Abstract
In the urinary tract, the innate immune system detects conserved bacterial components and responds to infection by activating the proinflammatory transcription factor NF-κB, resulting in cytokine secretion and neutrophil recruitment. Uropathogenic Escherichia coli (UPEC), however, has been shown to evade the host innate immune response by suppressing NF-κB activation in urothelial cells, which results in decreased cytokine secretion and increased urothelial apoptosis. To understand the molecular basis of UPEC modulation of inflammation, we performed a genetic screen with UPEC strain NU14 to identify genes which are required for modulation of urothelial cytokine secretion. Disruption of ampG (peptidoglycan permease), waaL (lipopolysaccharide O antigen ligase), or alr (alanine racemase) resulted in increased urothelial interleukin-8 (IL-8) and IL-6 release from urothelial cell cultures. Targeted deletion of these genes also resulted in elevated urothelial cytokine production during UPEC infection. Conditioned media from bacterial cultures of NU14 ΔampG and NU14 ΔwaaL contained a heat-stable factor(s) which stimulated greater urothelial IL-8 secretion than that in NU14-conditioned medium. In a mouse model of urinary tract infection, NU14 ΔampG, NU14 ΔwaaL, and NU14 Δalr were attenuated compared to wild-type NU14 and showed reduced fitness in competition experiments. Instillation of NU14 ΔampG or NU14 ΔwaaL increased bladder neutrophil recruitment, indicating that enhanced urothelial cytokine secretion during urinary tract infection results in an altered host response. Thus, UPEC evasion of innate immune detection of bacterial components, such as lipopolysaccharide and peptidoglycan fragments, is likely an important factor in the ability of UPEC to colonize the urinary tract.
The host innate immune system utilizes a family of pattern recognition receptors (PRRs) that detect invading pathogens by recognition of evolutionarily conserved components. Recognition of these pathogen-associated molecular patterns (PAMPs) through PRR family members, including the Toll-like receptors (TLRs) and nucleotide binding and oligomerization domain-like receptors, results in activation of antimicrobial responses, such as the production of antimicrobial peptides and secretion of proinflammatory cytokines (18, 29, 45). Many bacterial pathogens, however, have evolved strategies for subverting the host innate immune system by evading detection by PRRs and/or disruption of the downstream cellular signaling pathways (8, 30, 38).
Infection of the urinary tract by uropathogenic Escherichia coli (UPEC), the most frequent cause of urinary tract infection (UTI), is associated with a robust innate immune response characterized by the production of inflammatory cytokines and chemokines by the urothelium. The production of inflammatory cytokines and chemokines results in the rapid recruitment of neutrophils into the bladder lumen and in bacterial clearance (21, 22, 41). The activation of the innate immune response in the urinary tract is dependent upon PRR recognition of UPEC PAMPs, which include lipopolysaccharide (LPS), flagella, type 1 pili, and pap pili, as well as an unknown antigen recognized by TLR11 in mice but not humans (3, 19, 23, 51).
Some UPEC isolates, however, can suppress the activation of components of the innate immune response in the urinary tract. The cystitis isolates UPEC strains NU14 and CFT073 suppress NF-κB activation and interleukin-6 (IL-6) and IL-8 secretion in both urothelial cell cultures and nonurothelial cell types (10, 13, 24, 26, 32). Suppression of NF-κB activation by UPEC results in enhanced type I pilus-mediated apoptosis of urothelial cultures and in decreased levels of inflammatory cytokine production and neutrophil recruitment in vivo compared to those in nonsuppressor E. coli strains (10, 26, 31, 32). TLR4-dependent detection of UPEC LPS, initially by urothelial cells and later by neutrophils, is essential for urothelial IL-6 and IL-8 secretion and bacterial clearance (4, 24, 41, 42). Hunstad et al. recently identified several genes required for suppression of cytokine secretion in UPEC isolate UTI89, including the rfa and rfb gene clusters, which encode LPS biosynthesis enzymes, and the surA gene, which encodes a periplasmic prolyl isomerase (26). Recently, Cirl et al. reported that a newly described secreted toxin in CFT073, TcpC, contains a Toll/IL-1 receptor domain and inhibits MyD88-dependent cytokine secretion in urothelial cell cultures (13). RS218, a neonatal meningitis E. coli isolate closely related to UPEC strain NU14, requires OmpA expression to block NF-κB activation and the secretion of Mip-1β, IL-1β, and IL-8 from brain microvascular cells (44).
In order to identify potential novel urothelial innate immune pathways capable of detecting UPEC and to further elucidate the mechanisms by which UPEC evades recognition, we employed a genetic screen to isolate NU14 mutants with enhanced urothelial IL-8 secretion. We identified three genes, ampG, waaL, and alr, which encode proteins that function in the synthesis of outer membrane and peptidoglycan layer components, common targets for surveillance by PRRs. Furthermore, ampG and waaL were required for efficient colonization of the mouse bladder. Our data suggest that UPEC evasion of urothelial innate immune responses is dependent upon minimizing the production or recognition of components which are targets of PRRs.
MATERIALS AND METHODS
Bacterial strains and culture.
The strains and plasmids used in this study are shown in Table 1. All E. coli strains were cultured at 37°C in LB-Miller broth under static conditions for 48 h to promote the surface expression of type 1 pili (36). MG1655 is a fecal isolate, and NU14 (O18:K1:H7) is a streptomycin-resistant E. coli strain from the B2 clonal group isolated from a cystitis patient (11, 25). NU14-1 is a chloramphenicol-resistant fimH mutant of NU14 and is defective in type 1 pilus-mediated adherence (33). Antibiotics were added at the following appropriate concentrations: 100 μg/ml for streptomycin, 30 μg/ml for chloramphenicol, and 100 μg/ml for kanamycin. The deletion mutant strains were examined for growth defects in LB broth and minimal medium, and the presence of functional type 1 pili was confirmed by mannose-sensitive hemagglutination of guinea pig erythrocytes (17, 25). Bacterial culture supernatants were prepared by filtration of 40 ml of bacteria culture grown in EpiLife medium (Invitrogen) overnight through a 0.22-μm nylon filter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Plasmid or strain | Characteristics | Reference or source |
|---|---|---|
| Plasmids | ||
| pKD3 | PCR template for generation of chloramphenicol resistance cassette for λ red deletion mutagenesis | 14 |
| pKD46 | Expression of λ red recombinase for homologous recombination in E. coli | 14 |
| pFLAG-MAC | IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression of N-terminal FLAG fusion proteins | Sigma-Aldrich Co. |
| pampG | Derivative of pFLAG-MAC expressing AmpG | This study |
| pwaaL | Derivative of pFLAG-MAC expressing WaaL | This study |
| E. coli strains | ||
| NU14 | Clinical UPEC isolate (urine) | 25 |
| NU14 ΔampG | Derived from NU14 with targeted deletion of ampG | This study |
| NU14 ΔwaaL | Derived from NU14 with targeted deletion of waaL | This study |
| NU14 Δalr | Derived from NU14 with targeted deletion of alr | This study |
| NU14-1 | Derived from NU14 with insertion into fimH; defective type 1 pilus-mediated adherence | 33 |
| MG1655 | Fecal E. coli K-12 isolate | 11 |
| 15E6 | Derived from NU14 by random mutagenesis via conjugative transposition | This study |
| 22H12 | Derived from NU14 by random mutagenesis via conjugative transposition | This study |
| 25D9 | Derived from NU14 by random mutagenesis via conjugative transposition | This study |
| 40G5 | Derived from NU14 by random mutagenesis via conjugative transposition | This study |
Primers and cloning.
Primers (Integrated DNA Technologies) were designed using Lasergene software (DNASTAR) (Table 2). All DNA sequence analyses were performed by the Northwestern University Biotechnology Laboratory. AmpliTaq Gold (Applied Biosystems Inc.) was used to amplify all PCR products according to the manufacturer's recommended protocols. All PCRs were performed using an MJ Research PTC-200 thermocycler, and PCR products were visualized by running the products in 1% Tris-acetate-EDTA-agarose gels containing ethidium bromide.
TABLE 2.
PCR primers used in this study
| Primer and use | Sequence (5′→3′)a |
|---|---|
| Arbitrary PCR | |
| P2 | ATGACAAGATGTGTATCCACC |
| P11 | CTTGACGAGTTCTGAGCGGG |
| P12 | CAGCGCATCGCCTTCTATCGC |
| P20 | CCGCGGTGGAGCTCC |
| ARB1 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT |
| ARB2 | GGCCACGCGTCGACTAGTAC |
| ARB6 | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC |
| λ red deletion | |
| delampG forward | ATGTCCAGTCAATATTTACGTATTTTTCAACAGCCGCGTTTGTGAGGCTGGAGCTGCTTC |
| delampG reverse | TTACGTCAGATGTGTTTTTCGTAGCGCCAGATAATCCAGCCATATGAATATCCTCCTTAG |
| delwaaL forward | ATGTCGTTTTGTTGGAATGAAATTAACTCTGGTGTCAAGTTGTGAGGCTGGAGCTGCTTC |
| delwaaL reverse | TTACTTATCTAATAAACATTGGTCCGATTGTACTTTAAAACATATGAATATCCTCCTTAG |
| delalr forward | ATGCAAGCGGCAACTGTTGTGATTAACCGCCGCGCTCTGCTGTGAGGCTGGAGCTGCTTC |
| delalr reverse | TTAATCCACGTATTTCATCGCGACTCTTGAGGTCAGGCGCCATATGAATATCCTCCTTAG |
| λ red deletion verification | |
| ampG upstream forward | GCGCGTTAATTTCTGCCCTCTGG |
| ampG upstream reverse | CTTGCCCGCCTGATGAATGCTC |
| ampG downstream forward | AGGCGGGCAAGAATGTGAATAAAG |
| ampG downstream reverse | CAAACGCTAAGCCGCACAAAAGAA |
| waaL upstream forward | TGTTTCCATTGGCCTTTCTTCTTT |
| waaL upstream reverse | ATCCGGCCTTTATTCACATTCTTG |
| waaL downstream forward | AAACTGCCGGAAATCGTCGTGGTA |
| waaL downstream reverse | TCCGGGATTAATTGGTGGGTATGG |
| alr upstream forward | GATAAAGCGAGGGGGTGAGGTAAT |
| alr upstream reverse | GGCAGGGCGGGGCGTAAGG |
| alr downstream forward | GCCCCGCCCTGCCACTCA |
| alr downstream reverse | GTAACCGCCTGCCACGACGACA |
| Cloning | |
| ampG forward | CGAAAGCTTTCCAGTCAATATTTACGTATTTTTCAACAGC |
| ampG reverse | CGAGAATTCTCCATCAGATGTGTTTTTCGTAGCGCC |
| waaL forward | CGAAAGCTTTCGTTTTGTTGGAATGAAATTAACTCTGG |
| waaL reverse | CGAGAATTCTCTTACTTATCTAATAAACATTGGTCCGATTGTACTT |
N = any nucleotide.
Cell lines and culture.
The urothelial cell line TEU-1 was established by generating a primary epithelial culture from a healthy human ureter, followed by immortalization, as previously described (32). TEU-1 cells were maintained in EpiLife medium supplemented with human keratinocyte growth supplement (Invitrogen) and T24 human bladder carcinoma cells (ATCC) were grown in RPMI (MediaTech) with 10% fetal bovine serum (HyClone) and penicillin-streptomycin (MediaTech) in a 5% CO2 and 37°C atmosphere.
Transposon mutant library generation.
A library of NU14 insertional mutants was generated by transposition via conjugation with S17-1 λpir/pBSL180 (1). The plasmid contains a mini-Tn10-based transposon carrying a kanamycin resistance marker used for random mutagenesis in gram-negative bacterial species. NU14 was transformed with pUC18, which carries an ampicillin resistance marker to serve as a counterselection marker for conjugation. After conjugation of NU14/pUC18 and S17-1 λpir/pBSL180, the resulting transconjugates were isolated by being plated under selection on LB agar containing streptomycin and kanamycin. Colonies were picked, grown in 96-well polypropylene plates (Nunc) overnight, and stored as 30% glycerol stocks at −80°C. The library of approximately 5,600 mutants was screened in groups of 8 to 12 plates. The library was grown overnight in 300-μl U-bottomed 96-well plates, and 10 μl of each bacterial culture was added to 96-well plates containing TEU-1 cells in 90 μl of EpiLife medium. The infections were carried out for 4 hours, culture supernatants were harvested, and IL-8 secretion was measured by enzyme-linked immunosorbent assay (ELISA) (R&D Systems). Mutants which induced IL-8 levels that were more than twice the standard deviation above the plate average of IL-8 levels were selected for additional infection experiments in six-well plates to verify the phenotype. Ultimately, four NU14 mutants were identified (15E6, 22H12, 25D9, and 40G5) which induced significantly elevated IL-8 secretion from urothelial cells.
Arbitrary PCR and transposon site identification.
Arbitrary PCR was employed to identify the genomic location of the mini-Tn10 insertion site in the four identified NU14 mutants (49). Genomic DNAs were isolated from the four NU14 transposon mutants which showed increased stimulation of urothelial IL-8 secretion compare to wild-type NU14. Genomic DNA was used as the template for the arbitrary PCR protocol. The resulting PCR products were isolated by electrophoresis in a 1% Tris-acetate-EDTA-agarose gel followed by purification of the excised gel slices (Millipore). The PCR products were sequenced using the corresponding arbitrary PCR primers, and the resulting sequence information was used in nBLAST searches of the NCBI and University of Wisconsin-Madison (http://www.genome.wisc.edu/) databases for similar DNA sequences. The identification of the transposon insertion site was confirmed by PCR, using genomic DNAs from the mutants and both primers flanking the insertion site and primer sets where one primer was specific for the transposon and the other was specific for a region immediately surrounding either the 5′ or 3′ insertion site.
Targeted deletion mutagenesis.
The lambda red homologous recombination system was adapted to generate site-directed deletion mutants of UPEC strain NU14 (14). NU14 was transformed with pKD46, a plasmid which encodes the high-efficiency recombination system derived from lambda phage. PCR products were generated using PuReTaq Ready-to-Go PCR beads (Pharmacia), with primers containing 40 bp of sequence homologous to the 5′ and 3′ ends of the coding region of the gene targeted for deletion and with pKD3 as the template. The resulting PCR products were isolated using a Qiaquick PCR purification kit (Qiagen), transformed into electrocompetent NU14/pKD46 cells, and plated on LB agar containing chloramphenicol to select for the genomic recombination resulting in the deletion mutation. All deletion mutants were confirmed by PCR, using primers which were designed to amplify sequences flanking the 5′ and 3′ ends of the marker insertion and the chloramphenicol resistance marker.
Generation of complementation constructs.
Genomic DNA was isolated from NU14 and used as a template for PCR. The resulting PCR products for the ampG and waaL genes were isolated using a Qiaquick PCR purification kit (Qiagen), digested with HindIII and EcoRI (New England Biolabs), purified, and ligated with HindIII/EcoRI-digested pFLAG-MAC plasmid (Sigma) overnight at 14°C, using T4 ligase (New England Biolabs). The ligation mixture was transformed into XL-10 Gold electrocompetent E. coli cells (Stratagene) according to the manufacturer's recommendations and plated on selective medium. Insertion of the PCR products was confirmed by enzymatic digestion and gel electrophoresis, and the plasmid constructs were transformed into the corresponding deletion mutant strains.
Cytokine secretion assays.
TEU-1 cells were cultured in six-well plates in medium without antibiotics prior to infection with bacteria. We prepared bacterial suspensions in phosphate-buffered saline and used them to create inocula containing equivalent numbers of E. coli cells in antibiotic-free medium, determined by measuring the absorbance at 600 nm. The adjusted bacterial suspensions were added to each well, and cells were then returned to the incubator. Urothelial IL-8 and IL-6 secretion was determined following 4 h of treatment at a multiplicity of infection of 250:1 (bacteria to cells). Bacterial culture supernatants were added to cell cultures after 1:4 dilution in fresh EpiLife growth medium. Where noted, the medium was supplemented with 10 ng/ml recombinant human tumor necrosis factor alpha (TNF-α) (Biosource), 1 μg/ml O55:B5 LPS (Sigma), or 2.5 ng/ml of human recombinant IL-1β (Calbiochem) as a control. Culture supernatants were collected, cleared by centrifugation, and assayed for secreted IL-8 or IL-6 by ELISA (R&D Systems).
Bacterial colonization experiments.
Six- to 8-week-old female C57BL/6 mice were instilled while under isoflurane (Baxter Inc.) anesthesia via transurethral catheter with a volume of 10 μl containing 1 × 108 CFU of the indicated bacterial strain. For determination of bacterial colonization following infection, bladders were harvested, homogenized, and plated on eosin-methylene blue (EMB) agar with the appropriate antibiotics. The competitive index was determined by plating the mouse bladder homogenates on both EMB agar with streptomycin and EMB agar with streptomycin and chloramphenicol. All experiments were approved by the animal care and use committee at Northwestern University.
Neutrophil MPO ELISA.
Mice were instilled with 10 μl of saline or 1 × 108 CFU of either NU14, NU14 ΔampG, or NU14 ΔwaaL. At 6 h postinfection, urines and bladders were harvested. Bladder homogenates and urines were prepared according to the manufacturer's recommendations and assayed for neutrophil myeloperoxidase (MPO) by ELISA (HyCult Biosciences).
Statistical analysis.
Results were analyzed with the Student t test or one-way analysis of variance followed by Dunnet's multiple comparison posttest, using Prism software from GraphPad, Inc., as appropriate. Differences between groups of data were considered significant for P values of ≤0.05.
RESULTS
Transposon mutagenesis of NU14 identifies mutants which stimulate enhanced urothelial IL-8 secretion.
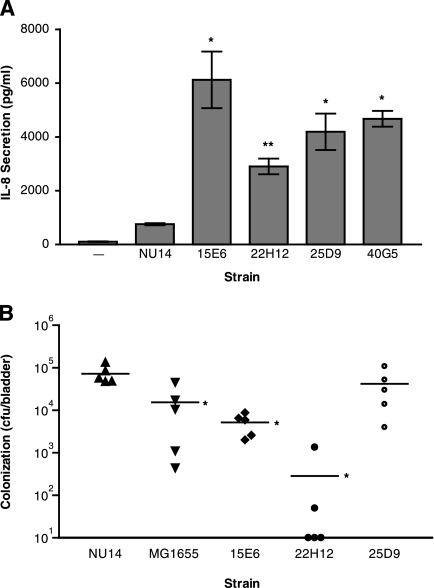
We conducted a genetic screen by transposon mutagenesis of UPEC strain NU14 to identify genes required for the modulation of IL-8 secretion in urothelial cells. A library of approximately 5,600 transposon mutants was screened for a significant increase in the stimulation of urothelial IL-8 secretion. This library represents approximately 1× genome coverage given the estimated genome size of 5.2 Mb (27). Our screen identified four NU14 transposon mutants, 15E6, 22H12, 25D9, and 40G5, which stimulated significantly higher mean levels of urothelial IL-8 secretion than that in NU14 (Fig. 1A) (P < 0.05). By employing an arbitrary PCR strategy, we identified the site of transposon insertion in each of these four mutants (Table 3). The 15E6 insertion was found within ampG, a gene encoding a muropeptide permease required for recycling peptidoglycan fragments and precursors from the periplasm back into the cytoplasm (12). The insertion mutant 22H12 was found to have a transpositional disruption of the rfa operon, specifically in waaL (rfaL), the gene encoding the O antigen ligase of the LPS biosynthetic gene cluster (50). The 25D9 and 40G5 mutants were both found to have insertions in or near the alanine racemase I gene, alr, whose product converts l-alanine into d-alanine for peptidoglycan synthesis (16). Thus, all four mutants contain disruptions of genes mediating PAMP biosynthesis.
FIG. 1.
Four transposon insertion mutants of NU14 induced significantly increased IL-8 secretion from urothelial cell cultures and had different phenotypes in a mouse model of UTI. (A) TEU-1 cell cultures were treated with NU14 and four transposon mutants, 15E6, 22H12, 25D9, and 40G5, for 4 hours, and IL-8 secretion was determined by ELISA. The data represent the means ± standard deviations for three samples, and each experiment was performed in duplicate. (B) Mice (n = 5) were instilled via catheter with either NU14, MG1655, 15E6, 22H12, or 25D9, and bladder colonization was determined at 24 h postinfection. The asterisks (*, P < 0.05; and **, P < 0.01) indicate statistically significant differences between NU14 and each of the transposon mutants.
TABLE 3.
Transposon insertion site identification
| Transposon mutant | Transposon insertion location | Genomic location (nt)a | ORF transposon insertion site (nt)a,b | Gene product | Gene function |
|---|---|---|---|---|---|
| 15E6 | ampG ORF | 466893 | 771 | Muropeptide permease | Peptidoglycan synthesis/recycling |
| 22H12 | waaL ORF | 4058764 | 11 | O antigen ligase | LPS biosynthesis |
| 25D9 | alr-nmn intergenic region | 4521498 | Intergenic | Alanine racemase I | Peptidoglycan synthesis |
| 40G5 | alr ORF | 4521455 | 195 | Alanine racemase I | Peptidoglycan synthesis |
The genomic location and open reading frame (ORF) location were determined using UTI89 and RS218 genome information.
Distance in nucleotides from coding region start site.
Phenotypes of transposon mutants in a murine model of UTI.
Our previous work indicated that modulation of innate immune responses in the urinary tract by NU14 plays a role in colonization of the mouse bladder in a model of UTI (10). In order to determine whether the transposon mutants were attenuated in vivo, we infected mice with either NU14, MG1655, 15E6, 22H12, or 25D9 (Fig. 1B). Infection with MG1655 resulted in approximately fivefold less bladder colonization at 24 h postinfection than that for infection with NU14 (15,500 ± 8,515 versus 72,200 ± 15,890 CFU/bladder). The bladders of mice instilled with 15E6 and 22H12 also had significantly fewer bacteria than did those of NU14-infected mice (5,159 ± 1,261 and 286.0 ± 266.1 CFU/bladder, respectively; P < 0.05). Infection with the transposon mutant 25D9 resulted in bladder colonization equivalent to that of wild-type NU14. Overall, the magnitude of the enhanced stimulation of urothelial cytokine secretion in vitro does not appear to correspond with the severity of the defects in mouse bladder colonization.
Targeted deletion mutagenesis recapitulates the phenotypes of insertional mutants.
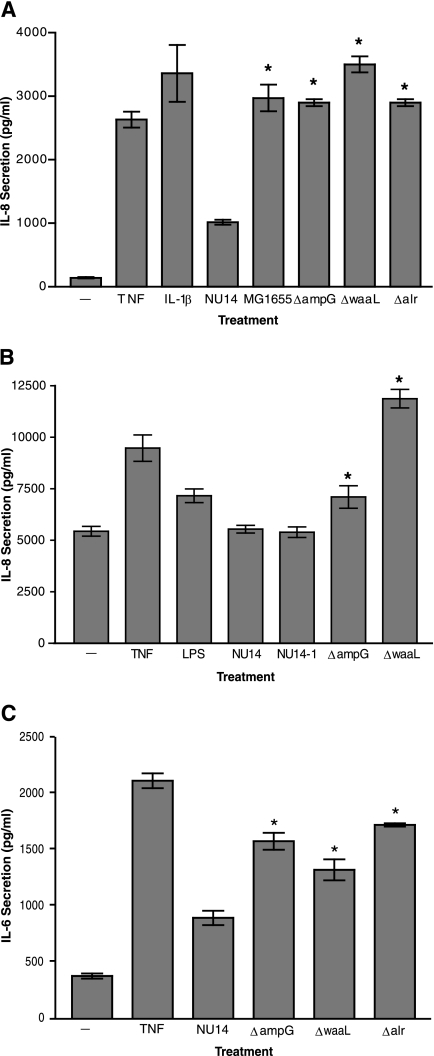
Insertional mutagenesis identified three genes which were required for evasion of urothelial innate immune surveillance. We constructed targeted deletion mutants of NU14 in each of these genes, i.e., ampG, waaL and alr, to confirm their role in modulation of host responses. Treatment with TNF-α and IL-1β, known activators of NF-κB and IL-8 secretion, induced elevated IL-8 secretion from TEU-1 cell cultures relative to that from untreated cultures or cultures treated with wild-type NU14 (Fig. 2A). As previously shown, infection of TEU-1 cells with MG1655 induced greater IL-8 secretion than did infection with NU14, at levels which approximated those induced by the targeted deletion mutants (Fig. 2A) (10). The enhancement of IL-8 secretion from urothelial cultures cannot be attributed to general growth defects or type 1 piliation, as the deletion mutants did not exhibit differences in growth in minimal medium or mannose-sensitive hemagglutination compared to wild-type NU14 (data not shown). The magnitude of the increase in IL-8 secretion from the TEU-1 cells stimulated by the deletion mutants did not always parallel the IL-8 levels induced by corresponding transposon mutants, possibly due to the imprecise nature of the defects induced by insertional mutagenesis. NU14 ΔampG and NU14 ΔwaaL also induced higher levels of IL-8 secretion in cultures of T24 cells, another urothelial cell line (Fig. 2B). The effects of NU14 mutants on urothelial cytokine secretion were not specific to IL-8 alone, for the mutants also induced significantly greater IL-6 secretion than did NU14 (Fig. 2C). Therefore, the deletion of ampG, waaL, or alr results in an increase in the bacterial stimulation of urothelial cytokine production.
FIG. 2.
Targeted deletion mutants confirm the phenotypes of the transposon mutants. (A) TEU-1 cell cultures were treated for 4 hours with TNF, IL-1β, NU14, MG1655, NU14 ΔampG, NU14 ΔwaaL, or NU14 Δalr, and IL-8 secretion was determined by ELISA. (B) T24 cultures were treated for 4 hours with TNF, LPS (O55:B5), NU14, NU14-1, NU14 ΔampG, or NU14 ΔwaaL, and IL-8 secretion was determined by ELISA. (C) IL-6 secretion from TEU-1 cultures was determined after treatment with TNF, NU14, NU14 ΔampG, NU14 ΔwaaL, or NU14 Δalr. The data represent the means ± standard deviations for three samples, and each experiment was performed in duplicate. The asterisks indicate statistically significant differences (P < 0.05) between samples treated with NU14 and the targeted deletion mutants.
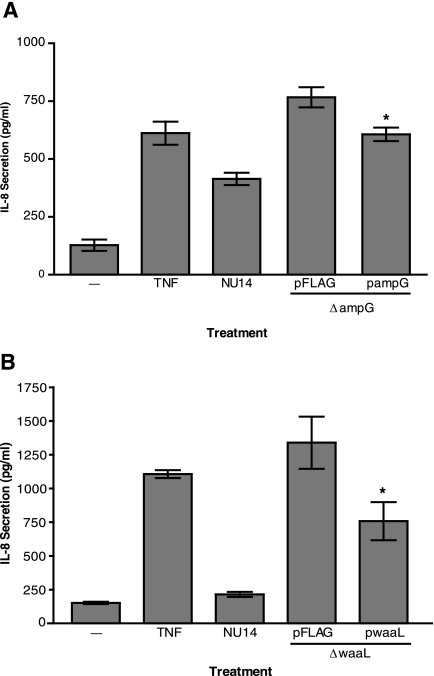
Complementation of NU14 ΔampG and NU14 ΔwaaL restores the capacity to modulate urothelial cytokine secretion.
We cloned a wild-type copy of either NU14 ampG or waaL to complement the defects created by the targeted deletion of these genes in NU14. The complemented strain NU14 ΔampG/pampG induced less IL-8 secretion from urothelial cultures than did the empty vector control strain NU14 ΔampG/pFLAG (Fig. 3A). Similarly, NU14 ΔwaaL/pwaaL induced less IL-8 secretion than did the empty vector control strain NU14 ΔwaaL/pFLAG (Fig. 3B). The complementation was not complete, as these constructs were unable to restore to these mutants the same ability as that of wild-type NU14 to cause diminished urothelial IL-8 secretion compared to the mutant strains carrying the empty vector. Nonetheless, these data support the hypothesis that ampG and waaL are essential for the ability of NU14 to evade the host innate immune response.
FIG. 3.
Deletion mutant phenotypes can be complemented through expression of the deleted gene products from plasmids. (A) TEU-1 cells were treated with TNF or infected with NU14, NU14 ΔampG/pFLAG, or NU14 ΔampG/pampG. Cell culture supernatants were harvested after 4 h of infection, and IL-8 secretion was determined by ELISA. (B) IL-8 secretion levels were determined after 4 hours of treatment of TEU-1 cultures with either TNF, NU14, NU14 ΔwaaL/pFLAG (empty vector), or NU14 ΔwaaL/pwaaL. The data represent the means ± standard deviations for three samples, and each experiment was performed in duplicate. The asterisks indicate statistically significant differences (P < 0.05) between samples treated with the deletion mutant strains carrying the empty vector control plasmid and the strains carrying the complementation construct plasmids.
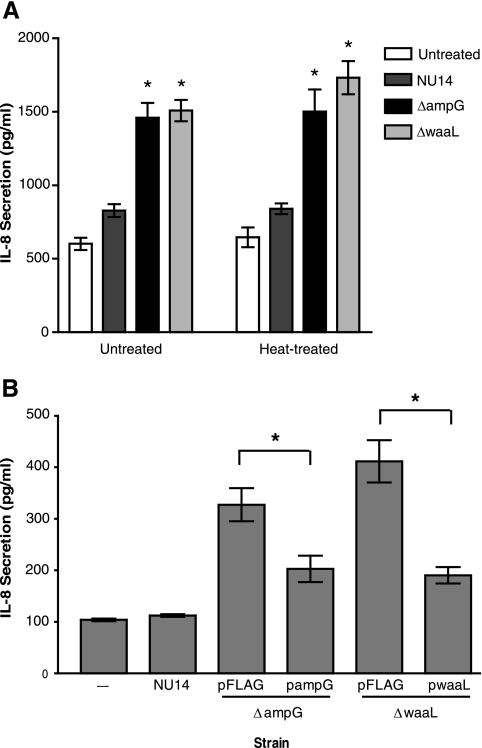
Conditioned media from NU14 ΔampG and NU14 ΔwaaL stimulate urothelial IL-8 secretion.
Our analysis of the functions of the genes responsible for the modulation of cytokine secretion from urothelial cells suggested that differences in bacterial membrane structural components may be involved. Previous studies have indicated that ampG mutants of Shigella flexneri release increased levels of peptidoglycan fragments into conditioned culture media (35). These secreted muropeptides stimulated NF-κB activation and IL-8 secretion via Nod1 (35). In addition, prior studies indicated that LPS structure, specifically lipid A acetylation and O antigen structure, can affect the ability of LPS to stimulate IL-6 secretion from urothelial cells (6). We examined the possibility that components which could stimulate urothelial IL-8 secretion may accumulate in conditioned media from NU14 ΔampG and NU14 ΔwaaL cultures. TEU-1 cells secreted high levels of IL-8 when they were treated with sterile filtered conditioned media from both NU14 ΔampG and NU14 ΔwaaL cultures but not with conditioned medium from NU14 cultures (Fig. 4A). Furthermore, conditioned media from the mutants retained stimulatory capacity after heat treatment (Fig. 4A). Conversely, conditioned media from the complemented mutant strains, NU14 ΔampG/pampG and NU14 ΔwaaL/pwaaL, showed marked reductions in stimulation of IL-8 secretion from TEU-1 cell cultures compared to the empty vector control strains (Fig. 4B) (P < 0.05). These data indicate that a heat-stable secreted factor(s) present in the culture supernatants from the mutants, but not wild-type NU14, stimulates urothelial cytokine production.
FIG. 4.
Conditioned media from NU14 ΔampG and NU14 ΔwaaL cultures contain heat-stable factors which stimulate urothelial IL-8 secretion. (A) Conditioned cell culture media were prepared from cultures of NU14, NU14 ΔampG, and NU14 ΔwaaL grown overnight. TEU-1 cells were then treated with the filtered conditioned media or with conditioned media which had been heat treated at 65°C for 10 min, and IL-8 secretion was determined by ELISA. (B) Conditioned medium from NU14, NU14 ΔampG/pFLAG, NU14 ΔampG/pampG, NU14 ΔwaaL/pFLAG, or NU14 ΔwaaL/pwaaL culture was used to treat TEU-1 cultures, and the resulting IL-8 secretion was determined. The data represent the means ± standard deviations for three samples, and each experiment was performed in duplicate. The asterisks indicate statistically significant differences (P < 0.05) between samples.
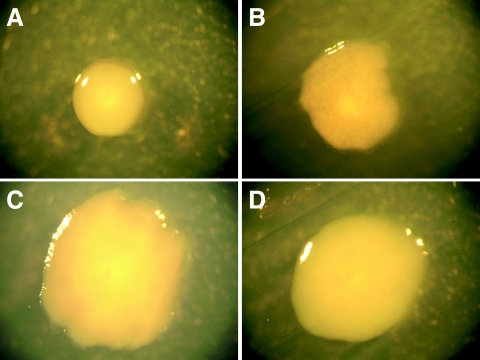
E. coli strains that lack the O antigen component of LPS display altered colony morphology.
NU14, like most clinical pathogenic E. coli isolates, has LPS which contains O antigen (smooth-type LPS), which results in a colony morphology characterized by a domed shape and smooth edges (Fig. 5A). Conversely, the deletion of waaL causes a change in colony morphology consistent with rough-type LPS, namely, flat colonies with irregular edges (Fig. 5B). Complementation of NU14 ΔwaaL by transformation with the plasmid pwaaL, creating strain NU14 DwaaL/pwaaL (Fig. 5C), results in a restoration of the wild-type colony morphology that is not restored by the empty vector control strain NU14 ΔwaaL/pFLAG (Fig. 5D). The deletion of waaL results in a colony morphology consistent with conversion of smooth-type LPS to the rough phenotype due to the loss of O antigen ligase activity encoded by waaL.
FIG. 5.
Deletion of waaL results in rough-type bacterial colony morphology. NU14 (A), NU14 ΔwaaL (B), NU14 ΔwaaL/pFLAG (C), and NU14 ΔwaaL/pwaaL (D) were grown on LB agar plates containing the appropriate antibiotics overnight at 37°C, and colony morphology was photographed at a magnification of ×4, using a Leica S8AP0 microscope.
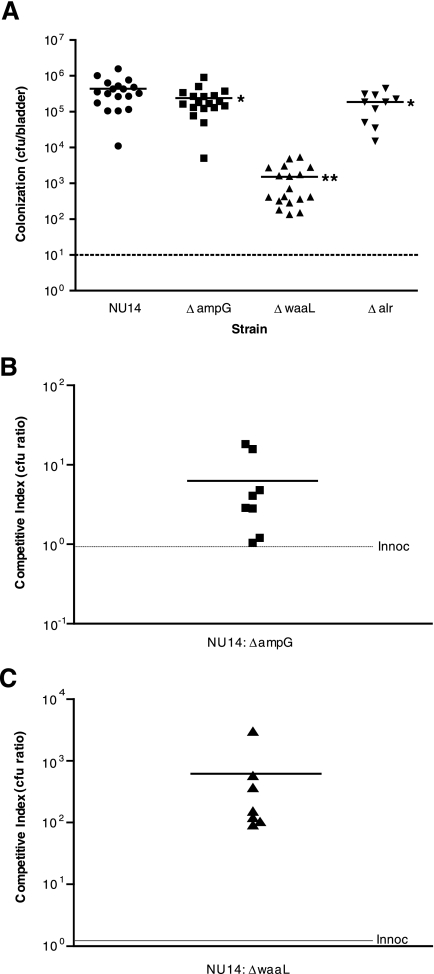
NU14 ΔampG and NU14 ΔwaaL are attenuated in the ability to colonize the mouse urinary tract.
In order to verify the phenotypes of the transposon insertion mutants in the murine model of UTI, we compared bacterial bladder colonization in mice infected with NU14, NU14 ΔampG, NU14 ΔwaaL, and NU14 Δalr. Like 15E6, NU14 ΔampG showed reduced bladder colonization after 24 h of infection relative to the wild type (Fig. 6A) (438,111 ± 378,255 versus 238,833 ± 206,604 CFU/bladder; P < 0.05). This reduction was similar to levels of bladder colonization in mice infected with the nonsuppressor strain MG1655 (Fig. 1B). Comparatively, infection with NU14 ΔwaaL resulted in greatly reduced colonization compared to infection with NU14 (Fig. 6A) (1,504 ± 1,637; P < 0.0001). The decreased colonization by NU14 ΔwaaL was equivalent to colonization by the corresponding insertional mutant 22H12 (Fig. 1B). The corresponding transposon insertion mutant in the alr region, 25D9, was not deficient in bladder colonization compared to NU14 (Fig. 1B). However, bladder colonization by NU14 Δalr was also significantly lower than that of the wild type (186,000 ± 137,311; P < 0.05). Thus, the phenotypes of the targeted deletion mutants verify a role for these genes in UPEC virulence.
FIG. 6.
NU14 ΔampG and NU14 ΔwaaL are deficient in colonization of the mouse bladder. (A) Mice were infected via instillation into the bladder with either NU14 (n = 18), NU14 ΔampG (n = 18), NU14 ΔwaaL (n = 18), or NU14 Δalr (n = 10), and bacterial colonization was measured at 24 h postinfection. The asterisks (*, P < 0.05; and **, P < 0.0001) indicate statistically significant differences. (B and C) Mice (n = 8) were inoculated via catheter with both NU14 and NU14 ΔampG (B) or NU14 and NU14 ΔwaaL (C), and colonization of each strain was determined by differential colony counts on selective agar medium. Each data point represents the competitive index ratio, which describes the numerical ratio of NU14 to the deletion mutant strain CFU present in each mouse bladder at 24 h postinfection. The initial inoculation ratio (Innoc) is represented by a dashed line.
We also compared the colonization of these strains in a competition assay with NU14. A mixed infection with NU14 ΔampG and NU14 resulted in a mean competitive index ratio (NU14 to NU14 ΔampG) of 6.79 (Fig. 6B). This index is consistent with the magnitude of the defects in bladder colonization of NU14 ΔampG in nonmixed infection and of the transposon mutant 15E6 (Fig. 1B and 6A). Strain NU14 ΔwaaL also showed reduced colonization upon coinfection with NU14, with a competitive index ratio (NU14 to NU14 ΔwaaL) of 619 (Fig. 6C). These data are consistent with the data for the corresponding transposon mutant, 22H12, and NU14 ΔwaaL infection (Fig. 1B and 6A). Each of the strains showed no significant differences in colonization in the mixed infection compared with colonization in the nonmixed infection, indicating that the colonization defects could not be complemented in trans by wild-type NU14. Also, colonization by NU14 was not affected by the presence of either strain during coinfection. This suggests that the defects of NU14 ΔampG and NU14 ΔwaaL are intrinsic to these strains' ability to persist in the bladder.
NU14 ΔampG and NU14 ΔwaaL induce greater levels of neutrophil recruitment to the mouse bladder than does the wild type.
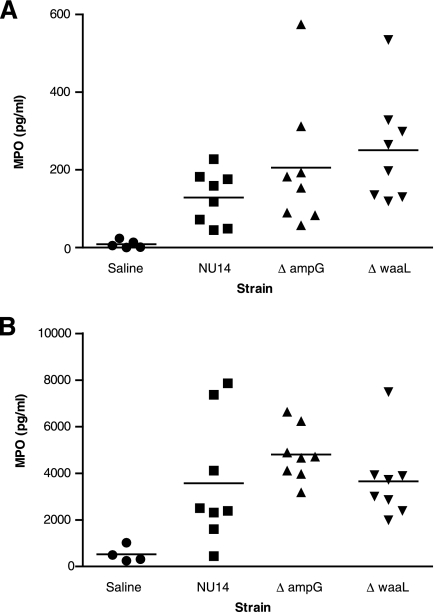
We previously observed that in a murine model of early UTI, instillation of NU14 into the mouse bladder results in lower levels of neutrophil MPO accumulation in the bladder and urine than does instillation of the nonsuppressor strain MG1655, indicating differential neutrophil recruitment (10). Therefore, we used this early infection model to compare MPO levels in mice infected with NU14, NU14 ΔampG, and NU14 ΔwaaL. Six hours after infection, mouse bladder homogenates from mice infected with either NU14 ΔampG or NU14 ΔwaaL had significantly greater levels of MPO than those induced by installation with saline (Fig. 7A) (205.6 ± 169.6, 250.8 ± 140.0, and 8.600 ± 9.883 pg/ml, respectively; P < 0.05). Infection with NU14 induced a small increase in bladder MPO levels that was not statistically different from the instillation of saline (Fig. 7A) (128.5 ± 68.16 pg/ml). Urines from mice infected with NU14 ΔampG had significantly higher MPO levels than those from mice infected with either NU14 or NU14 ΔwaaL (Fig. 7B) (4,800 ± 1,151, 3,575 ± 2,698, and 3,656 ± 1,702 pg/ml, respectively; P < 0.01). Infection with NU14 ΔwaaL enhanced bladder but not urine MPO levels. This may be due to the enhanced clearance of NU14 ΔwaaL relative to that after infections with NU14 and NU14 ΔampG (Fig. 6A to C). These results indicate that during UTI, NU14 ΔampG and NU14 ΔwaaL stimulate enhanced host responses which correlate with decreased colonization.
FIG. 7.
Infection with NU14 ΔampG and NU14 ΔwaaL stimulates enhanced MPO in mouse bladder homogenates and urines during UTI. Mice were instilled with saline or infected with NU14, NU14 ΔampG, or NU14 ΔwaaL via catheter (for bladder experiments, five saline control mice and eight infected mice were used; and for urine experiments, four control mice and eight infected mice were used). Urine samples were collected, and the bladders were removed and homogenized at 6 h postinfection. Bladder (A) and urine (B) MPO levels were determined by ELISA. Asterisks indicate statistically significant differences (P < 0.05) between saline-instilled controls and samples from mice infected with bacterial strains.
DISCUSSION
In this study, we identified three genes required for evasion of innate immune responses which lead to urothelial cytokine secretion. Our screen indicates that the functions of the genes we identified are important for the ability of UPEC to mask and/or prevent the release of PAMPs which activate PRRs in the urinary tract. While the insertion and targeted deletion mutants stimulated enhanced urothelial IL-8 and IL-6 secretion at similar levels in vitro, the mutants differed in the ability to colonize the mouse bladder during infection. These data suggest that in the murine UTI model, UPEC modulation of cytokine secretion plays an important role but is not the sole factor responsible for the defect in colonization of the ampG or waaL mutant. Furthermore, the ampG and waaL mutants differed substantially in the magnitude of the deficiency in bladder colonization, which suggests that these mutations may affect additional aspects of UPEC pathogenesis in addition to urothelial cytokine and chemokine secretion. It appears likely that UPEC bladder colonization is dependent upon a multifactorial strategy which also includes bacterial adherence and invasion, intracellular bacterial community/biofilm formation, induction of urothelial apoptosis, and nutrient acquisition (2, 28, 32, 46).
The bacterial cell wall, composed of cross-linked peptidoglycan, is continuously remodeled during growth and division. E. coli releases minimal amounts of peptidoglycan fragments during growth, suggesting that evolutionary pressure has selected for mechanisms which conserve these fragments or prevent their loss (37). AmpG is a transmembrane permease which transports the muropeptide fragments GlcNAc-anhMurNAc-tripeptide (tetrapeptide) from the periplasm back into the cytoplasm (12). For E. coli, ampG mutants secrete these peptidoglycan fragments into the culture medium but have no defects in growth (20). Recently, Nigro et al. found that sterile filtered culture supernatants from an ampG mutant of Shigella flexneri induced NF-κB activation and IL-8 secretion through Nod1. Furthermore, in vivo, the virulence of the ampG mutant was similar to that of the wild-type strain and the mutant did not increase the production of IL-6 or gamma interferon (35). There are several candidates for PRR recognition of peptidoglycan fragments in the urinary tract, including TLR1/2, the nucleotide-binding oligomerization domain-containing protein 1/2 (NOD1/2), and peptidoglycan recognition proteins (5, 15, 39).
TLR4-mediated detection of E. coli LPS is a well-appreciated aspect of UPEC-host interactions. Urothelial recognition of UPEC LPS through the TLR4-CD14-MD2 complex results in NF-κB activation and cytokine production (4, 5, 7, 40, 43). Hunstad et al. performed a similar screen of UPEC strain UTI89 and identified the rfa-rfb LPS biosynthetic operons required for suppression of IL-6 secretion from urothelial cultures (26). Our work demonstrates the specific requirement for the ligation of O antigen onto the lipid A core subunit for evasion of urothelial cytokine secretion and survival in the mouse urinary tract. This is consistent with previous observations that smooth-type LPS isolated from strains with O antigen, such as NU14, is 100 times less stimulatory than rough-type LPS lacking O antigen ligation on the lipid A core (6). Furthermore, LPS isolated from another smooth-type UPEC isolate induced significantly less IL-6 secretion from T24 cells than did the rough-type LPS purified from strain MG1655 (24). Thus, NU14 ΔwaaL may effectively convert the LPS of NU14 from a smooth form into a rough form, as indicated by the change in colony morphology that we observed (Fig. 5). These observations also suggest that UPEC may specifically modify its LPS structure by expressing O antigen to circumvent surveillance mediated by TLR4/CD14 in the urinary tract.
The mechanisms which underlie the active suppression of urothelial innate immune responses by UPEC remain poorly understood. Several UPEC isolates have been shown to actively suppress activation of NF-κB signaling pathway components via IκB stabilization as well as through blockade of the mitogen-activated protein kinases extracellular signal-regulated kinase 1/2 and p38 (9, 32, 44). TcpC, a toxin expressed by UPEC strain CFT073, actively suppresses NF-κB pathway activation and IL-6 secretion induced by stimuli which activate Toll/IL-1 receptor-dependent signaling through the adaptor MyD88. This toxin, however, has not yet been detected in the genomes of other UPEC isolates (such as 536, UTI89, RS218, or any E. coli isolate). UPEC also suppresses the cytokine secretion-inducing activity of a wide variety of stimuli, including whole bacteria, purified LPS, poly(I:C), FSL-1, and in some cases, TNF-α, but does not suppress IL-1β responses (10, 24, 26, 32). Furthermore, this suppression mechanism requires intact live bacteria and functions of urothelial and nonurothelial origin (9, 26, 32, 44). These observations suggest that UPEC strains may utilize several as yet undiscovered means to modulate host responses in several cell types.
The innate immune response is essential for the clearance of UPEC during UTI (34). Our data suggest that UPEC avoids PRR recognition and urothelial inflammatory cytokine and chemokine secretion during the early stages of infection (10). This results in a delayed or decreased recruitment of neutrophils to the bladder lumen and an increased opportunity for invasion of urothelial cells (10). UPEC modulation of the innate immune response may also affect other urothelial responses, including apoptosis and the newly described TLR4-dependent signaling pathways involved in intracellular invasion, through common signaling intermediates, such as cyclic AMP and calcium (31, 47, 48). Furthermore, the diminished innate immune responses elicited by UPEC strains such as NU14 could constitute a means to subvert the development of protective adaptive immune responses in the urinary tract against subsequent infections. UPEC evasion of PRR recognition of PAMPs, such as LPS and peptidoglycan, may therefore constitute an essential event in colonization of the urinary tract.
Acknowledgments
We thank Jose Puente for technical assistance with lambda red-mediated targeted deletion mutagenesis. We thank Fred Blattner and Guy Plunkett III for providing access to RS218 sequence data from the University of Wisconsin E. coli Genome Project (http://www.genome.wisc.edu) prior to publication. We also thank Bethany Boardman and Karla Satchell for providing the pBSL180 conjugative transposition system and for assistance with arbitrary PCR. We thank members of the lab for discussions and suggestions.
This work was supported by NIDDK award RO1 DK04648 (A.J.S.).
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 16 June 2008.
REFERENCES
- 1.Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene 16059-62. [DOI] [PubMed] [Google Scholar]
- 2.Alteri, C. J., and H. L. Mobley. 2007. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect. Immun. 752679-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen-Nissen, E., T. R. Hawn, K. D. Smith, A. Nachman, A. E. Lampano, S. Uematsu, S. Akira, and A. Aderem. 2007. Cutting edge: Tlr5−/− mice are more susceptible to Escherichia coli urinary tract infection. J. Immunol. 1784717-4720. [DOI] [PubMed] [Google Scholar]
- 4.Backhed, F., L. Meijer, S. Normark, and A. Richter-Dahlfors. 2002. TLR4-dependent recognition of lipopolysaccharide by epithelial cells requires sCD14. Cell. Microbiol. 4493-501. [DOI] [PubMed] [Google Scholar]
- 5.Backhed, F., S. Normark, and A. Richter-Dahlfors. 2002. TLR4-dependent lipopolysaccharide signalling in epithelial cells is independent of extracellular protease activity. Cell. Microbiol. 4297-303. [DOI] [PubMed] [Google Scholar]
- 6.Backhed, F., S. Normark, E. K. Schweda, S. Oscarson, and A. Richter-Dahlfors. 2003. Structural requirements for TLR4-mediated LPS signalling: a biological role for LPS modifications. Microbes Infect. 51057-1063. [DOI] [PubMed] [Google Scholar]
- 7.Backhed, F., M. Soderhall, P. Ekman, S. Normark, and A. Richter-Dahlfors. 2001. Induction of innate immune responses by Escherichia coli and purified lipopolysaccharide correlate with organ- and cell-specific expression of Toll-like receptors within the human urinary tract. Cell. Microbiol. 3153-158. [DOI] [PubMed] [Google Scholar]
- 8.Bhavsar, A. P., J. A. Guttman, and B. B. Finlay. 2007. Manipulation of host-cell pathways by bacterial pathogens. Nature 449827-834. [DOI] [PubMed] [Google Scholar]
- 9.Bhushan, S., S. Tchatalbachev, J. Klug, M. Fijak, C. Pineau, T. Chakraborty, and A. Meinhardt. 2008. Uropathogenic Escherichia coli block MyD88-dependent and activate MyD88-independent signaling pathways in rat testicular cells. J. Immunol. 1805537-5547. [DOI] [PubMed] [Google Scholar]
- 10.Billips, B. K., S. G. Forrestal, M. T. Rycyk, J. R. Johnson, D. J. Klumpp, and A. J. Schaeffer. 2007. Modulation of host innate immune response in the bladder by uropathogenic Escherichia coli. Infect. Immun. 755353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, Q., and J. T. Park. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 1846434-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirl, C., A. Wieser, M. Yadav, S. Duerr, S. Schubert, H. Fischer, D. Stappert, N. Wantia, N. Rodriguez, H. Wagner, C. Svanborg, and T. Miethke. 2008. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 14399-406. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delbridge, L. M., and M. X. O'Riordan. 2007. Innate recognition of intracellular bacteria. Curr. Opin. Immunol. 1910-16. [DOI] [PubMed] [Google Scholar]
- 16.de Roubin, M. R., D. Mengin-Lecreulx, and J. van Heijenoort. 1992. Peptidoglycan biosynthesis in Escherichia coli: variations in the metabolism of alanine and d-alanyl-d-alanine. J. Gen. Microbiol. 1381751-1757. [DOI] [PubMed] [Google Scholar]
- 17.Folkesson, A., S. Eriksson, M. Andersson, J. T. Park, and S. Normark. 2005. Components of the peptidoglycan-recycling pathway modulate invasion and intracellular survival of Salmonella enterica serovar Typhimurium. Cell. Microbiol. 7147-155. [DOI] [PubMed] [Google Scholar]
- 18.Franchi, L., J. H. Park, M. H. Shaw, N. Marina-Garcia, G. Chen, Y. G. Kim, and G. Nunez. 2008. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell. Microbiol. 101-8. [DOI] [PubMed] [Google Scholar]
- 19.Frendeus, B., C. Wachtler, M. Hedlund, H. Fischer, P. Samuelsson, M. Svensson, and C. Svanborg. 2001. Escherichia coli P fimbriae utilize the Toll-like receptor 4 pathway for cell activation. Mol. Microbiol. 4037-51. [DOI] [PubMed] [Google Scholar]
- 20.Goodell, E. W., and U. Schwarz. 1985. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J. Bacteriol. 162391-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hang, L., B. Frendeus, G. Godaly, and C. Svanborg. 2000. Interleukin-8 receptor knockout mice have subepithelial neutrophil entrapment and renal scarring following acute pyelonephritis. J. Infect. Dis. 1821738-1748. [DOI] [PubMed] [Google Scholar]
- 22.Hang, L., M. Haraoka, W. W. Agace, H. Leffler, M. Burdick, R. Strieter, and C. Svanborg. 1999. Macrophage inflammatory protein-2 is required for neutrophil passage across the epithelial barrier of the infected urinary tract. J. Immunol. 1623037-3044. [PubMed] [Google Scholar]
- 23.Hedlund, M., B. Frendeus, C. Wachtler, L. Hang, H. Fischer, and C. Svanborg. 2001. Type 1 fimbriae deliver an LPS- and TLR4-dependent activation signal to CD14-negative cells. Mol. Microbiol. 39542-552. [DOI] [PubMed] [Google Scholar]
- 24.Hilbert, D. W., K. E. Pascal, E. K. Libby, E. Mordechai, M. E. Adelson, and J. P. Trama. 2008. Uropathogenic Escherichia coli dominantly suppress the innate immune response of bladder epithelial cells by a lipopolysaccharide- and Toll-like receptor 4-independent pathway. Microbes Infect. 10114-121. [DOI] [PubMed] [Google Scholar]
- 25.Hultgren, S. J., W. R. Schwan, A. J. Schaeffer, and J. L. Duncan. 1986. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect. Immun. 54613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunstad, D. A., S. S. Justice, C. S. Hung, S. R. Lauer, and S. J. Hultgren. 2005. Suppression of bladder epithelial cytokine responses by uropathogenic Escherichia coli. Infect. Immun. 733999-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, J. R., S. J. Weissman, A. L. Stell, E. Trintchina, D. E. Dykhuizen, and E. V. Sokurenko. 2001. Clonal and pathotypic analysis of archetypal Escherichia coli cystitis isolate NU14. J. Infect. Dis. 1841556-1565. [DOI] [PubMed] [Google Scholar]
- 28.Justice, S. S., C. Hung, J. A. Theriot, D. A. Fletcher, G. G. Anderson, M. J. Footer, and S. J. Hultgren. 2004. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA 1011333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai, T., and S. Akira. 2007. SnapShot: pattern-recognition receptors. Cell 1291024. [DOI] [PubMed] [Google Scholar]
- 30.Kelly, D., and S. Conway. 2005. Bacterial modulation of mucosal innate immunity. Mol. Immunol. 42895-901. [DOI] [PubMed] [Google Scholar]
- 31.Klumpp, D. J., M. T. Rycyk, M. C. Chen, P. Thumbikat, S. Sengupta, and A. J. Schaeffer. 2006. Uropathogenic Escherichia coli induces extrinsic and intrinsic cascades to initiate urothelial apoptosis. Infect. Immun. 745106-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klumpp, D. J., A. C. Weiser, S. Sengupta, S. G. Forrestal, R. A. Batler, and A. J. Schaeffer. 2001. Uropathogenic Escherichia coli potentiates type 1 pilus-induced apoptosis by suppressing NF-kappaB. Infect. Immun. 696689-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langermann, S., S. Palaszynski, M. Barnhart, G. Auguste, J. S. Pinkner, J. Burlein, P. Barren, S. Koenig, S. Leath, C. H. Jones, and S. J. Hultgren. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276607-611. [DOI] [PubMed] [Google Scholar]
- 34.Mulvey, M. A., J. D. Schilling, J. J. Martinez, and S. J. Hultgren. 2000. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc. Natl. Acad. Sci. USA 978829-8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nigro, G., L. L. Fazio, M. C. Martino, G. Rossi, I. Tattoli, V. Liparoti, C. De Castro, A. Molinaro, D. J. Philpott, and M. L. Bernardini. 2008. Muramylpeptide shedding modulates cell sensing of Shigella flexneri. Cell. Microbiol. 10682-695. [DOI] [PubMed] [Google Scholar]
- 36.Old, D. C., and J. P. Duguid. 1970. Selective outgrowth of fimbriate bacteria in static liquid medium. J. Bacteriol. 103447-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, J. T. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 17421-426. [DOI] [PubMed] [Google Scholar]
- 38.Roy, C. R., and E. S. Mocarski. 2007. Pathogen subversion of cell-intrinsic innate immunity. Nat. Immunol. 81179-1187. [DOI] [PubMed] [Google Scholar]
- 39.Royet, J., and R. Dziarski. 2007. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat. Rev. 5264-277. [DOI] [PubMed] [Google Scholar]
- 40.Samuelsson, P., L. Hang, B. Wullt, H. Irjala, and C. Svanborg. 2004. Toll-like receptor 4 expression and cytokine responses in the human urinary tract mucosa. Infect. Immun. 723179-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schilling, J. D., S. M. Martin, C. S. Hung, R. G. Lorenz, and S. J. Hultgren. 2003. Toll-like receptor 4 on stromal and hematopoietic cells mediates innate resistance to uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 1004203-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schilling, J. D., S. M. Martin, D. A. Hunstad, K. P. Patel, M. A. Mulvey, S. S. Justice, R. G. Lorenz, and S. J. Hultgren. 2003. CD14- and Toll-like receptor-dependent activation of bladder epithelial cells by lipopolysaccharide and type 1 piliated Escherichia coli. Infect. Immun. 711470-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schilling, J. D., M. A. Mulvey, C. D. Vincent, R. G. Lorenz, and S. J. Hultgren. 2001. Bacterial invasion augments epithelial cytokine responses to Escherichia coli through a lipopolysaccharide-dependent mechanism. J. Immunol. 1661148-1155. [DOI] [PubMed] [Google Scholar]
- 44.Selvaraj, S. K., and N. V. Prasadarao. 2005. Escherichia coli K1 inhibits proinflammatory cytokine induction in monocytes by preventing NF-kappaB activation. J. Leukoc. Biol. 78544-554. [DOI] [PubMed] [Google Scholar]
- 45.Sirard, J. C., C. Vignal, R. Dessein, and M. Chamaillard. 2007. Nod-like receptors: cytosolic watchdogs for immunity against pathogens. PLoS Pathog. 3e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 726373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song, J., B. L. Bishop, G. Li, M. J. Duncan, and S. N. Abraham. 2007. TLR4 initiated and cAMP mediated abrogation of bacterial invasion of the bladder. Cell Host Microbe 1287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song, J., M. J. Duncan, G. Li, C. Chan, R. Grady, A. Stapleton, and S. N. Abraham. 2007. A novel TLR4-mediated signaling pathway leading to IL-6 responses in human bladder epithelial cells. PLoS Pathog. 3e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitfield, C., P. A. Amor, and R. Koplin. 1997. Modulation of the surface architecture of gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol. Microbiol. 23629-638. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, D., G. Zhang, M. S. Hayden, M. B. Greenblatt, C. Bussey, R. A. Flavell, and S. Ghosh. 2004. A Toll-like receptor that prevents infection by uropathogenic bacteria. Science 3031522-1526. [DOI] [PubMed] [Google Scholar]