Abstract
Alteration of surface lipoprotein profiles is a key strategy that the Lyme disease pathogen, Borrelia burgdorferi, has evolved to be maintained within its enzootic cycle between arthropods and mammals. Accumulated evidence indicates that the central regulatory pathway controlling differential gene expression by B. burgdorferi is the RpoN-RpoS pathway (the σ54-σS sigma factor cascade). It was previously shown that activation of the RpoN-RpoS pathway is controlled by Rrp2, a two-component response regulator and σ54-dependent transcriptional activator. The role of Rrp2 in the infectious cycle of B. burgdorferi has not been determined heretofore. In this report, we demonstrate that an rrp2 mutant defective in activating σ54-dependent transcription was unable to establish infection in mice, but the rrp2 mutant was capable of surviving within ticks during and after tick feeding. Because the rrp2 mutant was defective in the production of OspC, an outer surface lipoprotein essential for mammalian host infection, we further examined whether the loss of infectivity of the rrp2 mutant was solely due to the inability to produce OspC. While transformation with a shuttle vector carrying ospC under the control of a constitutive flaB promoter restored infection to an ospC mutant in immunodeficient SCID mice, it could not rescue the avirulent phenotype of the rrp2 mutant. These data indicate that, in addition to controlling OspC, Rrp2 controls another factor(s) essential for B. burgdorferi to establish infection in mammals. Furthermore, microarray analyses revealed that 125 and 19 genes were positively and negatively regulated, respectively, by Rrp2, which provides a foundation for future identification of additional Rrp2-dependent virulence determinants in B. burgdorferi.
Borrelia burgdorferi, the causative agent of Lyme disease, is maintained in nature in an enzootic cycle involving ticks (Ixodes scapularis) and mammals (Peromyscus leucopus) (4, 24, 47). B. burgdorferi adapts to diverse host environments by coordinately regulating the expression of numerous genes, many of which encode Borrelia surface lipoproteins (2, 33, 38, 42, 45, 51). In the past few years, efforts toward elucidating the underlying mechanisms of Borrelia differential gene expression have led to the identification of a novel regulatory pathway, the RpoN-RpoS pathway (also called the σ54-σS sigma factor cascade), which is central to the infectious cycle of B. burgdorferi (5, 7, 8, 16, 20, 25, 28, 46, 56). In this pathway, the two-component response regulator Rrp2, along with the alternative sigma factor RpoN (σ54 or σN), directly activates transcription of rpoS, which encodes another alternative sigma factor, RpoS (σS). RpoS functions as a global regulator that controls the expression of more than 145 Borrelia genes (8, 16). Many RpoS-activated genes appeared to be differentially expressed during tick feeding, and some, including ospC, dbpAB, BBK32, oppA5, BBA64, and BBA66, have been shown to be required for or associated with mammalian host infection (10, 14, 15, 19, 21, 22, 29, 30, 43, 44). In addition, an increased level of RpoS leads to the repression of a group of genes that are associated with spirochetal colonization and survival in ticks, including ospA and BB0365 (6, 8, 34).
The finding that the RpoN-RpoS pathway activates the transcription of ospC and ospC-like genes while repressing ospA and ospA-like genes implies that this pathway is not operative in flat ticks and is activated when ticks take a blood meal. Indeed, a recent report by Caimano et al. showed that rpoS expression is upregulated during tick feeding (8). It has been postulated that RpoS functions as a gatekeeper that modulates differential gene expression during the process of tick feeding which ensures the successful establishment of infection within the mammalian host (8). Both RpoN and RpoS are essential for the infectious cycle of B. burgdorferi; neither an rpoN nor an rpoS mutant was able to establish infection in mammalian hosts (7, 16). The rpoN mutant also failed to enter the tick salivary glands (16). The avirulent phenotype of the rpoN and rpoS mutants in mammals is consistent with the fact that both mutants were unable to produce OspC, a virulence factor essential for B. burgdorferi to establish infection in the mammalian host (22, 50) and possibly for spirochetal transmission from the tick gut to the salivary glands (13, 35). However, it remains unclear whether the loss of infectivity of the rpoN and rpoS mutants is due solely to the abrogation of OspC or is also related to the loss of additional virulence determinants.
The upstream activator of the RpoN-RpoS pathway, Rrp2, is predicted to comprise three functional domains: an N-terminal receiver domain typical of a two-component response regulator, a central σ54-dependent activation domain, and a C-terminal DNA-binding domain. Multiple attempts to inactivate rrp2 have not been successful (5, 56), suggesting that the abrogation of rrp2 may be deleterious to cell survival. However, successful generation of an rrp2 mutant encoding an Rrp2 variant with a point mutation of G239C in the central activation domain provided genetic evidence that Rrp2 is a σ54-dependent activator and controls the activation of the RpoN-RpoS pathway (56). In addition, Burtnick et al. recently reported that unlike other σ54-dependent activators that require an enhancer-binding site for activation, Rrp2 was capable of activating rpoS in an enhancer-independent manner (5).
In contrast to RpoN and RpoS, the role of Rrp2 in the infectious cycle of B. burgdorferi has not been examined due to the inability to generate any rrp2 mutant and the isogenic complemented strain from an infectious strain of B. burgdorferi. Herein, we report the successful construction of an rrp2 mutant from a virulent strain of B. burgdorferi and a corresponding complemented clone that retains full virulence. With these strains, we demonstrated that Rrp2 is required for mammalian infection but not for spirochetal survival in ticks. Furthermore, we show that constitutive expression of ospC could not rescue the avirulent phenotype of the rrp2 mutant, indicating that Rrp2 controls additional virulence determinants essential for B. burgdorferi to establish infection in mammals. Lastly, as an initial approach to identify Rrp2-dependent virulence factors, we performed microarray analyses to determine the global influence of Rrp2 on gene expression in B. burgdorferi.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. burgdorferi strains used in this study are listed in Table 1. Strain 5A4NP1 (a gift of H. Kawabata and S. Norris at the University of Texas Health Science Center at Houston) is a B31 clone that contains all essential endogenous plasmids and is infectious in mice and ticks (26, 36). It has a kanamycin resistance marker inserted in the restriction modification gene BBE02 on plasmid lp25, resulting in increased transformation efficiency and allowing selection for clones that retain the essential plasmid lp25. Strain 13A is a B31 5A13 derivative that is missing plasmids lp25 and lp56 (36, 52). Borreliae were cultivated in vitro in modified Barbour-Stoenner-Kelly medium (BSK-H; Sigma, St. Louis, MO) supplemented with 6% normal rabbit serum (Pel Freez Biologicals, Rogers, AR) or BSK-H complete medium at 35°C unless indicated otherwise.
TABLE 1.
B. burgdorferi strains used in this study
| Strain name | Description | Reference or source |
|---|---|---|
| 5A4NP1a | BBE02::Kanr, cp9− | 26 |
| 5A4NP1 rrp2(G239C) | Same as 5A4NP1, except rrp2 was replaced with rrp2(G239C)-ermC | This study |
| 5A4NP1 rrp2(wt) | Same as 5A4NP1 rrp2(G239C), except rrp2(G239C)-ermC was replaced with rrp2(wt)-aadA | This study |
| 13Aa | lp25−, lp56− | 52 |
| 13A ospC::Genr | Same as 13A, except ospC::Genr | 52 |
| 13A ospC::Genr/pBBE22-flaBp-ospC | Same as 13A ospC::Genr, except carrying a shuttle vector with a copy of wild-type BBE22 and constitutive flaBp-ospC | This study |
| 13A rrp2(G239C) | Same as 13A, except rrp2 was replaced with rrp2(G239C)-ermC and lp28-4 was lost | This study |
| 13A rrp2(G239C)/pBBE22-flaBp-ospC | Same as 13A rrp2(G239C), except carrying a shuttle vector with a copy of wild-type BBE22 and constitutive flaBp-ospC | This study |
| 13A rrp2(wt) | Same as 13A rrp2(G239C), except rrp2(G239C)-ermC was replaced with rrp2(wt)-aadA | This study |
| 13A rrp2(wt)/pBBE22-ospC | Same as 13A rrp2(wt), except carrying a shuttle vector with a copy of wild-type BBE22 and constitutive flaBp-ospC | This study |
A clonal isolate of strain B31.
Generation of transformants.
Electrocompetent B. burgdorferi cells were prepared and transformed as previously described (40, 58). Briefly, 20 to 50 μg of plasmid DNA was used in each transformation. After electroporation, the culture was incubated at 34°C overnight to allow recovery. Relevant antibiotics were then added to the cultures in the following final concentrations: 50 μg/ml for gentamicin, 50 ng/ml for erythromycin, 50 μg/ml for streptomycin, and 300 μg/ml for kanamycin. Cultures were then aliquoted into 96-well tissue culture plates (230 μl/well) for colony selection of transformants (58) instead of using semisolid medium as previously described (40). Two weeks after plating, wells containing positive cultures were identified by a color change in the medium, and the presence of viable spirochetes was verified by dark-field microscopy. To create the rrp2(G239C) mutant, 5A4NP1 and 13A were transformed with the suicide vector pXY201A containing the mutated rrp2 gene linked to an ermC marker (41, 56). To confirm marker exchange, PCR was performed on whole-cell lysates of transformants and resulting PCR products were then subjected to DNA sequence analysis to verify the presence of the mutation corresponding to G239C. The resulting strains were designated 5A4NP1 rrp2(G239C) and 13A rrp2(G239C), respectively (Table 1). To generate the corresponding complemented strains, 5A4NP1 rrp2(G239C) and 13A rrp2(G239C) were transformed with the suicide vector pXY206A, which harbors a wild-type copy of rrp2 linked to an aadA marker (17, 56). PCR and sequencing analyses were then performed to confirm the replacement of the mutant rrp2 with a wild-type copy of rrp2. The resulting strains were designated 5A4NP1 rrp2(wt) and 13A rrp2(wt), respectively.
To generate strains with constitutive expression of ospC, strains 13A ospC::Genr (53), 13A rrp2(G239C), and 13A rrp2(wt) were transformed with the shuttle vector pBBE22-flaBp-ospC (53), resulting in 13A ospC::Genr/pBBE22-flaBp-ospC, 13A rrp2(G239C)/pBBE22-flaBp-ospC, and 13A rrp2(wt)/pBBE22-ospC, respectively. Transformants were confirmed by the constitutive OspC expression via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. The plasmid contents of transformants were surveyed by PCR (27). In addition to lp25 and lp56, which are not present in parental strain 13A (53), strains 13A rrp2(G239C)/pBBE22-flaBp-ospC and 13A rrp2(wt)/pBBE22-ospC are missing lp28-4, which is not necessary for infectivity for the inoculum dose used in this study (105 spirochetes/mouse), despite the fact that it can partially contribute to infectivity; loss of lp28-4 resulted in an increase of the 50% infective dose of B. burgdorferi by approximately 1 log following intradermal inoculation (27, 36).
B. burgdorferi infection of mice via needle inoculation.
For mammalian infection studies, 3- to 4-week-old C3H/HeJ mice (Jackson Laboratory, Bar Harbor, ME) or C3H/SCID mice (Harlan, Indianapolis, IN) were inoculated intradermally with 1 × 105 spirochetes. At 10 to 14 days postinoculation, ear punch biopsy samples (and heart, spleen, and joint tissues for tick-infected mice) were collected and spirochetes were cultured in BSK-H medium supplemented with 1× Borrelia antibiotic mixture (Sigma). After 1 to 2 weeks, dark-field microscopy was used to examine cultures for the presence of motile spirochetes: a single growth-positive culture was used as the criterion for infection of each mouse. All animal and tick experimentation was approved by the University of Laboratory Animal Care Committee at Indiana University.
Microinjection of B. burgdorferi into ticks.
Pathogen-free Ixodes scapularis nymphs were obtained from the Tick Rearing Facility at Oklahoma State University. Microinjection was used to introduce spirochetes into the guts of I. scapularis nymphs as previously described (35, 58). Briefly, each B. burgdorferi variant was cultivated under normal conditions in BSK-H medium in the presence of selective antibiotics. Bacteria were harvested by centrifugation and concentrated in phosphate-buffered saline (PBS) to a density of 108 spirochetes per ml. Ten microliters of the cell suspension was then loaded into a 1-mm-diameter glass capillary needle (World Precision Instruments Inc., Sarasota, FL) by use of a microloader (Eppendorf AG, Westbury, NY). The bacterial suspension was then injected into the rectal apertures of unfed nymphal ticks by use of a FemtoJet microinjector system (Eppendorf AG). The parameters for injection were a pressure of 1,000 hPa, an injection time of 0.1 s, and a compensation pressure of 0 hPa, which delivered an average volume of 0.15 μl (1 × 104 to 2 × 104 spirochetes).
Transmission of B. burgdorferi to mice via tick bite.
After microinjection, ticks were allowed to recover for 2 to 4 h and then placed on C3H/HeJ mice (∼15 ticks/mouse). To confine the infected ticks to the mammalian host, nymphs were placed in containment capsules as previously described (55). To construct the capsules, the bottoms of screw-cap microcentrifuge tubes were cut 5 mm below the screw threads and attached to the shaved backs of mice with a melted mixture (weight/weight) of 4 parts rosin and 1 part beeswax. Mesh was placed over a hole made in the screw-cap lid and secured with Super Glue (Loctite). Ticks were then placed in the containment unit, which was quickly closed with the screw cap. Ticks were allowed to feed to repletion (4 to 5 days) and then collected.
Immunofluorescence assays.
The entire contents of the fed nymphs were placed onto silylated microscope slides (CEL Associates, Pearland, TX). Slides were allowed to air dry before being placed on a 65°C heating block for 25 min, followed by submersion in acetone for 5 min to complete fixation. Slides were incubated at 37°C for 1 h with blocking solution (PBS-Tween 20 with 5% goat serum) in a humid chamber. The blocking solution was replaced with BacTrace fluorescein isothiocyanate-conjugated goat anti-B. burgdorferi antibody (Kirkegaard and Perry Laboratories, Gaithersburg, MD) at a 1:100 dilution in blocking solution. The slides then were incubated for 1 h at 37°C in a dark, humid chamber. Slides then were washed twice in PBS-Tween 20 and counterstained with 20 μg/ml propidium iodide in PBS for 3 min. Slides were washed twice with PBS-Tween 20 and then mounted with antifade light mounting medium (Molecular Probes, Eugene, OR). Samples were observed for B. burgdorferi by using an Olympus BX50 fluorescence microscope with a 40× objective equipped with a charge-coupled-device camera (CCD-100S; DAGE-MTI, Michigan City, IN) and Olympus DP Controller software.
SDS-PAGE and immunoblotting.
SDS-PAGE and immunoblotting were carried out as previously described (55). Cells were loaded in gel lanes at 5 × 107 cells per lane. For immunoblotting, SDS-PAGE gels were transferred to nitrocellulose in a Bio-Rad Mini Trans-Blot transfer cell at room temperature. Protein bands were detected using a 1:50 dilution of mixed monoclonal antibodies against FlaB and OspC (54, 57) and a 1:1,000 anti-mouse immunoglobulin G peroxidase conjugate (Jackson ImmunoResearch Laboratories, West Grove, PA) secondary antibody and developed with 4-chloro-1-naphthol as the substrate.
RNA extraction, microarray, and quantitative reverse transcription-PCR (qRT-PCR) analyses.
RNA was extracted from three biological replicates of 5A4NP1 and 5A4NP1 rrp2(G239C) by use of Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Digestion of contaminating genomic DNA in the RNA samples was performed using RNase-free DNase I (GenHunter Technology, Nashville, TN), and removal of DNA was confirmed by PCR amplification using primers specific for the B. burgdorferi flaB gene. RNA quality was determined using the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA).
Oligonucleotides representing 1,723 putative open reading frames of B. burgdorferi B31MI and 19 random-sequence 70-mer negative controls were synthesized by Qiagen-Operon (Alameda, CA) as described previously (49). The oligonucleotides were resuspended in 150 mM sodium phosphate (pH 8.5; Microarrays Inc., Nashville, TN) to a concentration of 40 μM and printed on CodeLink activated slides (Amersham Biosciences, Piscataway, NJ) by use of a custom arrayer (Microarrays Inc.). Each oligonucleotide was printed in quadruplicate on each array. Arrays were blocked postprinting per CodeLink instructions with 50 mM ethanolamine. The attachment of probe DNA was confirmed by Microarrays' proprietary Veriprobe assay (Microarrays Inc.).
cDNA was synthesized and labeled with Cy3 or Cy5 by use of the Amersham postlabeling kit according to the manufacturer's instructions, with minor modifications (Amersham Biosciences). Briefly, 10 μg of total RNA was converted to cDNA by use of CyScript RT in the presence of 1 μl of random nanomers (Amersham Biosciences) and 4.5 μg of random hexamers (Invitrogen). Each cDNA sample was labeled with Cy3 and Cy5 separately. Cy3- or Cy5-labeled cDNA from parental B31 RNA was then combined with the Cy5- or Cy3-labeled cDNA from the mutants. Labeled probes were purified using the Qiaquick PCR purification kit (Qiagen, Valencia, CA) and then applied in the microarray experiment. With three pairs of samples plus dye switching, we made a total of six hybridized slides. Hybridized slides were then scanned on an Axon 4000B microarray scanner using GenePix Pro 6.1 (Molecular Devices, Sunnyvale, CA). The image was analyzed using the GenePix program, and data were then analyzed with Acuity 4.0 (Molecular Devices) by using the ratio-based normalization method and a cutoff value of a threefold change. Statistical analyses were performed using the one- and two-sample significance test (P < 0.05) in the Acuity program.
qRT-PCR was performed with RNA samples used for microarray analysis. cDNA was synthesized using the ThermoScript RT system (Invitrogen). qPCR was performed in triplicate on an ABI 7000 sequence detection system using Platinum SYBR green qPCR SuperMix (Invitrogen). Calculations of the relative expression of the gene of interest were normalized to flaB gene expression by using the threshold cycle (ΔΔCT) method. Comparisons of B. burgdorferi gene expression between the wild type and the rrp2 mutant were performed using the Student t test.
Microarray data accession number.
The array data have been deposited at http://www.ncbi.nlm.nih.gov/geo/ (accession number GSE11284).
RESULTS
Rrp2 is essential for mammalian host infection via needle or tick inoculation.
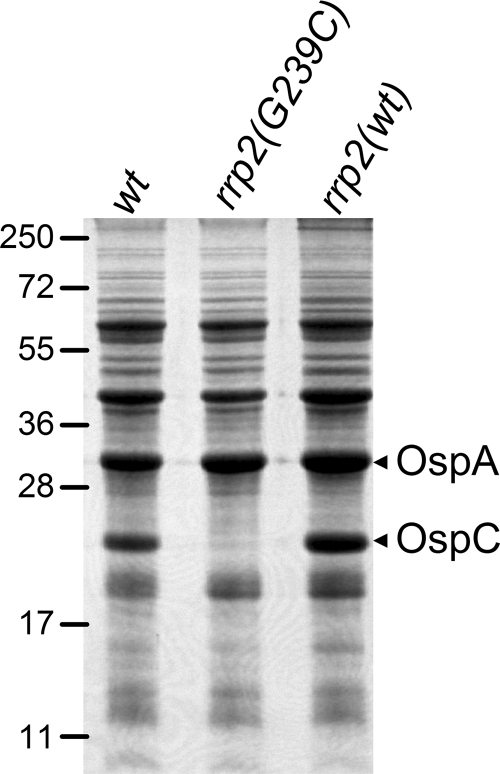
We previously showed that a point mutation (G239C) in the activation domain of Rrp2 abolishes temperature- and pH-induced activation of rpoS and ospC, providing genetic evidence that Rrp2 is the σ54-dependent activator that governs expression of rpoS and, consequently, the production of OspC and other RpoS-dependent proteins. Despite the fact that the previously constructed rrp2 mutant was generated from an infectious clone of B. burgdorferi strain 297, the corresponding cis-complemented strain (created upon replacing the mutant rrp2 with a wild-type copy of rrp2) was avirulent and therefore rendered the rrp2 mutant unsuitable for studying Rrp2 function in vivo (data not shown). To overcome this obstacle and to elucidate the role of Rrp2 in the infectious cycle of B. burgdorferi, a similar rrp2 mutant was generated from infectious clone 5A4NP1 of B. burgdorferi B31 by use of the previously reported strategy (56). One rrp2 mutant clone that has an endogenous plasmid profile identical to that of 5A4NP1 was subsequently selected for the generation of a complemented (replacement) strain by transforming a suicide vector carrying a wild-type rrp2 linked to an aadA marker, which confers streptomycin resistance in B. burgdorferi (17, 56). Consistent with previous findings for B. burgdorferi 297, the G239C mutation in Rrp2 abolished the production of OspC in B31 (Fig. 1).
FIG. 1.
Protein expression profiles of B. burgdorferi strains used for the infection and microarray analyses. Infectious clone 5A4NP1 [wild type (wt)], the isogenic rrp2 mutant [rrp2(G239C)], and the complemented rrp2 strain [rrp2(wt)] were cultivated in BSK-H medium and harvested at the late logarithmic phase of growth (5 × 107 spirochetes/ml), and whole-cell lysates (5 × 107 spirochetes/gel lane) were subjected to SDS-PAGE (Coomassie blue-stained gel). Numbers at the left denote protein molecular mass markers in kDa. The bands corresponding to OspC and OspA are indicated on the right.
To examine the role of Rrp2 in spirochetal infection in mammals, groups of C3H/HeJ mice were inoculated intradermally with 105 spirochetes of either wild-type strain 5A4NP1, rrp2 mutant 5A4NP1 rrp2 (G239C), or rrp2-complemented strain 5A4NP1 rrp2(wt). Two weeks after inoculation, ear punch biopsy samples were harvested and cultured in BSK-H medium. As shown in Table 2, none of the mice infected with the rrp2 mutant were culture positive for B. burgdorferi infection, suggesting that Rrp2 is required for mammalian infection. However, the rrp2-complemented strain was readily observed in all cultures of ear punch biopsy samples, indicating that the loss of virulence in the rrp2 mutant was solely due to the mutation in the rrp2 gene.
TABLE 2.
Rrp2 is required for mammalian infection
| Strain | No. of mice infected by indicated route/total no. of mice
|
|
|---|---|---|
| Needle inoculation | Tick bite | |
| 5A4NP1 | 5/5 | 4/4 |
| 5A4NP1 rrp2(G239C) | 0/5 | 0/7 |
| 5A4NP1 rrp2(wt) | 5/5 | 5/5 |
| 13A ospC::Genr/pBBE22-flaBp-ospC | 5/5 | NAa |
| 13A rrp2(G239C)/pBBE22-ospC | 0/6 | NA |
| 13A rrp2(wt)/pBBE22-ospC | 6/6 | NA |
NA, not available.
It has been reported that needle inoculation and tick challenge of mice may have profoundly different infection outcomes (23). Therefore, we further examined the infectivity of the rrp2 mutant via tick bite. To do so, we employed a previously developed microinjection technique to artificially inject spirochetes into sterile nymph guts (35, 58). This technique allows the delivery of equal numbers of mutant and wild-type spirochetes directly into the tick gut via the tick rectum with virtually 100% efficiency. Approximately 1.5 × 104 spirochetes of 5A4NP1, the rrp2 mutant, or the corresponding complemented strain were microinjected into flat I. scapularis nymphs. The injected ticks then fed on C3H/HeJ mice. Consistent with the results from needle inoculation, bacteria could not be recovered from any tissue (ear punch biopsy samples or heart, spleen, or joint tissue) of the mice challenged with the rrp2 mutant strain, whereas all mice challenged by ticks injected with wild-type or rrp2-complemented strains yielded spirochetes from cultivated tissues (Table 2). These data indicate that a functional Rrp2 is required for mammalian infection regardless of the infection route.
Rrp2 is not required for spirochetal survival during tick feeding.
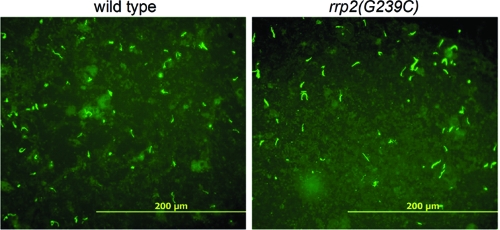
Because the rrp2 mutant was incapable of infecting mice, the entire infectious cycle of the rrp2 mutant between ticks and mammals could not be examined. However, the ability to generate artificially infected ticks allowed an examination of the mutant's behavior during tick feeding, the period when Rrp2 and the RpoN-RpoS pathway become activated (8). To do so, microinjected ticks carrying various B. burgdorferi strains fed on naïve mice and naturally detached ticks were collected. Fed nymphs were dissected and the tick smears were subjected to immunofluorescence assays. As shown in Fig. 2, both 5A4NP1 and the isogenic rrp2 mutant were readily detectable in ticks and no obvious difference in spirochetal numbers was observed. These data indicate that the rrp2 mutant is able to survive in the tick vector upon the intake of a blood meal, implying that Rrp2 does not control the production of molecules essential for spirochetal survival in the tick gut during host feeding.
FIG. 2.
Detection of wild-type or rrp2 mutant spirochetes in ticks by immunofluorescence assays. Unfed I. scapularis nymphs were microinjected with either wild-type 5A4NP1 or isogenic rrp2 mutant [rrp2(G239C)] spirochetes and allowed to feed to repletion on mice. After detachment, fed nymphs were dissected, and tick smear preparations were subjected to immunofluorescence assays with fluorescein isothiocyanate-labeled anti-B. burgdorferi antibody. Panels shown are representative images from three separate experiments.
Constitutive expression of ospC did not restore virulence in an rrp2 mutant.
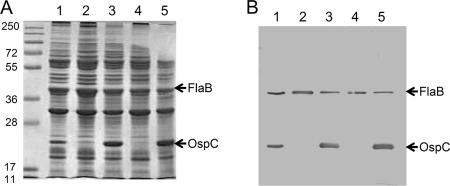
OspC production is controlled by the Rrp2-RpoN-RpoS regulatory cascade and is required for B. burgdorferi to infect mammals (5, 12, 20, 25, 46, 56, 57). A recent report showed that complementation of an ospC mutant with a copy of constitutively expressed ospC (under the flaB promoter of B. burgdorferi) restored infectivity in immunodeficient SCID mice, albeit the complemented strain did not sustain infection in immunocompetent mice (53). To determine whether the avirulence of the rrp2 mutant was solely due to the loss of OspC, we generated an rrp2(G239C) mutant from 13A, the strain that was used for the ospC deletion and complementation studies with flaBp-ospC (52, 53). The newly generated strain, namely, the 13A rrp2(G239C) mutant, along with the 13A ospC::Genr mutant, was then transformed with a shuttle vector, pBBE22-flaBp-ospC. A wild-type copy of the BBE22 gene was also included on the shuttle vector, because 13A has lost lp25, which contains the BBE22 gene essential for spirochete growth in mice (37). As shown in Fig. 3, transformation with pBBE22-flaBp-ospC restored expression of ospC to both the ospC and rrp2 mutants during in vitro growth.
FIG. 3.
Restoration of OspC expression with the pBBE22-flaBp-ospC plasmid. SDS-PAGE (Coomassie blue-stained gel) (A) and immunoblot (B) analysis of whole-cell lysates of various strains of spirochetes harvested at the late logarithmic phase of growth (5 × 107 spirochetes/ml). Lanes for both panels: 1, 13A (wild type); 2, 13A ospC::Genr; 3, 13A ospC::Genr/pBBE22-flaBp-ospC; 4, 13A rrp2(G239C); 5, 13A rrp2(G239C)/pBBE22-flaBp-ospC. For immunoblotting, a 1:10 dilution of the samples used for SDS-PAGE analysis was loaded and monoclonal antibodies directed against FlaB (loading control) and OspC were pooled. The bands corresponding to FlaB and OspC are indicated by arrows.
Groups of C3H/SCID mice were challenged with strains 13A ospC::Genr/pBBE22-flaBp-ospC and 13A rrp2(G239C)/pBBE22-flaBp-ospC or complemented strain 13A rrp2(wt)/pBBE22-ospC (Table 2). Whereas spirochetes were readily detected in tissues from mice infected with the ospC mutant carrying pBBE22-flaBp-ospC (five mice infected of five mice total), no spirochetes were detected in tissues from mice inoculated with 105 rrp2 mutant spirochetes harboring pBBE22-flaBp-ospC (zero mice infected of six mice total) (Table 2). Replacement of rrp2(G239C) with wild-type rrp2 restored infectivity (six mice infected of six mice total). These results showed that the avirulent phenotype of the rrp2 mutant was not solely due to the loss of OspC production, indicating that Rrp2 controls the expression of an additional virulence determinant(s) that is indispensable for B. burgdorferi to establish infection in mammalian hosts.
Genome-wide analyses of Rrp2-controlled genes.
As an initial approach to identify virulence determinants controlled by Rrp2, we performed microarray analyses of 5A4NP1 and the isogenic rrp2 mutant spirochetes harvested from late-logarithmic-phase cultures grown at 35°C in BSK-H medium. When a cutoff value of a threefold change was used as the gene selection criterion, 125 genes showed higher expression in wild-type spirochetes than in the rrp2 mutant (Table 3), suggesting that they are positively regulated by Rrp2. As expected, the expression of rpoS was 40-fold higher in the wild type than in the rrp2 mutant. Consistent with this finding, the expression of ospC, dbpBA, BBK32, oppA5, BBA64, and BBA66, which are well-described RpoS-dependent genes (10, 19, 21, 22, 29, 30, 43, 44, 50), showed greater dependence on Rrp2 (9- to 90-fold difference in expression). Notably, more than half of the Rrp2-activated genes (60 genes, or 52%) have previously been identified by microarray analyses as differentially regulated genes under different conditions varying in terms of temperature, pH, the addition of blood, or host adaptation (namely, a dialysis membrane chamber [DMC] implanted in the rat peritoneal cavity) (2, 8, 33, 38, 51). These findings further reinforce the notion that Rrp2 is the major transcriptional regulator controlling differential gene expression in B. burgdorferi.
TABLE 3.
Genes activated by Rrp2 (>3-fold)
| Gene | Function(s) | Fold change |
|---|---|---|
| BBA06 | Hypothetical protein | 3,629.0 |
| BBD07 | Hypothetical protein | 2,330.0 |
| BBA34 | OppAV, oligopeptide ABC transporter | 542.8 |
| BBG27 | Conserved hypothetical protein | 428.0 |
| BBA72 | Hypothetical protein | 109.7 |
| BBB19 | OspC | 90.1 |
| BBA71 | Hypothetical protein | 78.0 |
| BBA37 | Hypothetical protein | 71.9 |
| BBG24 | Hypothetical protein | 62.7 |
| BBA66 | P35 | 56.9 |
| BBA25 | DbpB, decorin-binding protein | 51.0 |
| BBG26 | Hypothetical protein | 43.7 |
| BB0771 | RNA polymerase sigma factor RpoS | 40.8 |
| BB0844 | Hypothetical protein | 38.3 |
| BBA73 | Antigen, P35, putative | 35.3 |
| BBH41 | Conserved hypothetical protein | 34.9 |
| BBA36 | Lipoprotein | 30.6 |
| BBA05 | S1 antigen | 25.6 |
| BBD24 | Hypothetical protein | 24.4 |
| BBK32 | P35, fibronectin-binding protein | 23.5 |
| BBA07 | ChpAI protein, putative (homolog to Mlp) | 23.3 |
| BBA24 | DbpA, decorin-binding protein | 19.7 |
| BBM27 | Rev protein | 16.5 |
| BBP27 | Rev protein | 16.4 |
| BBM28 | MlpF, lipoprotein | 15.3 |
| BBJ001 | Conserved hypothetical protein, pseudogene | 14 |
| BBG25 | Conserved hypothetical protein | 13.0 |
| BBR29 | Conserved hypothetical protein | 12.9 |
| BBA65 | Hypothetical protein | 10.3 |
| BBJ01 | Hypothetical protein | 10.0 |
| BBD19 | Hypothetical protein | 9.9 |
| BBJ23 | Hypothetical protein | 9.3 |
| BBA64 | P35, antigen (Gilmore) | 9.2 |
| BBA26 | Hypothetical protein | 8.7 |
| BBJ02 | Hypothetical protein | 8.4 |
| BBM38 | ErpK protein | 7.2 |
| BBG22 | Hypothetical protein | 7.1 |
| BBA35 | Hypothetical protein | 7.0 |
| BBS41 | ErpG (OspG) | 6.8 |
| BBS42 | Associated protein A (BapA) | 6.7 |
| BBE31 | P35, putative | 6.5 |
| BB0680 | Methyl-accepting chemotaxis protein Mcp-4 | 6.2 |
| BBO39 | ErpL protein | 6.2 |
| BBK07 | Hypothetical protein | 6.0 |
| BBS28 | Hypothetical protein | 6.0 |
| BB0519 | GrpE protein | 5.6 |
| BBG10 | Hypothetical protein | 5.4 |
| BBA33 | Hypothetical protein | 5.4 |
| BBR44 | Hypothetical protein | 5.2 |
| BBJ26 | ABC transporter, ATP-binding protein | 4.8 |
| BBF01 | ErpD-like protein, putative | 4.8 |
| BB0681 | Methyl-accepting chemotaxis protein Mcp-5 | 4.7 |
| BBP28 | MlpA, lipoprotein | 4.5 |
| BBJ27 | Hypothetical protein | 4.5 |
| BBQ64 | Hypothetical protein | 4.5 |
| BBH09.1 | Conserved hypothetical protein, pseudogene | 4.4 |
| BBA17 | Hypothetical protein | 4.4 |
| BB0418 | Hypothetical protein | 4.3 |
| BBQ65 | Conserved hypothetical protein, pseudogene | 4.3 |
| BBJ25 | Hypothetical protein | 4.3 |
| BB0040 | Chemotaxis protein methyltransferase CheR-1 | 4.3 |
| BBH10 | Hypothetical protein | 4.2 |
| BB0509 | Hypothetical protein | 4.2 |
| BBD001 | Conserved hypothetical protein | 4.2 |
| BBN29 | Hypothetical protein, authentic point mutation | 4.2 |
| BBL29 | Conserved hypothetical protein | 4.2 |
| BBJ29 | Hypothetical protein | 4.2 |
| BBR40 | ErpH protein | 4.0 |
| BBG20 | Hypothetical protein | 4.0 |
| BBG16 | Hypothetical protein | 4.0 |
| BBJ28 | Hypothetical protein | 4.0 |
| BBJ48 | Hypothetical protein | 4.0 |
| BBH01 | Conserved hypothetical protein | 4.0 |
| BBK48 | P37, putative | 3.9 |
| BBG29 | Conserved hypothetical protein | 3.9 |
| BBD20 | Transposase-like protein, authentic frameshift | 3.9 |
| BBK52.1 | Conserved hypothetical protein, pseudogene | 3.9 |
| BBF11.1 | Conserved hypothetical protein, pseudogene | 3.8 |
| BBG19 | Hypothetical protein | 3.8 |
| BBJ24 | Hypothetical protein | 3.8 |
| BBL39 | ErpA protein | 3.8 |
| BBP26 | Conserved hypothetical protein | 3.8 |
| BBK53 | Outer membrane protein | 3.7 |
| BBQ63 | Hypothetical protein | 3.7 |
| BBR41 | Conserved hypothetical protein | 3.7 |
| BBO29 | Hypothetical protein | 3.7 |
| BBJ47 | Hypothetical protein | 3.6 |
| BBG17 | Hypothetical protein | 3.6 |
| BBL40 | ErpB2 protein | 3.6 |
| BBO40 | ErpM protein | 3.5 |
| BBM39 | Hypothetical protein | 3.5 |
| BBR15 | Hypothetical protein | 3.5 |
| BBI42 | Outer membrane protein, putative | 3.5 |
| BBG03 | Conserved hypothetical protein, authentic frameshift | 3.5 |
| BBR42 | ErpY (OspF) | 3.4 |
| BBA32 | Hypothetical protein | 3.4 |
| BBS27 | Hypothetical protein | 3.4 |
| BBH09 | Hypothetical protein | 3.3 |
| BBM26 | Conserved hypothetical protein | 3.3 |
| BBR36 | Conserved hypothetical protein | 3.3 |
| BBG14 | Hypothetical protein | 3.3 |
| BBG21 | Hypothetical protein | 3.3 |
| BBA57 | Hypothetical protein | 3.3 |
| BBL36 | Conserved hypothetical protein | 3.3 |
| BBH06 | Hypothetical protein, CspZ | 3.2 |
| BBG18 | Hypothetical protein | 3.2 |
| BBA01 | Conserved hypothetical protein | 3.2 |
| BBP40 | Hypothetical protein | 3.2 |
| BBK17 | Adenine deaminase, AdeC | 3.2 |
| BBB23 | Conserved hypothetical protein | 3.2 |
| BBM35 | Conserved hypothetical protein | 3.2 |
| BBK24.1 | Hypothetical protein | 3.1 |
| BBS38 | Conserved hypothetical protein | 3.1 |
| BBN35 | BdrD10 | 3.1 |
| BBO36 | Conserved hypothetical protein | 3.1 |
| BBB15 | Hypothetical protein | 3.1 |
| BBR43 | Hypothetical protein | 3.1 |
| BBN28 | MlpI lipoprotein | 3.1 |
| BBB20 | Hypothetical protein | 3.1 |
| BBK39 | Hypothetical protein | 3.1 |
| BBJ43 | Hypothetical protein | 3.0 |
| BBQ43 | Conserved hypothetical protein | 3.0 |
| BBE11 | Hypothetical protein | 3.0 |
| BBJ46 | Hypothetical protein | 3.0 |
| BBH18 | Hypothetical protein | 3.0 |
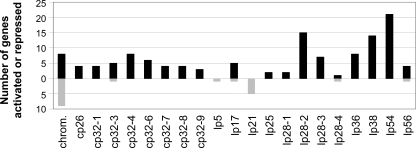
The genome distribution of the Rrp2-activated genes revealed that 94% of these genes are located on plasmids, mainly on linear plasmids lp54, lp28-2, and lp38, as well as on the cp32 circular plasmids (Fig. 4). This is consistent with the notion that the plasmids harbor the majority of differentially regulated genes (2, 32, 39, 51). Because most of the plasmid-located genes are annotated either as hypothetical or as conserved hypothetical genes, they provided limited clues about the physiological processes regulated by Rrp2. However, one prominent feature was that 26% of the Rrp2-activated genes encode known or putative surface lipoproteins, suggesting their potential roles in pathogen-host interactions, as has been demonstrated for dbpBA and BBK32 (14, 15, 43, 44).
FIG. 4.
Summary of genes activated or repressed by Rrp2. Numbers represent the total numbers of genes activated (black bars) or repressed (gray bars) for each plasmid. Plasmid cp9 is absent from the parental strains and is not presented in the diagram. No genes on lp5 or lp21 were found to be activated by Rrp2. Only genes on the chromosome (chrom.), lp17, lp28-4, lp56, cp32-3, lp5, and lp21 were found to be repressed by Rrp2.
In contrast to the large number of plasmid-contained genes controlled by Rrp2, there are only eight chromosomal genes shown to be positively regulated by Rrp2. In addition to rpoS, the chromosomal gene that displayed the greatest dependence on Rrp2 was BB0844 (38-fold difference in expression), which encodes a putative lipoprotein that shares no homology with any other protein in the database. Interestingly, BB0844 is one of the most highly regulated genes under every condition parameter tested, including temperature, pH, blood addition, and DMC, as revealed by previous microarray analyses (2, 33, 38, 51). The finding of its dependence on Rrp2 is also consistent with recent microarray and qRT-PCR analyses showing that BB0844 expression is RpoS dependent (8). Aside from BB0844, three out of eight total Rrp2-dependent chromosomal genes are related to chemotaxis (mcp4, mcp5, and cheR1). Although they present moderate differences in expression (four- to sixfold), further qRT-PCR analysis confirmed that the influence on expression of these genes by Rrp2 was significant (Fig. 5). This result is also consistent with the recent microarray findings that expression of these genes was influenced by temperature, blood, or DMC conditions (2, 32, 39, 51) and affected by the inactivation of rpoS (8).
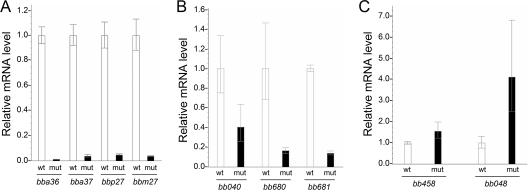
FIG. 5.
qRT-PCR analysis of representative genes regulated by Rrp2. mRNA levels of four plasmid-contained, Rrp2-activated genes (A), three chromosome-contained, Rrp2-activated, chemotaxis-related genes (B), and two Rrp2-repressed genes (C) in the 5A4NP1 rrp2(G239C) mutant (mut) relative to the levels in the 5A4NP1 wild type (wt) (value = 1). The level of flaB mRNA was used for normalization of the relative mRNA level of each gene. Data were calculated from three independent cultures, and the differences in mRNA levels between the wild type and the rrp2 mutant are statistically significant (P < 0.05).
In addition to Rrp2-activated genes, 19 genes showed a level of expression in the wild-type strain that was decreased by more than threefold compared to that in the rrp2 mutant, suggesting that the expression of these genes is negatively regulated by Rrp2 (i.e., Rrp2-repressed genes) (Table 4). However, the overall range of the change in expression for this group of genes was much less (3- to 6.3-fold) than what was observed for Rrp2-activated genes (3- to 3,629-fold). Among them, five repressed genes were located on lp21, a plasmid not essential for mammalian infection. Interestingly, eight of the Rrp2-repressed genes belong to paralogous gene family 57, whose members are located on various plasmids (9). In addition, there are nine chromosomal genes that showed mild but statistically significant repression by Rrp2 (3.2- to 4.3-fold), two of which were confirmed by qRT-PCR analysis (Table 4 and Fig. 5C). Some of the Rrp2-repressed genes encode proteins with putative functions, including DNA helicase (BB010), putative flagellum protein (BB0180), asparaginyl-tRNA synthetase (BB101), alanine racemase (BB0160), and holo-acyl-carrier protein synthase (BB010) (18).
TABLE 4.
Genes repressed by Rrp2 (>3-fold)
| Gene | Function(s) | Fold change |
|---|---|---|
| BB0762 | Hypothetical protein | 6.3 |
| BBI02.2 | Brute force ORFa | 5.3 |
| BBU04 | Conserved hypothetical protein | 5.0 |
| BBU07 | Conserved hypothetical protein | 4.8 |
| BBU10 | Hypothetical protein | 4.8 |
| BB0773 | Hypothetical protein | 4.3 |
| BBD14 | Conserved hypothetical protein | 3.8 |
| BBU06 | Conserved hypothetical protein | 3.7 |
| BB0111 | Replicative DNA helicase | 3.7 |
| BBQ38 | Conserved hypothetical protein | 3.7 |
| BB0180 | Flagellar protein, putative | 3.4 |
| BB0128 | Cytidylate kinase | 3.4 |
| BB0101 | Asparaginyl-tRNA synthetase | 3.4 |
| BBU05 | Plasmid partition protein, putative | 3.3 |
| BB0010 | Holo-acyl-carrier protein synthase, putative | 3.3 |
| BB0048 | Hypothetical protein | 3.3 |
| BBT04 | Conserved hypothetical protein | 3.3 |
| BB0160 | Alanine racemase | 3.2 |
| BBS33 | Conserved hypothetical protein | 3.0 |
ORF, open reading frame.
DISCUSSION
Two-component systems are a mainstay of signal transduction pathways in bacteria (48). In contrast to what is the case for free-living bacteria such as Escherichia coli, which has more than 30 two-component systems, the B. burgdorferi genome encodes only two sets of two-component systems, Hk1-Rrp1 and Hk2-Rrp2, in addition to the CheY-CheA systems involved in chemotaxis (18, 31). We and others have previously shown that Rrp2 controls the activation of the RpoN-RpoS regulatory pathway, which in turn governs the expression of numerous B. burgdorferi genes (5, 7, 8, 16, 25, 56). In this report, we further show that Rrp2 is indispensable for spirochetes to establish infection in the mammalian host and that the avirulent phenotype of the rrp2 mutant is not solely due to the abrogation of OspC. Thus, Rrp2 likely controls additional factors essential for mammalian host infection of B. burgdorferi.
The avirulent phenotype of the rrp2 mutant, which is consistent with the phenotypes of the rpoN and rpoS mutants, was anticipated, since the mutant could no longer produce OspC, a virulence factor essential for mammalian host infection. However, it was not clear heretofore whether the loss of virulence in an rrp2, rpoN, or rpoS mutant was solely due to the loss of OspC. In addition to OspC, three Rrp2-dependent lipoproteins, DbpA, DbpB, and the BBK32 protein, have been shown to contribute to mammalian infection to various degrees (1, 11, 43, 44). Disruption of the dbpBA locus or the BBK32 gene exhibited a 4-log or 1-log decrease in infectivity with needle inoculation, respectively. However, both the dbpBA and the BBK32 mutants were capable of infecting mice through the natural route, i.e., tick bite (1, 44). Thus, the lack of DbpA, DbpB, and the BBK32 protein could not fully account for the avirulent phenotype of the 13A rrp2(G239C)/pBBE22-ospC strain (Table 2). Future work is needed to determine the infectivity of an rrp2 mutant with simultaneous constitutive expression of ospC, dbpBA, and BBK32. Nevertheless, the result that constitutive expression of ospC in the rrp2 mutant did not restore the infectivity in SCID mice indicates that the Rrp2-RpoN-RpoS pathway controls an additional yet-to-be-identified virulence determinant(s) important to the mammalian infection.
In addition to the role of Rrp2 in mammals, its potential function in ticks was also examined in this study. Upon artificially delivering spirochetes into tick guts via microinjection, we were able to examine the mutant's phenotype during tick feeding, the time when Rrp2 is predicted to become activated and when spirochetes encounter the most dramatic environmental changes in ticks. Our data showed that the rrp2 mutant was able to survive in ticks upon feeding. This result is consistent with previous findings for the rpoN mutant (16). Interestingly, the rpoN mutant was shown to be incapable of entering tick salivary glands (16). Further studies are needed to determine whether the rrp2 mutant shares a similar defect. In addition, the ability of the rrp2 mutant to survive through the tick molting process also warrants further study.
Global analyses of gene expression revealed numerous genes controlled by Rrp2, which provides a foundation for the further identification of new virulence determinants. In this study, we focused only on the genes that showed greater-than-threefold transcriptional changes, which should have reduced the number of genes that are false positives. Our analyses revealed that 125 and 19 genes appeared to be activated and repressed by Rrp2, respectively. Because Rrp2 and RpoN act in concert to activate transcription, we anticipate that the rrp2 and rpoN mutants affect similar groups of genes. In this regard, Fisher et al. showed that inactivation of rpoN (ntrA) affected a much higher number of genes (305 genes) (16). This is most likely due to the fact that the previous study included all genes that had statistically significant changes in expression (i.e., without a minimal cutoff of change level). It is noteworthy that both studies showed that a large number of Rrp2- and RpoN-influenced genes are located on lp54 and cp32s (16).
Since rpoS is the only gene that has been experimentally demonstrated to have a σ54 promoter and to be directly activated by Rrp2 and RpoN (5, 25, 28, 46, 56), one would expect that genes dependent on RpoS should also be dependent on Rrp2. The influence of RpoS on global gene expression in both strains B31 and 297 has been analyzed under the in vitro standard cultivation condition as well as under the host adaptation DMC condition (8, 16). A comparison of the Rrp2-dependent and the RpoS-dependent gene profiles revealed that 46 of the top 50 RpoS-activated genes were shown to be Rrp2 dependent, further reinforcing the Rrp2-RpoS linear relationship. On the other hand, nearly half of the top 50 Rrp2-activated genes (22 genes, or 44%) were not present in the RpoS-activated gene profile under either in vitro or DMC conditions. One possibility is that the expression of these genes may be dependent on Rrp2 but not on RpoS. It is also possible that they are RpoS dependent but were missed in the analysis when the B31-based microarray was used for analyzing transcription in strain 297, which is the case for at least one of the genes, ospC (8).
In contrast to the Rrp2-activated genes, which largely overlap with the RpoS-activated genes, none of the 19 Rrp2-repressed genes were shown to be repressed by RpoS (8). Similarly, no RpoS-repressed genes identified in the DMC condition were repressed by Rrp2 in vitro. This is not surprising, since the RpoS-repressed genes, such as ospA and lp6.6, are known to be downregulated under the DMC condition but not under the standard in vitro culture condition that was used in the current study (35°C, pH 7.5). Therefore, they were not expected to be identified as Rrp2-repressed genes by this study. Future work is needed to identify Rrp2-repressed genes under the DMC growth condition.
One caveat of the current study is that a mutant strain carrying the Rrp2 variant with only a G239C point mutation was used rather than an Rrp2-deficient strain. Consequently, this study was limited to elucidating the roles of the central activation domain of Rrp2, but not the entire Rrp2 protein, in the infectious cycle of B. burgdorferi. As pointed out earlier, generating an Rrp2-deficient mutant was not achievable despite multiple attempts (5, 56). It has been postulated that full-length Rrp2 is required for cell survival during in vitro cultivation. For instance, in addition to being a σ54-dependent transcriptional activator, Rrp2 may function as a repressor via its C-terminal putative DNA-binding domain, which suppresses genes that are otherwise lethal for spirochetal growth in vitro. This and other potential functions of Rrp2 remain to be elucidated.
The major temporal environmental change B. burgdorferi encounters during its infectious cycle is the process of tick feeding. During this process, B. burgdorferi in the tick midgut faces a sudden influx of mammalian blood as well as increased tick components (e.g., secreted proteases). In order to adapt to such dramatic changes and prepare for successful transmission and establishment of mammalian infection, spirochetes must evolve strategies that provide quick and global responses to environmental stimuli. In this regard, B. burgdorferi deploys a two-component system, Hk2-Rrp2, to modulate both of the alternative sigma factors present in the genome, σ54 and σS, to fulfill this crucial task. The unique feature of such a regulatory system is that while the activation of Rrp2 allows a quick and tight response to environmental signals (3), having Rrp2 and RpoN (σ54) control the production of the global regulator RpoS (σS), primarily through transcriptional activation (5, 8, 28, 56), provides a global influence on gene expression. Rrp2, as the key regulator of this pathway, plays a central role in the infectious cycle of B. burgdorferi, and the current study further supports this notion. This study also provides a foundation for further identification of the unknown virulence determinants controlled by Rrp2. In addition, the importance of Rrp2 warrants further studies to elucidate the upstream signaling events that lead to the activation of Rrp2 and the RpoN-RpoS pathway (5).
Acknowledgments
We thank S. Norris, H. Kawabata, and F. Liang for generously supplying the 5A4NP1 and 13A ospC::Genr mutant strains and the pBBE22-flaBp-ospC plasmid, Z. Xu for the use of his microscope, and M. Bauer for comments on the manuscript.
The work was supported in part by grants from NIH-National Institute of Arthritis and Musculoskeletal and Skin Disease and by an American Heart Association scientist development grant and the Indiana INGEN grant of Indiana University, funded by the Lilly Endowment, Inc. B.K.B. is supported by the NIH T32 training grant.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 23 June 2008.
REFERENCES
- 1.Blevins, J., K. E. Hagman, and M. V. Norgard. 2008. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 713371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck, M., M.-T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 1824129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease, a tick-borne spirochetosis? Science 2161317-1319. [DOI] [PubMed] [Google Scholar]
- 5.Burtnick, M. N., J. S. Downey, P. J. Brett, J. A. Boylan, J. G. Frye, T. R. Hoover, and F. C. Gherardini. 2007. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol. Microbiol. 65277-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caimano, M. J., C. H. Eggers, C. A. Gonzalez, and J. D. Radolf. 2005. Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J. Bacteriol. 1877845-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caimano, M. J., C. H. Eggers, K. R. O. Hazlett, and J. D. Radolf. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 726433-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caimano, M. J., R. Iyer, C. H. Eggers, C. Gonzalez, E. A. Morton, M. A. Gilbert, I. Schwartz, and J. D. Radolf. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 651193-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35490-516. [DOI] [PubMed] [Google Scholar]
- 10.Clifton, D. R., C. L. Nolder, J. L. Hughes, A. J. Nowalk, and J. A. Carroll. 2006. Regulation and expression of bba66 encoding an immunogenic infection-associated lipoprotein in Borrelia burgdorferi. Mol. Microbiol. 61243-258. [DOI] [PubMed] [Google Scholar]
- 11.Coburn, J., J. R. Fischer, and J. M. Leong. 2005. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol. Microbiol. 571182-1195. [DOI] [PubMed] [Google Scholar]
- 12.Eggers, C. H., M. J. Caimano, and J. D. Radolf. 2004. Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J. Bacteriol. 1867390-7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fingerle, V., G. Goettner, L. Gern, B. Wilske, and U. Schulte-Spechtel. 2007. Complementation of a Borrelia afzelii OspC mutant highlights the crucial role of OspC for dissemination of Borrelia afzelii in Ixodes ricinus. Int. J. Med. Microbiol. 29797-107. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, J. R., K. T. LeBlanc, and J. M. Leong. 2006. Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect. Immun. 74435-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, J. R., N. Parveen, L. Magoun, and J. M. Leong. 2003. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 1007307-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher, M. A., D. Grimm, A. K. Henion, A. F. Elias, P. E. Stewart, P. A. Rosa, and F. C. Gherardini. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl. Acad. Sci. USA 1025162-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank, K. L., S. F. Bundle, M. E. Kresge, C. H. Eggers, and D. S. Samuels. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J. Bacteriol. 1856723-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390580-586. [DOI] [PubMed] [Google Scholar]
- 19.Gautam, A., M. Hathaway, N. McClain, G. Ramesh, and R. Ramamoorthy. 2008. Analysis of the determinants of bba64 (P35) gene expression in Borrelia burgdorferi using a gfp reporter. Microbiology 154275-285. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert, M. A., E. A. Morton, S. F. Bundle, and D. S. Samuels. 2007. Artificial regulation of ospC expression in Borrelia burgdorferi. Mol. Microbiol. 631259-1273. [DOI] [PubMed] [Google Scholar]
- 21.Gilmore, R. D., Jr., R. R. Howison, V. L. Schmit, A. J. Nowalk, D. R. Clifton, C. Nolder, J. L. Hughes, and J. A. Carroll. 2007. Temporal expression analysis of the Borrelia burgdorferi paralogous gene family 54 genes BBA64, BBA65, and BBA66 during persistent infection in mice. Infect. Immun. 752753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimm, D., K. Tilly, R. Byram, P. E. Stewart, J. G. Krum, D. M. Bueschel, T. G. Schwan, P. F. Policastro, A. F. Elias, and P. A. Rosa. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. USA 1013142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagman, K. E., X. Yang, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 2000. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 684759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hengge, U. R., A. Tannapfel, S. K. Tyring, R. Erbel, G. Arendt, and T. Ruzicka. 2003. Lyme borreliosis. Lancet Infect. Dis. 3489-500. [DOI] [PubMed] [Google Scholar]
- 25.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 9812724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawabata, H., S. J. Norris, and H. Watanabe. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect. Immun. 727147-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lybecker, M. C., and D. S. Samuels. 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 641075-1089. [DOI] [PubMed] [Google Scholar]
- 29.Maruskova, M., M. D. Esteve-Gassent, V. L. Sexton, and J. Seshu. 2008. Role of the BBA64 locus of Borrelia burgdorferi in early stages of infectivity in a murine model of Lyme disease. Infect. Immun. 76391-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medrano, M. S., Y. Ding, X. G. Wang, P. Lu, J. Coburn, and T. T. Hu. 2007. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J. Bacteriol. 1892653-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motaleb, M. A., M. R. Miller, C. Li, and N. W. Charon. 2007. Phosphorylation assays of chemotaxis two-component system proteins in Borrelia burgdorferi. Methods Enzymol. 422438-447. [DOI] [PubMed] [Google Scholar]
- 32.Ojaimi, C., C. Brooks, D. Akins, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katonah, J. Radolf, M. Caimano, J. Skare, K. Swingle, S. Sims, and I. Schwartz. 2002. Borrelia burgdorferi gene expression profiling with membrane-based arrays. Methods Enzymol. 358165-177. [DOI] [PubMed] [Google Scholar]
- 33.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 711689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal, U., J. Dai, X. Li, G. Neelakanta, P. Luo, M. Kumar, P. Wang, X. Yang, J. Anderson, and E. Fikrig. 2008. A differential role for BB0365 in the persistence of Borrelia burgdorferi in mice and ticks. J. Infect. Dis. 197148-155. [DOI] [PubMed] [Google Scholar]
- 35.Pal, U., X. Yang, M. Chen, L. K. Bockenstedt, J. F. Anderson, R. A. Flavell, M. V. Norgard, and E. Fikrig. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Investig. 113220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 9713865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. K. Howell, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48753-764. [DOI] [PubMed] [Google Scholar]
- 38.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 991562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Revel, A. T., J. S. Blevins, C. Almazan, L. Neil, K. M. Kocan, J. de la Fuente, K. E. Hagman, and M. V. Norgard. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. USA 1026972-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi, p. 253-259. In J. A. Nickoloff (ed.), Methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PMC free article] [PubMed]
- 41.Sartakova, M., E. Dobrikova, and F. C. Cabello. 2000. Development of an extrachromosomal cloning vector system for use in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 974850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 922909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seshu, J., M. D. Esteve-Gassent, M. Labandeira-Rey, J. H. Kim, J. P. Trzeciakowski, M. Hook, and J. T. Skare. 2006. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol. Microbiol. 591591-1601. [DOI] [PubMed] [Google Scholar]
- 44.Shi, Y., Q. Xu, K. McShan, and F. T. Liang. 2008. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect. Immun. 761239-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh, S. K., and H. J. Girschick. 2004. Molecular survival strategies of the Lyme disease spirochete Borrelia burgdorferi. Lancet Infect. Dis. 4575-583. [DOI] [PubMed] [Google Scholar]
- 46.Smith, A. H., J. S. Blevins, G. N. Bachlani, X. F. Yang, and M. V. Norgard. 2007. Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN). J. Bacteriol. 1892139-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steere, A. C., J. Coburn, and L. Glickstein. 2004. The emergence of Lyme disease. J. Clin. Investig. 1131093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 49.Terekhova, D., R. Iyer, G. P. Wormser, and I. Schwartz. 2006. Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J. Bacteriol. 1886124-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tilly, K., J. G. Krum, A. Bestor, M. W. Jewett, D. Grimm, D. Bueschel, R. Byram, D. Dorward, M. J. VanRaden, P. Stewart, and P. Rosa. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 743554-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 725419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, Q., K. McShan, and F. T. Liang. 2007. Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi. Mol. Microbiol. 64220-231. [DOI] [PubMed] [Google Scholar]
- 53.Xu, Q., S. V. Seemanapalli, K. McShan, and F. T. Liang. 2006. Constitutive expression of outer surface protein C diminishes the ability of Borrelia burgdorferi to evade specific humoral immunity. Infect. Immun. 745177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 371470-1479. [DOI] [PubMed] [Google Scholar]
- 55.Yang, X., T. G. Popova, K. E. Hagman, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 1999. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect. Immun. 676008-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, X. F., S. M. Alani, and M. V. Norgard. 2003. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 10011001-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, X. F., M. C. Lybecker, U. Pal, S. M. Alani, J. Blevins, A. T. Revel, D. S. Samuels, and M. V. Norgard. 2005. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J. Bacteriol. 1874822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, X. F., U. Pal, S. M. Alani, E. Fikrig, and M. V. Norgard. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]