage phagocytosis is more efficient for Candida hyphal-form than yeast-form cells.
Morphological plasticity is one of the hallmarks of the human fungal pathogen Candida albicans (80). When C. albicans cells are incubated in macrophage medium at 37°C with 5% CO2, their phenotype switches from the yeast to the hyphal form. Hyphal forms of C. albicans appear to express molecules that allow for their recognition by mouse macrophages; as shown in Fig. 2, yeast cells were poorly recognized by these macrophages relative to hyphal cells. CAI4-GFP yeast or hyphal cells were fixed with paraformaldehyde to maintain their respective morphologies and incubated at an MOI of 5 with mouse BMDM for the times indicated in Fig. 2. They were then washed and stained with an anti-Candida polyclonal antibody, as described in Materials and Methods. Engulfed Candida, protected from primary antibody binding, remained green, whereas nonphagocytosed Candida became stained in red (or yellow in overlay with the GFP signal). Eighty percent of the BMDM contained at least one Candida hyphal cell at the 30-min time point, whereas only 10% contained yeast cells. A plateau was reached at the 60-min time point, with 90% phagocytosis. Phagocytosis of the yeast form was slower and proceeded at a linear rate of about 10% every 30 min. After a 3-h incubation, only 50% of BMDM contained yeast forms of Candida. This differential phagocytosis was also observed with the RAW 264.7 cell line (data not shown).
FIG. 2.
Differential phagocytosis of yeast and hyphal forms of Candida albicans. (A) CAI4-GFP yeast or hyphal forms were fixed and incubated with BMDM at an MOI of 5 for the indicated times. They were then stained with an anti-Candida polyclonal antibody (red), as described in Materials and Methods, and visualized at ×400 magnification. Engulfed Candida, protected from primary antibody binding, remained green, whereas nonphagocytosed Candida became yellow-red. Macrophage nuclei stained with Hoechst 33342 appear blue. (B) Percentages of BMDM containing at least one Candida cell at the indicated times.
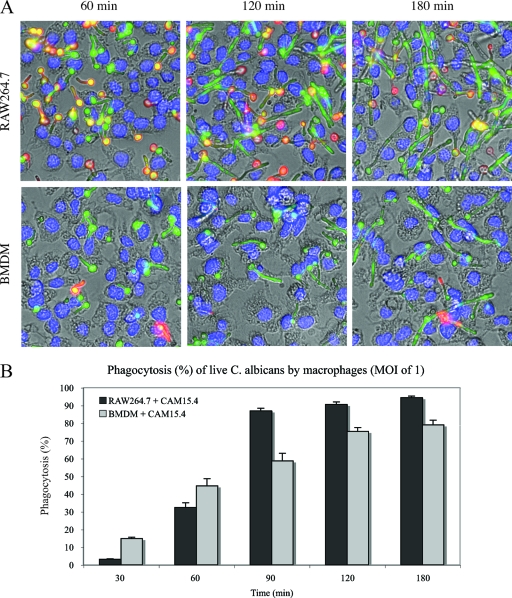
This distinction between yeast and hyphal cells was also observed for living cells. Figure 3 shows phagocytosis of live Candida at an MOI of 1 while cells were undergoing the morphological switch. The rate of phagocytosis was low at 30 min (Fig. 3B). BMDM phagocytosed Candida at early time points (15% and 45% at 30 and 60 min, respectively) more efficiently than did RAW 264.7 macrophages (4% and 32%, respectively). The number of macrophages containing Candida cells increased rapidly with time, to reach a plateau at 1.5 h for RAW 264.7 macrophages and at 2 h for BMDM. Beyond these time points, Candida cells escaped macrophages and were phagocytosed by neighboring macrophages, as shown in Fig. 3A (180-min time point).
FIG. 3.
Phagocytosis of live Candida by the RAW 264.7 macrophage line or primary mouse BMDM. (A) GFP-expressing Candida strain CAI4-GFP was grown in the presence of RAW 264.7 macrophages or BMDM at an MOI of 1 for the indicated times. They were stained as described in the legend for Fig. 2 and visualized at ×400 magnification. (B) Percentages of macrophages containing at least one CAI4-GFP cell at the indicated times.
Changes in C. albicans transcript abundance upon phagocytosis by primary and immortalized mouse macrophages.
The interaction between the fungal pathogen Candida albicans and cells of the innate immune system is associated with significant changes in gene expression in both fungal and mammalian cells (24, 26, 35, 46, 69). We used microarray analysis to investigate the transcriptional consequences of the engulfment of cells of the SC5314 strain of C. albicans by both primary macrophages (BMDM) and the macrophage line RAW 264.7. Because the number of phagocytosed Candida cells was still low after 30 min of incubation (Fig. 3B), we collected transcriptional profiling data after 1, 2, and 4 h. Overnight cultures of C. albicans were washed, counted, and diluted at the required concentration in the appropriate culture medium supplemented with 10% heat-inactivated FBS. The C. albicans cells were incubated at 37°C in the presence of 5% CO2 with or without macrophages at an MOI of 1. Preliminary studies using luciferase-expressing C. albicans strains indicated that these culture conditions allowed the analysis of the fungal cells responding to phagocytosis without severe nutrient limitation (unpublished observations). Transcriptional profiles from at least four independent experiments for each condition were analyzed and compared to profiles of four independent cultures of C. albicans grown alone under the same conditions.
As seen in Fig. 4, primary macrophages (BMDM) were much more effective than RAW 264.7 cells in eliciting a transcriptional response in the C. albicans cells. Nevertheless, both types of macrophage elicited similar responses in C. albicans, as evidenced by the significant overlap between the respective lists of significantly modulated genes (see Table S1 in the supplemental material for a list of annotated genes). Our results, especially those obtained with the BMDM, are similar to those previously observed in the studies of Lorenz et al. (46) and Fradin et al. (25) (see PMN in Fig. S1B in the supplemental material). Notably, many of the upregulated genes are associated with carbohydrate transport/metabolism/fermentation (HGT2, HGT12, HGT18, IFE2, ARO10, GAL1, GLK1, GLK4, ADH5, ICL1, SDH2, INO1, etc.) and oxidative stress (CIP1, CAT1, YHB1, CCP1, SOD3, etc.) (18). Only a few modulated genes were associated with the yeast-to-hypha transition transcription profile (54), since the control cells were also undergoing the yeast-to-hypha transition. Principal-component analysis (see Fig. S1B in the supplemental material) also shows significant correlations with other transcriptional profiles, including cyclic AMP-dependent stress responses and treatments with oxidative agents, high salt, and hydroxyurea, as well as downregulation of the Cdc5p Polo-like kinase (6, 18, 33). We also see significant correlations with the responses observed for mutants of the Gal4p and Tac1p transcription factors (43, 49). Interestingly, while we observe a downregulation of genes whose products are localized to the ribosomes, nucleolus, Golgi apparatus, and endoplasmic reticulum, phagocytosis induces an upregulation of peroxisomal and lysosomal gene products (see Fig. S1C in the supplemental material). After a 4-h incubation, upon pathogen escape from the macrophage phagosome, the majority of the modulated transcripts have started to recover their normal basal levels of expression, which probably reflects rebalancing of the cellular components as the pathogen returns to a more normal growth state.
FIG. 4.
Transcriptional profiling analysis. (A) Venn diagram showing the overlap between the numbers of genes with a statistically significant change in transcript abundance (>1.5-fold, P value of <0.05) following the coincubation of C. albicans with either BMDM or RAW 264.7 macrophages. (B) C. albicans genes (n = 1,129) that were significantly modulated following coincubation with macrophages at the indicated times (in hours) were separated by hierarchical clustering. Upregulated genes are colored in red and downregulated genes in green. Numbers in the color legend represent change (n-fold) in transcript abundance.
PRA1 transcripts are strongly induced upon phagocytosis.
Cell surface components (proteins, glycans, etc.) are molecules critical for the recognition of pathogens by immune cells. The modulation of confirmed or putative cell surface/membrane/secreted protein-encoding genes, such as PRA1, DDR48, CSH1, ECM4, HGT12, HGT18, CIP1, INO1, etc., can offer new insight into the molecular mechanisms involved in macrophage phagocytosis and/or Candida escape. One of the most highly upregulated genes identified in the pathogen upon engulfment by the macrophage was PRA1, a gene encoding a cell surface product that has been described as a fibrinogen binding protein (78). In vitro, the PRA1 (pH-regulated antigen 1) gene is activated, along with PHR1 (pH responsive) and its regulator RIM101 (regulator of IME2), when Candida cells are grown at an alkaline pH (15). Figure 5 shows a Northern blot analysis of C. albicans PRA1 and PHR1 and a control gene, TEF2. Panel A shows that when the SC5314 strain is grown in YPD at pH 6.0, the PRA1 and PHR1 mRNAs are not transcribed. However, when this C. albicans strain is grown in IMDM, pH 7.4 to 8.0, both transcripts are expressed. As shown in Fig. 2B, the PRA1 transcript was strongly upregulated upon phagocytosis by the RAW 264.7 mouse macrophage line (in D-10, pH 7.4), compared to Candida cells incubated in the same culture medium (D-10) without macrophages. In contrast, although transcription of the PHR1 gene was induced in macrophage culture medium, its expression remained unchanged during phagocytosis. This differential regulation of PRA1 and PHR1 in response to macrophages suggests that a regulatory event can activate PRA1 transcription specifically upon phagocytosis.
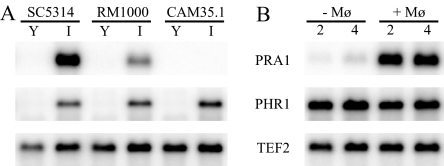
FIG. 5.
Northern blot analysis of PRA1, PHR1, and control gene TEF2. (A) Gene expression when Candida strains SC5314 (wild type), RM1000 (parent of the pra1-deleted strain), and CAM35.1 (pra1 null) were grown in YPD at pH 6.0 (Y) or in IMDM at pH 7.4 (I). (B) Gene expression of the SC5314 Candida strain grown in D10 medium, in the absence (−Mφ) or presence (+Mφ) of RAW 264.7 macrophages, for the indicated times (in hours).
Construction and characterization of a PRA1 knockout strain.
Pra1p, also described as mp58 or fibrinogen binding protein (Fbp1p) (45), is a candidate for binding to the Mac-1 macrophage receptor (CR3, CD11a/CD18) through fibrinogen, a known ligand for this macrophage receptor (81). Pra1p could then serve as a component that would facilitate C. albicans internalization and/or escape from macrophages. In order to study the role of this gene in the Candida-macrophage interaction, a pra1 knockout strain, CAM35.1, was constructed using a PCR-based cassette strategy, as described in Materials and Methods (Fig. 1A). The PRA1 deletion was confirmed by PCR analysis (Fig. 1B) and Northern blot analysis, as shown in Fig. 5A. The RM1000 parental strain and the CAM35.1 pra1 null mutant were grown in YPD or IMDM for 4 h and mRNA was isolated as described in Materials and Methods. The PRA1 and PHR1 genes were not expressed when both strains were grown in YPD liquid medium but were induced in RM1000 grown in IMDM with an alkaline pH. As expected, PHR1 but not PRA1 mRNA was expressed under alkaline conditions in the pra1-deleted strain CAM35.1.
Survival of the PRA1 null mutant in macrophages is similar to that of the wild-type strain in vitro.
Since PRA1 is highly upregulated upon phagocytosis, we tested whether the absence of this transcript/protein would change the fate of Candida survival in a macrophage end point dilution assay. The wild-type C. albicans strain SC5314 and the pra1 null strain CAM35.1 were serially diluted in the presence or absence of BMDM or the RAW 264.7 mouse macrophage line. Twenty-four hours later, colonies were counted and the survival rates were determined, as described previously (48). The survival rates of the pra1−/− strain CAM35.1 (60.5% ± 4.1% in BMDM and 37.1% ± 5.9% in RAW 264.7 cells) were not significantly different from the rates of the wild-type strain SC5314 (62.3% ± 6.3% in BMDM and 38.5% ± 3.1% in RAW 264.7 cells) (data represent percentages of survival ± standard deviations and are expressed as the numbers of colonies in the presence of BMDM [n = 8] or RAW 264.7 macrophages [n = 3] divided by the number of colonies in the absence of macrophages). The end point dilution assay measures survival at a low MOI; somewhat surprisingly, the survival of these Candida strains was higher in primary macrophages than in the RAW 264.7 cell line, perhaps as a consequence of the relative confluencies of the different macrophage monolayers.
Pra1p is not a major fibrinogen binding protein.
We used AF546-labeled fibrinogen as a tracer to detect Pra1p binding to fibrinogen. The direct binding of fibrinogen can then be followed by microscopy; this approach avoids the possible cross-reactivity of secondary antibodies often observed with indirect immunofluorescence staining techniques applied to C. albicans cells (personal observations). To test the contribution of Pra1p to fibrinogen binding, fibrinogen-binding quantification in the wild-type strain SC5314 was compared to that in the pra1 null strain CAM35.1, as described in Materials and Methods. Cells were grown for 2 h at 37°C in I-10 in the presence or absence of unlabeled fibrinogen, followed by AF546-labeled fibrinogen staining. As shown in Table 3, labeling was observed at similar levels in both strains, and a prior incubation of Candida cells with unlabeled human fibrinogen decreased the overall fluorescence intensity similarly in both strains, the difference between the two strains being statistically nonsignificant. Thus, the contribution of Pra1p to fibrinogen binding appears minor under these culture conditions.
TABLE 3.
AF546-fibrinogen binding quantification in Candida cells in the absence or presence of unlabeled fibrinogen
| Strain (genotype) | AF546-fibrinogen binding (RFU)a
|
Binding inhibition (%)b | |
|---|---|---|---|
| Without unlabeled fibrinogen | With unlabeled fibrinogen | ||
| SC5314 (wild type) | 4,407 ± 879 | 1,174 ± 295 | 73.4 ± 10.0 |
| CAM35.1 (pra1/pra1) | 4,056 ± 512 | 1,323 ± 197 | 67.4 ± 7.9 |
Data were calculated from 10 individual Candida hyphal tips for each of 10 microscopic fields (100 cells) per condition and are expressed as relative fluorescence units (RFU) ± standard deviations.
The percentage of inhibition by unlabeled fibrinogen was calculated as [1 − (mean RFU of cell with unlabeled fibrinogen/mean RFU of cells without unlabeled fibrinogen)] × 100.
Pra1p localization using a GFP-tagged protein.
CAM38.3, a strain expressing a C-terminal GFP-tagged Pra1p under its own promoter, was generated as described in Materials and Methods. Under inducing conditions of a 6- to 20-h incubation in IMDM at 30°C, Pra1p-GFP localized to the periphery of the cell, consistent with a cell membrane/wall localization (Fig. 6A, top right panel). Cell wall localization has already been reported through biochemical analysis of cell wall extracts (12). It should be noted that cell morphology is pseudohyphal when grown in this medium, compared to the yeastlike morphology when cells are grown in YPD at the same temperature (not shown). Although the mRNA is expressed early under inducing conditions (Fig. 5A), the Pra1-GFP protein could be observed microscopically from only 5 to 6 h postinduction (data not shown). As well, although the PRA1 transcript is rapidly and highly upregulated during Candida phagocytosis by RAW 264.7 macrophages, attempts to visualize Pra1-GFP fluorescence during macrophage phagocytosis were not successful, due to macrophage autofluorescence combined with the delay between mRNA transcription and protein detection. We have previously shown that, at an MOI of 1 under these culture conditions, macrophages die at 9 h postinfection (48).
FIG. 6.
Expression of Pra1p-GFP in the Candida strain CAM38.3. (A) CAM38.3 cells were grown at 30°C for 16 h in IMDM (pH 7.4), diluted, and then grown for an additional 4 h with (+FBS) or without FBS. Fluorescence was visualized at ×1,000 magnification. Arrows indicate representative freshly growing hyphal tips. (B) Percentages of cells expressing membrane-bound GFP. A total of 697 cells without FBS (−FBS) and 929 cells with FBS (+FBS) were counted.
Pra1-GFP expression is inhibited in the presence of serum.
CAM38.3 Candida cells were incubated overnight in IMDM at 30°C to prime Pra1-GFP expression. They were then diluted in fresh medium with or without FBS and incubated for an additional 4 h. As shown in Fig. 6A, membrane-localized GFP expression at the growing tips of Candida hyphae could be observed only in cells grown in IMDM alone. This expression was abrogated on newly formed cells grown in I-10. As shown in Fig. 6B, 90% of the cells grown in IMDM without FBS had membrane-localized Pra1-GFP expression, compared to 40% of the cells grown in the presence of FBS (including starting cells), although the addition of serum did not interfere with the pH of the medium, as monitored with the phenol red pH indicator included in the assay.
Colocalization of Pra1-GFP with labeled fibrinogen is a minor event.
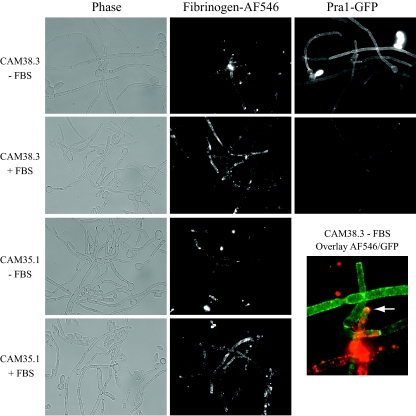
To test directly for Pra1p binding to labeled fibrinogen, we colocalized the GFP signal from the Pra1-GFP-expressing strain CAM38.3 with AF546-labeled fibrinogen. The GFP-expressing strain CAM38.3 and the pra1 null strain CAM35.1 were grown overnight at 37°C in IMDM supplemented or not supplemented with FBS and further incubated on ice with AF546-labeled human fibrinogen, washed, fixed, and mounted as described in Materials and Methods. As shown in Fig. 7, in the presence of FBS both the CAM38.3 wild-type strain and the CAM35.1 pra1 null mutant strain were similarly labeled by fibrinogen; both strains exhibited patchlike staining at the hyphal part of the cells. This staining was still observable in cells grown without FBS, although to a smaller extent. When cells were grown at a lower temperature (30°C) without FBS, labeled fibrinogen binding was rarely observed (data not shown). As shown earlier (Fig. 6), the presence of FBS abrogated the Pra1-GFP signal. Although some colocalization events between Pra1-GFP and labeled fibrinogen can occur (Fig. 7, overlay), each signal is generally unique. Fibrinogen staining is almost exclusively hyphal, while the Pra1-GFP signal is distributed extensively under inducing conditions.
FIG. 7.
Colocalization of Pra1p-GFP and AF546-fibrinogen. Strains CAM38.3 (Pra1p-GFP expressing) and CAM35.1 (pra1 null) were grown overnight at 37°C in IMDM (without FBS [−FBS]) or I-10 (with FBS [+FBS]) and then stained with AF546-labeled fibrinogen as described in Materials and Methods and visualized at ×630 magnification. The arrow indicates colocalization.
Live Candida cells inhibit phagosomal acidification.
During the engulfment of C. albicans cells, we detected induction of the PRA1 transcript, which in vitro is triggered by alkaline conditions, suggesting a possible block in phagosome acidification during maturation. To follow phagosome maturation, BMDM were preincubated with LysoTracker red DND-99, a fluorescent probe that labels and traces acidic organelles, mainly lysosomes, in living cells. Macrophages were then incubated with live cells or paraformaldehyde-fixed hyphal cells of the GFP-expressing C. albicans strain CAI4-GFP, and the fluorescence was monitored with time-lapse microscopy in order to follow lysosome fusion to the phagosomes. We chose paraformaldehyde fixation as this method is likely to preserve surface antigenicity. The majority (78%) of the live-Candida-containing phagosomes did not associate with LysoTracker red organelles (22.1% ± 9.6% did associate). As a consequence, Candida cells could escape from macrophages without apparent damage (see Movie S1 in the supplemental material). However, the majority (70.7% ± 8.1%) of the fixed-Candida-containing phagosomes did associate with the acidic compartment, leading to both a strong decrease in GFP fluorescence, which is sensitive to a pH lower than 5.5 (39), and the appearance of red fluorescent C. albicans cells due to the LysoTracker signal (see Movie S2 in the supplemental material). (Data were compiled from 16 individual 6-h time-lapse experiments [n = 3] for each of the Candida forms and represent the percentages of phagolysosomes [±standard deviations] containing GFP-expressing Candida cells tracked using LysoTracker red DND-99 fluorescent lysosome probe.)
Candida escapes from macrophages at the primary/early endosome stage.
Since Candida-containing phagosomes do not fuse with lysosomes before the pathogen is able to escape from the primary macrophages, we investigated the stage of the phagosome maturation during C. albicans engulfment. The transferrin receptor (CD71) is a marker of sorting/early endosomes. It binds iron-loaded transferrin at the cell surface and transports it into the cell. Under mildly acidic conditions (such as the early endosome), iron is released from transferrin and the CD71-apotransferrin complex recycles back to the outer cell membrane, where apotransferrin is released (65). Live or hyphal fixed C. albicans SC5314 cells and BMDM were incubated for the times indicated in Fig. 8 and then fixed, permeabilized, blocked, and stained with PE-labeled anti-mouse CD71, as described in Materials and Methods. As shown in Fig. 8A, the transferrin receptor was associated with the live-Candida-containing phagosome very early in the phagocytic process (2 h postinfection). It remained associated with the Candida-containing phagosome during the process of C. albicans hyphal elongation (4 h postinfection) and even during escape from the macrophage (6 h postinfection). C. albicans hyphal cells that had escaped from macrophages were still macrophage-CD71 coated, suggesting that phagosome maturation was blocked at an early stage. Candida cells seemed to sequester CD71-containing macrophage membranes, as the CD71 staining was concentrated around the Candida phagosome. Candida cells that were not phagocytosed were not stained by the anti-CD71 antibody (Fig. 8A). The association of CD71 with fixed Candida cells was minor (Fig. 8B).
DISCUSSION
Candida albicans is primarily a commensal organism of humans, and there is normally a balance between immune response and tolerance in normal immunocompetent individuals. However, when those defense mechanisms are perturbed there is a potential for host infection. Candida albicans cells can use transcriptional regulation to rapidly adapt to the hostile environment created by variations in pH and oxidative state caused by attack from immune cells. This adaptation is accompanied by posttranslational modifications, including those of cell surface components such as proteins and glycan structures that play important roles in the recognition of the pathogen by immune cells and thus in the outcome of the interaction.
Hyphal-form cells of Candida are rapidly phagocytosed by macrophages.
It has previously been shown that Candida yeast- or hyphal-form cells can be differentially recognized by host cells via distinct receptors (reviewed in reference 57). For example, dectin-1 is involved mainly in the recognition of β-glucans present in the yeast form of C. albicans (27, 30), whereas dectin-2 was shown to be involved in the recognition of its hyphal form (70). Under our experimental conditions, hyphal forms of Candida are phagocytosed more rapidly by macrophages (Fig. 2) than are yeast forms. As macrophages are known to bear both dectin-1 and -2 receptors at their cell surface, further experiments will be necessary to establish if relative receptor levels or perhaps other receptors or factors are responsible for the differential rates of engulfment.
Transient gene expression profile.
We have shown previously that, in vitro, cells of C. albicans strain SC5314 could survive and escape from phagocytosis by RAW 264.7 mouse macrophages (48). To clarify some of the mechanisms involved in Candida survival, we have investigated the effects of this interaction through gene expression profile analysis of two different types of mouse macrophages. We also compared the resulting transcriptional profiles with similar experiments produced by others. Upon phagocytosis, there is transient upregulated expression of genes associated with both carbohydrate metabolism/fermentation and oxidative stress. The majority of these transcripts dropped toward control levels by 4 h postinteraction. The transient nature of this gene expression is consistent with previously published data on C. albicans engulfed by macrophages (46) and reflects the rapid adaptation of Candida cells to a hostile environment. BMDM proved especially adept at eliciting a transcriptional response by the engulfed C. albicans cells, suggesting that the RAW 264.7 cell line may have lost some capacity.
We focused on one of the upregulated genes that was observed in all of these studies: PRA1, coding for a cell wall/membrane-associated protein known to be expressed at an alkaline pH in vitro (15, 71) (Fig. 5A) and to be positively regulated by the Rim101p transcription factor (15). The PRA1 transcript was strongly induced upon RAW 264.7 mouse macrophage phagocytosis of C. albicans cells (Fig. 5B), while the PHR1, RIM101, RIM8, and PHR2 genes, which are part of a high-pH-induced regulon that includes PRA1, were not measurably modulated upon RAW 264.7 phagocytosis (Table 4). However, in BMDM the regulon of PRA1, RIM101, RIM8, and PHR2 is upregulated (Table 4), consistent with the generally enhanced transcriptional response elicited from the pathogen by the BMDM compared to that elicited by the RAW 264.7 cells. Intriguingly, the PHR1 gene, which is normally coregulated with PRA1, is not upregulated in either macrophage line. Fradin et al. (25) derived similar conclusions about the distinct behaviors of PRA1 and PHR1 in vivo, in this case from the observation of a downregulation of PRA1 transcripts but not PHR1 transcripts after Candida exposure to human blood; this transcriptional response appears driven by granulocytes (24). This differential response of PRA1 to macrophage and granulocyte engulfment, as well as the upregulation in granulocytes of PHR2, an acidic-pH-regulated gene, might be explained by the efficient acidification of the Candida-containing granulocyte phagosomes in contrast to the response of macrophage phagosomes.
TABLE 4.
Change in selected transcripts associated with the pH response in macrophages
| Gene name | Fold change
|
|||||
|---|---|---|---|---|---|---|
| BMDM
|
RAW 264.7
|
|||||
| 1 h | 2 h | 4 h | 1 h | 2 h | 4 h | |
| PRA1 | 1.2 | 6.1 | 17.8 | 2.9 | 7.3 | 15.0 |
| PHR1 | 1.2 | 1.1 | 1.1 | 1.0 | 1.0 | 1.0 |
| PHR2 | 3.0 | 2.4 | 3.2 | 1.2 | 1.1 | 1.1 |
| RIM101 | 1.4 | 1.5 | 1.7 | 1.0 | 1.1 | 1.2 |
| RIM8 | 1.6 | 1.6 | 1.8 | 1.1 | 1.1 | 1.1 |
| RIM20 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
Pra1p is not a major fibrinogen binding protein.
Fibrinogen binding to C. albicans germ tubes was discovered 20 years ago (9), and further characterization led to the identification of a 68-kDa binding factor associated with the fungus (5). Later, a 58-kDa C. albicans component with affinity to fibrinogen was identified (10); polyclonal antibodies raised against this protein could block fibrinogen binding and cross-reacted with a cDNA product called FBP1 (3), which had a sequence identical to that of the PRA1 gene (45, 71). Therefore, it was suggested that Pra1p binds fibrinogen, but in the present study, we found that AF546-labeled human fibrinogen could still bind to a pra1 null mutant to the same extent as to the SC5314 wild-type strain (Table 3). Using a GFP-tagged Pra1p-expressing strain, we confirmed Pra1p membrane/cell wall localization, but this localization was inconsistent with fibrinogen binding. Pra1-GFP protein expression was induced in vitro under alkaline-pH conditions (IMDM) and could be visualized at the surfaces of both yeast cells and hyphae (Fig. 6 and 7), but expression was significantly abrogated in the presence of serum. However, under these culture conditions (IMDM), AF546-labeled human fibrinogen binding was observed only on the elongated, hyphal part of the yeast, with the extent of labeling increasing when C. albicans was actively grown in serum containing IMDM (Fig. 7). Fibrinogen binding was also observed in acidic media, such as YPD supplemented with FBS or Lee's medium supplemented or not supplemented with FBS (data not shown). Overall, the binding pattern was patchlike and resembled actin patch distribution, consistent with previous microscopic experiments (10, 75). Colocalization of Pra1-GFP with AF546-fibrinogen was rarely observed (Fig. 7, overlay). It is therefore unlikely that Pra1p is a major fibrinogen binding protein. Our data suggest that fibrinogen binding to molecules present on hyphal cells occurs and that the binding capacity is augmented upon FBS addition. This binding could be mediated by the 68-kDa moiety identified earlier (5) or through an O-linked oligosaccharide structure, as this possibility was not excluded by Casanova et al. (10). A possible cross-reactivity of the anti-mp58 antiserum with oligosaccharides might therefore explain the misidentification; removal of the fibrinogen binding protein (FBP1) annotation to the PRA1 transcript may thus be warranted. Because PRA1 is induced under alkaline conditions or macrophage engulfment and its protein expression is repressed in the presence of serum, a possible role for this cell membrane protein may be to act as a “protector” for Candida cell integrity. Recently, it was shown that soluble Pra1p could bind to the CD11b/CD18 (Mac-1) receptor on leukocytes, so that pra1 knockout cells were resistant to phagocytosis and killing by human polymorphonuclear leukocytes, and that addition of soluble Pra1p could prevent killing of wild-type Candida (72). However, it is not clear whether the protein itself or other components of the described 250-kDa complex were responsible for this activity. It is interesting to note that this increased resistance of the pra1 null mutant was not observed when macrophages were the engulfing cells.
Live Candida inhibited phagosomal maturation.
The elevated expression of the PRA1 transcript during C. albicans phagocytosis was somewhat surprising, given that normal phagosome maturation involves organelle acidification through sequential fusion with early endosomes, late endosomes, and finally lysosomes (37). In order to investigate whether phagosome maturation was perturbed during C. albicans engulfment, we labeled lysosomes with LysoTracker red and followed by time-lapse microscopy their fusion to the phagosomes containing GFP-expressing C. albicans cells (see Movies S1 and S2 in the supplemental material). We found that BMDM lysosomes fused efficiently (70%) to phagosomes containing fixed C. albicans cells; these fusion events were reduced (22%) in BMDM phagosomes containing live Candida, and the growth of the remaining live Candida cells did not seem to be impaired by phagocytosis. The same results were observed during RAW 264.7 phagocytosis (data not shown). Newman et al. (61) have shown similar results by comparing levels of phagocytosis of live and heat-killed Candida cells by human macrophages; the percentage of lysosome fusion was less than 20% upon phagocytosis of live Candida cells, compared to 95% for heat-killed cells. They also showed an increase in lysosome fusion when they incubated macrophages on collagen gel prior to contact with C. albicans. In contrast, Kaposzta et al. (38) have shown through LAMP1 indirect staining or Texas red dextran-loaded lysosomes that, in mouse peritoneal macrophages, Candida phagocytosis was followed by a rapid recruitment of lysosomes. This difference might be attributed to the different cell sources, Candida strains, or experimental procedures used. We have also shown that the efficient lysosome fusion leads to Candida cell death (see Movie S1 in the supplemental material) and that this lysosomal fusion seems to be perturbed at an early stage of phagosome maturation, as Candida cells that escaped macrophages were embedded in CD71-labeled membranes/phagosomes (Fig. 8A). This transferrin receptor staining was also reported by Kaposzta et al. (38). As transferrin is an iron transporter for the macrophages, it is tempting to speculate that Candida might sequester transferrin to the detriment of macrophages. Our transcript profiling (upregulation of SFU1, YER67, and YHB1 and downregulation of FTH1, FET3, SIT1, FRP1, CFL2, and CFL5) also agrees with the engulfed Candida finding the macrophage a high-iron environment (41). As well, the apparent coating of the escaped C. albicans hyphae with a CD71-labeled membrane may aid the ability of Candida cells to escape macrophage through “immunoevasion.” Many pathogens have developed mechanisms to avoid phagocytic destruction, either by avoiding engulfment through signaling inhibition (Escherichia coli) or actin polymerization inhibition (Yersinia spp.) or by impairment of phagosome maturation (Brucella, Legionella, Coxiella, Chlamydia, Leishmania, and Mycobacterium spp.) (reviewed in references 4 and 50). Our results, together with others (61), indicate that this mechanism might also be used by Candida albicans.
The mechanisms by which Candida avoids lysosomal fusion are still unknown. However, it is likely that the process is mediated by de novo synthesis of molecules upon macrophage phagocytosis, as paraformaldehyde-fixed Candida hyphae did not block this phagosome-lysosome fusion. Although PRA1 expression is positively regulated during Candida phagocytosis, it did not seem to be implicated in this fusion inhibition process, as the null mutant could still escape macrophages (data not shown) and the survival of the pra1 null mutant in vitro (see Results) was not significantly different from that of the wild-type strain.
Many investigations of the interaction of phagocytic cells with Candida albicans have used heat-killed, fixed, or antibiotic-impaired microorganisms. This study emphasizes the importance of including live/nonimpaired pathogens in this type of study, as the dynamic interaction and adaptation of Candida to its host environment are important in the final outcome of these interactions.
Supplementary Material
Acknowledgments
We are grateful to U. Oberholzer, D. Harcus, D. Dignard, J. S. Deneault, C. Bachewich, and H. Hogues for advice and helpful discussions, M. Mercier and C. Hélie for technical assistance, L. Bourget for flow cytometry analysis, A. Descôteaux for providing the RAW 264.7 macrophage line, and A. P. Brown and J. Morschhauser for providing plasmids.
This is National Research Council publication 47557.
Editor: A. Casadevall
Footnotes
Published ahead of print on 14 July 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alarco, A. M., I. Balan, D. Talibi, N. Mainville, and M. Raymond. 1997. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 27219304-19313. [DOI] [PubMed] [Google Scholar]
- 2.Alarco, A. M., A. Marcil, J. Chen, B. Suter, D. Thomas, and M. Whiteway. 2004. Immune-deficient Drosophila melanogaster: a model for the innate immune response to human fungal pathogens. J. Immunol. 1725622-5628. [DOI] [PubMed] [Google Scholar]
- 3.Alloush, H. M., J. L. Lopez-Ribot, and W. L. Chaffin. 1996. Dynamic expression of cell wall proteins of Candida albicans revealed by probes from cDNA clones. J. Med. Vet. Mycol. 3491-97. [PubMed] [Google Scholar]
- 4.Amer, A. O., and M. S. Swanson. 2002. A phagosome of one's own: a microbial guide to life in the macrophage. Curr. Opin. Microbiol. 556-61. [DOI] [PubMed] [Google Scholar]
- 5.Annaix, V., J. P. Bouchara, G. Tronchin, J. M. Senet, and R. Robert. 1990. Structures involved in the binding of human fibrinogen to Candida albicans germ tubes. FEMS Microbiol. Immunol. 2147-153. [DOI] [PubMed] [Google Scholar]
- 6.Bachewich, C., A. Nantel, and M. Whiteway. 2005. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol. Microbiol. 57942-959. [DOI] [PubMed] [Google Scholar]
- 7.Bates, S., H. B. Hughes, C. A. Munro, W. P. Thomas, D. M. MacCallum, G. Bertram, A. Atrih, M. A. Ferguson, A. J. Brown, F. C. Odds, and N. A. Gow. 2006. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 28190-98. [DOI] [PubMed] [Google Scholar]
- 8.Blasi, E., A. Mucci, R. Neglia, F. Pezzini, B. Colombari, D. Radzioch, A. Cossarizza, E. Lugli, G. Volpini, G. Del Giudice, and S. Peppoloni. 2005. Biological importance of the two Toll-like receptors, TLR2 and TLR4, in macrophage response to infection with Candida albicans. FEMS Immunol. Med. Microbiol. 4469-79. [DOI] [PubMed] [Google Scholar]
- 9.Bouali, A., R. Robert, G. Tronchin, and J. M. Senet. 1986. Binding of human fibrinogen to Candida albicans in vitro: a preliminary study. J. Med. Vet. Mycol. 24345-348. [DOI] [PubMed] [Google Scholar]
- 10.Casanova, M., J. L. Lopez-Ribot, C. Monteagudo, A. Llombart-Bosch, R. Sentandreu, and J. P. Martinez. 1992. Identification of a 58-kilodalton cell surface fibrinogen-binding mannoprotein from Candida albicans. Infect. Immun. 604221-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 2183-84. [DOI] [PubMed] [Google Scholar]
- 12.Choi, W., Y. J. Yoo, M. Kim, D. Shin, H. B. Jeon, and W. Choi. 2003. Identification of proteins highly expressed in the hyphae of Candida albicans by two-dimensional electrophoresis. Yeast 201053-1060. [DOI] [PubMed] [Google Scholar]
- 13.Crowe, J. D., I. K. Sievwright, G. C. Auld, N. R. Moore, N. A. Gow, and N. A. Booth. 2003. Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol. Microbiol. 471637-1651. [DOI] [PubMed] [Google Scholar]
- 14.Cunnick, J., P. Kaur, Y. Cho, J. Groffen, and N. Heisterkamp. 2006. Use of bone marrow-derived macrophages to model murine innate immune responses. J. Immunol. Methods 31196-105. [DOI] [PubMed] [Google Scholar]
- 15.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desjardins, M., J. E. Celis, G. van Meer, H. Dieplinger, A. Jahraus, G. Griffiths, and L. A. Huber. 1994. Molecular characterization of phagosomes. J. Biol. Chem. 26932194-32200. [PubMed] [Google Scholar]
- 17.Dignard, D., and M. Whiteway. 2006. SST2, a regulator of G-protein signaling for the Candida albicans mating response pathway. Eukaryot. Cell 5192-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enjalbert, B., A. Nantel, and M. Whiteway. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 141460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filler, S. G. 2006. Candida-host cell receptor-ligand interactions. Curr. Opin. Microbiol. 9333-339. [DOI] [PubMed] [Google Scholar]
- 20.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsyth, C. B., and H. L. Mathews. 2002. Lymphocyte adhesion to Candida albicans. Infect. Immun. 70517-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forsyth, C. B., E. F. Plow, and L. Zhang. 1998. Interaction of the fungal pathogen Candida albicans with integrin CD11b/CD18: recognition by the I domain is modulated by the lectin-like domain and the CD18 subunit. J. Immunol. 1616198-6205. [PubMed] [Google Scholar]
- 23.Fortier, A. H., and L. A. Falk. 2001. Isolation of murine macrophages. Curr. Protoc. Immunol. 1411. [DOI] [PubMed] [Google Scholar]
- 24.Fradin, C., P. De Groot, D. MacCallum, M. Schaller, F. Klis, F. C. Odds, and B. Hube. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56397-415. [DOI] [PubMed] [Google Scholar]
- 25.Fradin, C., M. Kretschmar, T. Nichterlein, C. Gaillardin, C. d'Enfert, and B. Hube. 2003. Stage-specific gene expression of Candida albicans in human blood. Mol. Microbiol. 471523-1543. [DOI] [PubMed] [Google Scholar]
- 26.Fradin, C., A. L. Mavor, G. Weindl, M. Schaller, K. Hanke, S. H. Kaufmann, H. Mollenkopf, and B. Hube. 2007. The early transcriptional response of human granulocytes to infection with Candida albicans is not essential for killing but reflects cellular communications. Infect. Immun. 751493-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gantner, B. N., R. M. Simmons, and D. M. Underhill. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 241277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaur, N. K., and S. A. Klotz. 1997. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 655289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gola, S., R. Martin, A. Walther, A. Dunkler, and J. Wendland. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 201339-1347. [DOI] [PubMed] [Google Scholar]
- 30.Gow, N. A., M. G. Netea, C. A. Munro, G. Ferwerda, S. Bates, H. M. Mora-Montes, L. Walker, T. Jansen, L. Jacobs, V. Tsoni, G. D. Brown, F. C. Odds, J. W. Van der Meer, A. J. Brown, and B. J. Kullberg. 2007. Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 1961565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffiths, G. 2004. On phagosome individuality and membrane signalling networks. Trends Cell Biol. 14343-351. [DOI] [PubMed] [Google Scholar]
- 32.Hampton, M. B., A. J. Kettle, and C. C. Winterbourn. 1998. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 923007-3017. [PubMed] [Google Scholar]
- 33.Harcus, D., A. Nantel, A. Marcil, T. Rigby, and M. Whiteway. 2004. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell 154490-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heidenreich, F., and M. P. Dierich. 1985. Candida albicans and Candida stellatoidea, in contrast to other Candida species, bind iC3b and C3d but not C3b. Infect. Immun. 50598-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, Q., D. Liu, P. Majewski, L. C. Schulte, J. M. Korn, R. A. Young, E. S. Lander, and N. Hacohen. 2001. The plasticity of dendritic cell responses to pathogens and their components. Science 294870-875. [DOI] [PubMed] [Google Scholar]
- 36.Ibata-Ombetta, S., T. Idziorek, P. A. Trinel, D. Poulain, and T. Jouault. 2003. Role of phospholipomannan in Candida albicans escape from macrophages and induction of cell apoptosis through regulation of bad phosphorylation. Ann. N. Y. Acad. Sci. 1010573-576. [DOI] [PubMed] [Google Scholar]
- 37.Jutras, I., and M. Desjardins. 2005. Phagocytosis: at the crossroads of innate and adaptive immunity. Annu. Rev. Cell Dev. Biol. 21511-527. [DOI] [PubMed] [Google Scholar]
- 38.Kaposzta, R., L. Marodi, M. Hollinshead, S. Gordon, and R. P. da Silva. 1999. Rapid recruitment of late endosomes and lysosomes in mouse macrophages ingesting Candida albicans. J. Cell Sci. 1123237-3248. [DOI] [PubMed] [Google Scholar]
- 39.Kneen, M., J. Farinas, Y. Li, and A. S. Verkman. 1998. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 741591-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194398-405. [DOI] [PubMed] [Google Scholar]
- 41.Lan, C. Y., G. Rodarte, L. A. Murillo, T. Jones, R. W. Davis, J. Dungan, G. Newport, and N. Agabian. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 531451-1469. [DOI] [PubMed] [Google Scholar]
- 42.Levitin, A., A. Marcil, G. Tettweiler, M. J. Laforest, U. Oberholzer, A. M. Alarco, D. Y. Thomas, P. Lasko, and M. Whiteway. 2007. Drosophila melanogaster Thor and response to Candida albicans infection. Eukaryot. Cell 6658-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, T. T., S. Znaidi, K. S. Barker, L. Xu, R. Homayouni, S. Saidane, J. Morschhauser, A. Nantel, M. Raymond, and P. D. Rogers. 2007. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot. Cell 62122-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90939-949. [DOI] [PubMed] [Google Scholar]
- 45.Lopez-Ribot, J. L., P. Sepulveda, A. M. Cervera, P. Roig, D. Gozalbo, and J. P. Martinez. 1997. Cloning of a cDNA fragment encoding part of the protein moiety of the 58-kDa fibrinogen-binding mannoprotein of Candida albicans. FEMS Microbiol. Lett. 157273-278. [DOI] [PubMed] [Google Scholar]
- 46.Lorenz, M. C., J. A. Bender, and G. R. Fink. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 31076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magee, B. B., and P. T. Magee. 1997. WO-2, a stable aneuploid derivative of Candida albicans strain WO-1, can switch from white to opaque and form hyphae. Microbiology 143289-295. [DOI] [PubMed] [Google Scholar]
- 48.Marcil, A., D. Harcus, D. Y. Thomas, and M. Whiteway. 2002. Candida albicans killing by RAW 264.7 mouse macrophage cells: effects of Candida genotype, infection ratios, and gamma interferon treatment. Infect. Immun. 706319-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martchenko, M., A. Levitin, H. Hogues, A. Nantel, and M. Whiteway. 2007. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr. Biol. 171007-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meresse, S., O. Steele-Mortimer, E. Moreno, M. Desjardins, B. Finlay, and J. P. Gorvel. 1999. Controlling the maturation of pathogen-containing vacuoles: a matter of life and death. Nat. Cell Biol. 1E183-E188. [DOI] [PubMed] [Google Scholar]
- 51.Morschhauser, J., S. Michel, and J. Hacker. 1998. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol. Gen. Genet. 257412-420. [DOI] [PubMed] [Google Scholar]
- 52.Murciano, C., E. Villamon, D. Gozalbo, P. Roig, J. E. O'Connor, and M. L. Gil. 2006. Toll-like receptor 4 defective mice carrying point or null mutations do not show increased susceptibility to Candida albicans in a model of hematogenously disseminated infection. Med. Mycol. 44149-157. [DOI] [PubMed] [Google Scholar]
- 53.Murciano, C., E. Villamon, A. Yanez, J. E. O'Connor, D. Gozalbo, and M. L. Gil. 2006. Impaired immune response to Candida albicans in aged mice. J. Med. Microbiol. 551649-1656. [DOI] [PubMed] [Google Scholar]
- 54.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 133452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nantel, A., T. Rigby, H. Hogues, and M. Whiteway. 2007. Microarrays for studying pathogenicity in Candida albicans, p. 181-209. In K. Kavanagh (ed.), Medical mycology. Cellular and molecular techniques. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 56.Negredo, A., L. Monteoliva, C. Gil, J. Pla, and C. Nombela. 1997. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143297-302. [DOI] [PubMed] [Google Scholar]
- 57.Netea, M. G., G. D. Brown, B. J. Kullberg, and N. A. Gow. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 667-78. [DOI] [PubMed] [Google Scholar]
- 58.Netea, M. G., N. A. Gow, C. A. Munro, S. Bates, C. Collins, G. Ferwerda, R. P. Hobson, G. Bertram, H. B. Hughes, T. Jansen, L. Jacobs, E. T. Buurman, K. Gijzen, D. L. Williams, R. Torensma, A. McKinnon, D. M. MacCallum, F. C. Odds, J. W. Van der Meer, A. J. Brown, and B. J. Kullberg. 2006. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Investig. 1161642-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Netea, M. G., R. Sutmuller, C. Hermann, C. A. Van der Graaf, J. W. Van der Meer, J. H. van Krieken, T. Hartung, G. Adema, and B. J. Kullberg. 2004. Toll-like receptor 2 suppresses immunity against Candida albicans through induction of IL-10 and regulatory T cells. J. Immunol. 1723712-3718. [DOI] [PubMed] [Google Scholar]
- 60.Netea, M. G., C. A. Van Der Graaf, A. G. Vonk, I. Verschueren, J. W. Van Der Meer, and B. J. Kullberg. 2002. The role of Toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J. Infect. Dis. 1851483-1489. [DOI] [PubMed] [Google Scholar]
- 61.Newman, S. L., B. Bhugra, A. Holly, and R. E. Morris. 2005. Enhanced killing of Candida albicans by human macrophages adherent to type 1 collagen matrices via induction of phagolysosomal fusion. Infect. Immun. 73770-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pitt, A., L. S. Mayorga, P. D. Stahl, and A. L. Schwartz. 1992. Alterations in the protein composition of maturing phagosomes. J. Clin. Investig. 901978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poulain, D., and T. Jouault. 2004. Candida albicans cell wall glycans, host receptors and responses: elements for a decisive crosstalk. Curr. Opin. Microbiol. 7342-349. [DOI] [PubMed] [Google Scholar]
- 65.Richardson, D. R., and P. Ponka. 1997. The molecular mechanisms of the metabolism and transport of iron in normal and neoplastic cells. Biochim. Biophys. Acta 13311-40. [DOI] [PubMed] [Google Scholar]
- 66.Romani, L., F. Bistoni, and P. Puccetti. 2003. Adaptation of Candida albicans to the host environment: the role of morphogenesis in virulence and survival in mammalian hosts. Curr. Opin. Microbiol. 6338-343. [DOI] [PubMed] [Google Scholar]
- 67.Romani, L., F. Bistoni, and P. Puccetti. 2002. Fungi, dendritic cells and receptors: a host perspective of fungal virulence. Trends Microbiol. 10508-514. [DOI] [PubMed] [Google Scholar]
- 68.Romani, L., C. Montagnoli, S. Bozza, K. Perruccio, A. Spreca, P. Allavena, S. Verbeek, R. A. Calderone, F. Bistoni, and P. Puccetti. 2004. The exploitation of distinct recognition receptors in dendritic cells determines the full range of host immune relationships with Candida albicans. Int. Immunol. 16149-161. [DOI] [PubMed] [Google Scholar]
- 69.Rubin-Bejerano, I., I. Fraser, P. Grisafi, and G. R. Fink. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. USA 10011007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sato, K., X. L. Yang, T. Yudate, J. S. Chung, J. Wu, K. Luby-Phelps, R. P. Kimberly, D. Underhill, P. D. Cruz, Jr., and K. Ariizumi. 2006. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J. Biol. Chem. 28138854-38866. [DOI] [PubMed] [Google Scholar]
- 71.Sentandreu, M., M. V. Elorza, R. Sentandreu, and W. A. Fonzi. 1998. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans. J. Bacteriol. 180282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soloviev, D. A., W. A. Fonzi, R. Sentandreu, E. Pluskota, C. B. Forsyth, S. Yadav, and E. F. Plow. 2007. Identification of pH-regulated antigen 1 released from Candida albicans as the major ligand for leukocyte integrin {alpha}Mbeta2. J. Immunol. 1782038-2046. [DOI] [PubMed] [Google Scholar]
- 73.Sundstrom, P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4461-469. [DOI] [PubMed] [Google Scholar]
- 74.Szabo, I., L. Guan, and T. J. Rogers. 1995. Modulation of macrophage phagocytic activity by cell wall components of Candida albicans. Cell. Immunol. 164182-188. [DOI] [PubMed] [Google Scholar]
- 75.Tronchin, G., R. Robert, A. Bouali, and J. M. Senet. 1987. Immunocytochemical localization of in vitro binding of human fibrinogen to Candida albicans germ tube and mycelium. Ann. Inst. Pasteur Microbiol. 138177-187. [DOI] [PubMed] [Google Scholar]
- 76.Tushinski, R. J., I. T. Oliver, L. J. Guilbert, P. W. Tynan, J. R. Warner, and E. R. Stanley. 1982. Survival of mononuclear phagocytes depends on a lineage-specific growth factor that the differentiated cells selectively destroy. Cell 2871-81. [DOI] [PubMed] [Google Scholar]
- 77.Viudes, A., A. Lazzell, S. Perea, W. R. Kirkpatrick, J. Peman, T. F. Patterson, J. P. Martinez, and J. L. Lopez-Ribot. 2004. The C-terminal antibody binding domain of Candida albicans mp58 represents a protective epitope during candidiasis. FEMS Microbiol. Lett. 232133-138. [DOI] [PubMed] [Google Scholar]
- 78.Viudes, A., S. Perea, and J. L. Lopez-Ribot. 2001. Identification of continuous B-cell epitopes on the protein moiety of the 58-kilodalton cell wall mannoprotein of Candida albicans belonging to a family of immunodominant fungal antigens. Infect. Immun. 692909-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walther, A., and J. Wendland. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42339-343. [DOI] [PubMed] [Google Scholar]
- 80.Whiteway, M., and C. Bachewich. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61529-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yakubenko, V. P., D. A. Solovjov, L. Zhang, V. C. Yee, E. F. Plow, and T. P. Ugarova. 2001. Identification of the binding site for fibrinogen recognition peptide gamma 383-395 within the alpha(M)I-domain of integrin alpha(M)beta2. J. Biol. Chem. 27613995-14003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.