Abstract
Proteus mirabilis, a gram-negative bacterium, is a frequent cause of complicated urinary tract infections in those with functional or anatomical abnormalities or those subject to long-term catheterization. To systematically identify surface-exposed antigens as potential vaccine candidates, proteins in the outer membrane fraction of bacteria were separated by two-dimensional gel electrophoresis and subjected to Western blotting with sera from mice experimentally infected with P. mirabilis. Protein spots reactive with sera were identified by mass spectrometry, which in conjunction with the newly completed genome sequence of P. mirabilis HI4320, was used to identify surface-exposed antigens. Culture conditions that may mimic in vivo conditions more closely than Luria broth (growth in human urine and under iron limitation and osmotic stress) were also used. Thirty-seven antigens to which a humoral response had been mounted, including 23 outer membrane proteins, were identified. These antigens are presumably expressed during urinary tract infection. Protein targets that are both actively required for virulence and antigenic may serve as protective antigens for vaccination; thus, five representative antigens were selected for use in virulence studies. Strains of P. mirabilis with mutations in three of the corresponding genes (the PMI0047 gene, rafY, and fadL) were not attenuated in the murine model of urinary tract infection. Putative iron acquisition proteins PMI0842 and PMI2596, however, both contribute to fitness in the urinary tract and thus emerge as vaccine candidates.
Proteus mirabilis, a gram-negative bacterium, is among the most prevalent isolates from individuals with complicated urinary tract infections (cUTIs) (61). cUTIs affect patients whose urinary tracts are subjected to long-term catheterization or have functional or anatomic abnormalities, and such infections occur via the ascending route (6). Consequences of P. mirabilis cUTIs include catheter encrustation, the formation of urinary stones (urolithiasis), renal scarring, and progression to bacteremia (41). Catheter encrustation caused by these infections can block the flow of urine through the catheter. In addition to the possibility of causing permanent renal damage, the formation of stones, due to the action of the bacterial enzyme urease, may make infections difficult to clear due to the presence of bacteria within the stones, where P. mirabilis may be shielded from the action of antibiotics (35).
Catheter-associated cUTIs are the most commonly occurring nosocomial infections, with more than one million cases documented each year in the United States (57). The prevention of P. mirabilis-caused cUTIs would improve patient care and quality of life and reduce the substantial economic burden associated with the treatment of these infections. Since infection does not appear to protect fully against reinfection (27), a vaccine is a logical goal for several reasons. First, patients with known urinary tract abnormalities and patients at the onset of long-term catheterization could be specifically targeted for vaccination due to the high incidence of cUTIs in these populations. Second, P. mirabilis infections are very difficult to clear due to the presence of bacteria within the urinary stones (35). Third, based on studies of the urease and mrpH genes, there is evidence that virulence factor genes are well conserved among different strains of P. mirabilis, which supports the notion of a cross-protective vaccine (33, 40).
Immunization with heat-killed bacterial preparations or prior infection offers little protection (27, 34). Purified fimbrial preparations, however, protect mice from subsequent transurethral challenge (31, 33). Three different structural fimbrial proteins have also been used in vaccine studies, with various degrees of success: MrpA, UcaA, and PmfA (47). In a recent study, one of these structural proteins, MrpA, was expressed in the food-grade bacterium Lactococcus lactis (51). Mice were intranasally immunized with L. lactis expressing MrpA prior to transurethral challenge and had significantly lower levels of bacterial colonization in the kidneys than controls. In another study, an outer membrane vaccine significantly protected mice from death, renal colonization, and renal damage (39). One of the most promising vaccines to date consists of the N-terminal domain of MrpH (the tip adhesin of the MR/P fimbria) fused to domains of cholera toxin (32). Previously, a translational fusion of MrpH and the cholera toxin A2 subunit was coexpressed with the cholera toxin B subunit; the vector used for vaccine expression replaced the toxic A1 subunit of cholera toxin with MrpH (24). The result of the expression of this construct is the spontaneous assembly of a single chimeric protein that contains both an antigen (MrpH) and an adjuvant (cholera toxin), which mediates highly effective delivery to the systemic and mucosal immune systems (18). Although this vaccine was able to protect mice from infection, we have concerns about the efficacy of a vaccine targeting solely this adhesin since the mrp operon is capable of undergoing phase variation (64). For this reason, we believe it to be prudent to identify additional antigens for inclusion in a multivalent vaccine.
In this study, we used an immunoproteomic approach to identify additional surface-exposed antigens of P. mirabilis. Fractions enriched for outer membrane proteins were isolated from bacterial cells, and proteins were separated by two-dimensional (2D) gel electrophoresis. Immunoreactive proteins, identified by Western blotting using sera from mice with experimental P. mirabilis urinary tract infections, were submitted for mass spectrometry analysis. Using the recently completed genome sequence (46), this study identified 37 immunoreactive antigens, including 23 outer membrane proteins. Five antigens were assessed for their roles in virulence in the murine model of ascending urinary tract infection.
MATERIALS AND METHODS
Strain and culture conditions.
P. mirabilis HI4320 was cultured from the urine of a catheterized nursing home patient with bacteriuria (41). Luria broth (LB; 10 g of tryptone, 5 g of yeast extract, and 0.5 g of NaCl per liter) and nonswarming agar (10 g of tryptone, 5 g of yeast extract, 0.5 g of NaCl, and 15 g of agar per liter) were used to culture bacteria. Minimal medium (200 ml of 5× M9 salts [64 g of Na2HPO4·7H2O, 15 g of KH2PO4, 2.5 g of NaCl, and 5 g of NH4Cl per liter], 2 ml of 1 M MgSO4, 20 ml of 20% glucose, and 100 μl of CaCl2 per liter) was inoculated with a 1:100 dilution of P. mirabilis cultured overnight in LB. Iron limitation was achieved by the addition of 15 μM desferoxamine (M. Pearson and H. Mobley, unpublished data) to LB cultures. Osmotic stress was induced by the addition of 0.3 M NaCl to LB or minimal medium (3). Urine was collected from three healthy human donors, pooled, filter sterilized, and stored at −20°C until use. All cultures were incubated at 37°C with aeration (200 rpm) unless otherwise noted.
Isolation of outer membranes.
A modification of the method of Piccini et al. (48) was used for the isolation of outer membranes. Bacteria were harvested by centrifugation (10,000 × g for 15 min at 4°C) and washed twice in 10 mM HEPES, pH 7.4. Cells were lysed by two passes through a French pressure cell (manufactured by the American Instrument Company, a division of Travenol Laboratories Inc., Silver Spring, MD) at 20,000 lb/in2. Intact bacteria were cleared by centrifugation (10,000 × g for 15 min at 4°C). Supernatants were centrifuged (90,000 × g for 45 min at 4°C) to pellet membranes from the lysate. Membrane pellets were resuspended in a solution containing 10 mM HEPES (pH 7.4), 10 mM MgCl2, and 2% Triton X-100. Following an hour-long incubation at 37°C, Triton X-100-insoluble fractions (enriched for outer membrane proteins) were collected by centrifugation (90,000 × g for 45 min at 4°C). Pellets were washed in a solution containing 10 mM HEPES (pH 7.4), 10 mM MgCl2, and 2% Triton X-100. The resulting outer membrane-enriched pellets were resuspended in isoelectric focusing (IEF) solution (7 M urea, 2 M thiourea, 1% amidosulfobetaine-14, 40 mM Tris, 2 mM tributylphosphine, and 0.5% Bio-Lyte 3/10). Protein was quantified by using a 2D Quant kit (Amersham Biosciences).
2D gel electrophoresis.
The method of Molloy et al. (42) for 2D gel electrophoresis was adapted. A 17-cm pH 4 to 7 ReadyStrip IPG strip (Bio-Rad) was rehydrated for 16 to 24 h with 350 μl of IEF solution containing outer membrane protein. IEF was conducted under the following conditions in a Protean IEF cell (Bio-Rad): 250 V for 20 min, 10,000 V for 2.5 h, and 10,000 V for 40,000 V·h. Prior to the second dimension, strips were equilibrated for 20 min at room temperature by rocking in a solution of 0.15 M bis-Tris-0.1 M HCl, 6 M urea, 2% (wt/vol) sodium dodecyl sulfate (SDS), 20% (vol/vol) glycerol, 5 mM tributylphosphine, and 2.5% (wt/vol) acrylamide. Samples were run on 10% polyacrylamide gels, as described previously (42). The cathode buffer contained 0.2 M taurine, 25 mM Tris, and 0.1% (wt/vol) SDS, and the anode buffer contained 0.384 M glycine, 50 mM Tris, and 0.1% SDS. Gels were run at a constant current of 50 mA at room temperature until completion and then stained for 24 h in colloidal Coomassie blue G-250 (43). Unstained Precision Plus protein standards (Bio-Rad) were used as molecular weight standards. Proteins corresponding to immunoreactive spots on Western blots (described below) were cut from gels using a clean razor blade and submitted for mass spectrometry analysis.
Sera.
Sera were obtained from mice 42 days after experimental urinary tract infection with P. mirabilis as part of a previous study (27). In the present work, we used preimmune sera and postrechallenge sera from that study. Sera from 20 of the mice were individually screened for reactivity to P. mirabilis proteins by Western blotting (details are given below). The five sera with the strongest reaction to P. mirabilis lysate were pooled in equal amounts and used in all further studies.
Western blotting.
Proteins were transferred onto an Immobilon-P polyvinylidene difluoride membrane (Millipore) for 1 h at 400 mA and 4°C. After transfer, membranes were blocked with 5% milk in TBS-T (0.05% Tween, 100 mM Tris [pH 7.5], 9% NaCl). Membranes were incubated with shaking at room temperature in the presence of sera diluted in TBS-T. After one 15-min wash and three 5-min washes in TBS-T, secondary antibody was applied to the membrane. Goat anti-mouse immunoglobulin G (IgG) conjugated to horseradish peroxidase (1:100,000 dilution in TBS-T) was applied for 45 min with shaking at room temperature. Precision StrepTactin-horseradish peroxidase conjugate (Bio-Rad), used for ladder detection, was added to the secondary antibody incubation mixture. The wash procedure was repeated, and detection was performed with an Amersham ECL Plus Western blotting detection system by following the protocol recommended by the manufacturer (GE Healthcare). To screen sera from individual mice, a 1:1,000 dilution was used. After sera were combined, a 1:10,000 dilution of the pooled sera was used in all further experiments.
Mass spectrometry.
Proteins were identified either by peptide mass fingerprinting (PMF) or tandem mass spectrometry (MS/MS). PMF was performed by personnel at the University of Michigan Protein Structure Facility. Samples were digested with trypsin by using the Millipore Montage system, cocrystallized with alpha-cyano-hydroxy-cinnamic acid (sample/alpha-cyano-hydroxy-cinnamic acid ratio, 1:1), and analyzed via matrix-assisted laser desorption ionization-time of flight mass spectrometry. ProteinProspector software (http://prospector.ucsf.edu), along with the preliminary genome annotation (46), was used to identify proteins in PMF samples. MS/MS analysis was performed by the Michigan Proteomics Consortium according to the protocols described on the website (http://www.proteomeconsortium.org/protocols.html). Briefly, samples were digested with trypsin, and spectra were acquired by using a model 4700 proteomics analyzer (Applied Biosystems). The eight most intense peaks in each spectrum were selected for MS/MS. Ion scores are available upon request.
Construction of mutants.
Insertional mutants were constructed using the TargeTron system (Sigma), as described by Pearson and Mobley (45). Briefly, genes were disrupted by the insertion of an intron (containing a kanamycin resistance gene) which was targeted specifically to each gene of interest by using a set of three primers (designated by the gene name and the suffixes IBS, EBS1d, and EBS2, as listed in Table 1) in a mutagenic PCR. This mutated intron region was ligated into the vector pACD4K-C. Resultant plasmids were sequenced to confirm proper retargeting of the intron. Correctly retargeted plasmids were introduced into electrocompetent P. mirabilis HI4320 containing the helper plasmid pAR1219 (14) by electroporation. Transformants were selected on agar containing kanamycin and screened by PCR for an insertion in the appropriate gene by using the screening primers listed in Table 1.
TABLE 1.
Primers used to generate and confirm mutations in P. mirabilis HI4320
| Primer name | Sequence (5′ to 3′) |
|---|---|
| PMI0047-IBS | AAAAAAGCTTATAATTATCCTTAGTAACCCACCCAGTGCGCCCAGATAGGGTG |
| PMI0047-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCCACCCATATAACTTACCTTTCTTTGT |
| PMI0047-EBS2 | TGAACGCAAGTTTCTAATTTCGATTGTTACTCGATAGAGGAAAGTGTCT |
| PMI0288-IBS | AAAAAAGCTTATAATTATCCTTAGCAGTCATAGTTGTGCGCCCAGATAGGGTG |
| PMI0288-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCATAGTTTCTAACTTACCTTTCTTTGT |
| PMI0288-EBS2 | TGAACGCAAGTTTCTAATTTCGATTACTGCTCGATAGAGGAAAGTGTCT |
| PMI0842-IBS | AAAAAAGCTTATAATTATCCTTACCAGCCTCTGTTGTGCGCCCAGATAGGGTG |
| PMI0842-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCTCTGTTTCTAACTTACCTTTCTTTGT |
| PMI0842-EBS2 | TGAACGCAAGTTTCTAATTTCGGTTGCTGGTCGATAGAGGAAAGTGTCT |
| PMI1810-IBS | AAAAAAGCTTATAATTATCCTTAATAAGCTACCACGTGCGCCCAGATAGGGTG |
| PMI1810-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCTACCACCCTAACTTACCTTTCTTTGT |
| PMI1810-EBS2 | TGAACGCAAGTTTCTAATTTCGATTCTTATTCGATAGAGGAAAGTGTCT |
| PMI2596-IBS | AAAAAAGCTTATAATTATCCTTAGATGACTTGAGCGTGCGCCCAGATAGGGTG |
| PMI2596-EBS1d | AGATAAGTCTTGAGCGCTAACTTACCTTTCTTTGTCAGATTGTACAAATGTGGTGATAAC |
| PMI2596-EBS2 | TGAACGCAAGTTTCTAATTTCGATTTCATCTCGATAGAGGAAAGTGTCT |
| PMI0047screenFor | NNNNNNTTCGAAATGAACCATAAATATAAAC |
| PMI0047screenRev | NNGGTACCTTTACGAGTAATGTTGTTCG |
| PMI0288screenFor | NNNNNNTTCGAAATGAACATCAAACACCTTGCG |
| PMI0288screenRev | NNGGTAGCGAAGAAGTATTTAAAACGTGC |
| PMI0842screenFor | NNNNNTTCGAAATGGTAGTGTTAAAGAAAN |
| PMI0842screenRev | NNGGTACCGAAGTTGTAGTTTAATCCG |
| PMI1810screenFor | NNNNNNCATATGAACCGTAAAAACCTGTTTAC |
| PMI1810screenRev | NNNAGATCTGAAGCGATAGTTAAAGTTC |
| PMI2596screenFor | NNNNNNTTCGAAATGGAAAATAAAGGAAC |
| PMI2596screenRev | NNGGTACCAAAGTTAATACTTCCCTG |
CBA/J mouse model of urinary tract infection.
Cochallenges and independent challenges of CBA/J mice were carried out as described previously by using a modified Hagberg protocol (23, 28). Because bacteria are introduced via a catheter and because P. mirabilis urease catalyzes crystal and larger stone formation, this system can be classified as a model of cUTI. Briefly, single colonies were picked from a fresh plate and used to start overnight cultures in LB. On the day of inoculation, cultures were diluted with LB to an optical density at 600 nm of approximately 0.2. For independent challenges, mice were inoculated transurethrally with 50 μl (containing approximately 107 CFU). For cochallenge experiments, the dilute wild-type and mutant cultures were pooled in a 1:1 ratio and mice were inoculated with 50 μl of this mixture (containing approximately 107 CFU). After 7 days, urine was collected, mice were euthanized, and their bladders, kidneys, and spleens were harvested and transferred into sterile tubes containing phosphate-buffered saline (0.138 M NaCl, 0.0027 M KCl, pH 7.4). Tissues were homogenized (Omni TH homogenizer; Omni International) and plated onto agar plates by using a spiral plater (Autoplate 4000; Spiral Biotech). For independent challenges, all samples were plated onto LB agar. For cochallenges, all samples were plated onto both LB agar plates and LB agar plates containing 25 μM kanamycin. Since the mutant strains carried a kanamycin resistance gene, the colony counts from the kanamycin plates represent mutant bacteria only; LB agar plates allow the growth of both mutant and wild-type colonies. Statistical significance for independent and cochallenge experiments was assessed by using the Mann-Whitney test and the Wilcoxon matched-pairs test, respectively.
RESULTS
Mice with experimental P. mirabilis urinary tract infections have varied antibody responses.
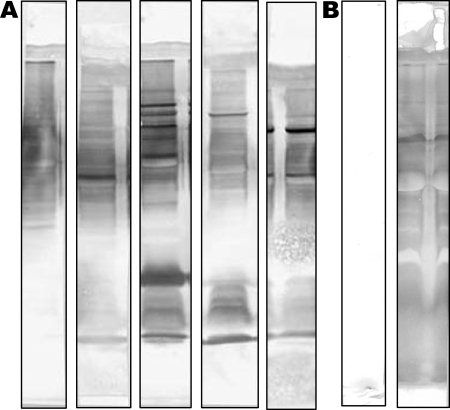
In our previous study, mice were transurethrally infected with 107 CFU of P. mirabilis HI4320 and, 4 weeks later, treated with antibiotics for 4 days to clear the infection. Following a 3-day washout period, the same mice were challenged with 106 CFU. Sera were collected 7 days after reinfection (42 days after the initial infection) (27). In the present study, sera from 20 of these mice were individually screened for reactivity to P. mirabilis proteins. Proteins in a P. mirabilis HI4320 cell lysate were separated by SDS-polyacrylamide gel electrophoresis, transferred onto a polyvinylidene difluoride membrane, and screened by Western blotting with a 1:1,000 dilution of sera from individual mice. Preimmune sera at a 1:1,000 dilution were also screened by Western blotting (data not shown). Sera showed some diversity in antigen recognition, which may reflect differences in the severity of the individual infections; all postinfection sera were reactive. Equal volumes of the five most strongly reacting sera were pooled and used for all further Western blot analyses (Fig. 1). The five corresponding preimmune sera were also pooled for use as a control.
FIG. 1.
Western blots of P. mirabilis HI4320 lysate reacting with sera from mice experimentally infected with P. mirabilis HI4320. (A) Western blots of P. mirabilis lysate probed with sera from infected mice. Each strip represents serum from one mouse. The five strips shown correspond to the five most strongly reacting sera. (B) Pooled pre- and postimmune sera reacting with P. mirabilis lysate by Western blot analysis.
Sera from infected mice recognize outer membrane antigens.
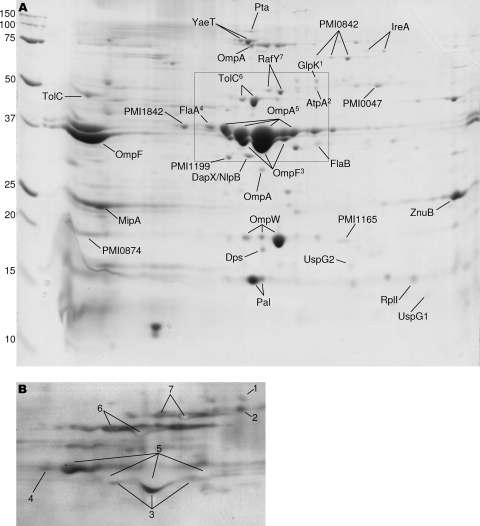
P. mirabilis was cultured in LB with aeration at 37°C, and fractions enriched for outer membranes were isolated by differential centrifugation and detergent solubilization. Proteins in the outer membrane-enriched fractions were separated by 2D gel electrophoresis (Fig. 2A). Proteins were screened by Western blotting using the pooled sera at a dilution of 1:10,000 (Fig. 2B). Antigens that reacted with sera were excised from a duplicate Coomassie blue-stained gel and identified by mass spectrometry.
FIG. 2.
Sera from infected mice recognize outer membrane antigens. (A) Coomassie blue-stained 2D gel loaded with 300 mg of Triton X-100-insoluble outer membrane-enriched protein from P. mirabilis HI4320 cultured in LB. Antigenic proteins are labeled. The boxed region shows an area of the gel that corresponds to the Western blot shown in panel B. The pH gradient ranged from 4 to 7, from left to right. Marker sizes in kilodaltons are shown to the left. (B) Western blot of a 2D gel from an analysis performed with a 1:100,000 dilution of pooled sera from infected mice (the sera used in the right Western blot shown in Fig. 1B). Numbers designate proteins labeled in panel A. White (negative) spots are most likely due to an overabundance of protein and a high antibody concentration, as suggested by the manufacturer of the detection system used (see Materials and Methods). Proteins that reacted with sera were cut from a duplicate gel and identified by mass spectrometry.
Proteins expressed in vivo are not necessarily expressed in rich culture medium. To identify antigens that may not be expressed in LB, we used additional culture conditions that may more closely mimic in vivo conditions during urinary tract infection. For example, P. mirabilis was cultured in pooled human urine and also in minimal medium. Since these media are presumably more nutrient limited than LB, bacteria may require the synthesis of additional proteins for growth that may not be required during growth in LB. The production of these additional proteins increases the number of potential antigens screened. A study of the transcriptome of uropathogenic Escherichia coli (UPEC) during mouse infection suggested that the urinary tract is iron limited and characterized by high osmolarity (55). Therefore, we also isolated protein from P. mirabilis HI4320 cultured in media that mimic these conditions: LB containing 15 μM desferoxamine (an iron chelator) and LB containing 0.3 M sodium chloride, respectively. The use of these culture conditions enabled the identification of nine additional antigens that were not expressed in LB, including four outer membrane proteins that appear to be related to iron acquisition (PMI0409, HmuR2, IreA, and PMI2596).
In total, 37 P. mirabilis antigens were identified. Twenty-three antigens are predicted to reside in the outer membrane (Table 2). An additional 14 proteins, not predicted to be present in the outer membrane, were also identified as immunoreactive antigens (Table 3).
TABLE 2.
P. mirabilis antigens predicted to be in the outer membrane
| Category | Gene no.a | Protein | Growth phase(s) in which protein was identifiedb in culture grown in:
|
Medium(a) in which protein was identifiedb in culture under osmotic stress | |||
|---|---|---|---|---|---|---|---|
| LB | Low-iron medium | Urine | MM | ||||
| Porin | PMI0288† | RafY | E (S) | E, S | (S) | (LB) | |
| PMI0765 | OmpF | E (S) | E (S) | S | E | (LB, MM) | |
| PMI0785 | OmpA | E, S | S (E) | S | (E) | LB (MM) | |
| PMI1017* | Putative exported protein | E | |||||
| PMI1350 | OmpW | E, S | S | (LB, MM) | |||
| Lipoproteins and associated | PMI0585 | Pal | E, S | S | (LB, MM) | ||
| proteins | PMI1563 | DapX/NlpB | E (S) | (S) | (LB, MM) | ||
| PMI1165 | Putative lipoprotein | S (E) | (S) | (LB) | |||
| PMI1842 | Putative lipoprotein | E (S) | (S) | (LB, MM) | |||
| PMI2277 | YaeT | E (S) | E, S | (S) | LB | ||
| Motility proteins | PMI1619 | FlaB | E | (LB, MM) | |||
| PMI1620 | FlaA | E, S | S (E) | S | (LB, MM) | ||
| PMI1651* | FlgE | E | |||||
| Iron acquisition proteins | PMI0409* | TonB-dependent receptor | E | ||||
| PMI0842† | TonB-dependent receptor | E, S | E (S) | S | (E) | MM (LB) | |
| PMI1426* | HmuR2 | S (E) | (E) | ||||
| PMI1945 | IreA | (E, S) | E, S | S | (E) | ||
| PMI2596†* | Putative siderophore TonB-dependent receptor | E, S | |||||
| Toxins | PMI2043* | Putative toxin | S | (LB) | |||
| PMI2341 | Pta | E (S) | S | (S) | (LB) | ||
| Other proteins | PMI1199 | Putative exported protein | S (E) | (S) | |||
| PMI1810†* | FadL | E (S) | (S) | (LB, MM) | |||
| PMI2349 | TolC | E (S) | E, S | S | (E) | (LB, MM) | |
†, assessed for contribution to virulence (Table 4); *, corresponding protein is not on gel shown in Fig. 2 but was identified from a subsequent gel.
Bold letters correspond to proteins identified directly by mass spectrometry. Letters in parentheses correspond to proteins identified by the comparison of electrophoretic mobilities to those of identified proteins on other gels. E, exponential phase; S, stationary phase; MM, minimal medium.
TABLE 3.
Soluble and inner membrane proteins identified as P. mirabilis antigens
| Gene no.a | Protein | Growth phase(s) in which protein was identifiedb in culture grown in:
|
Medium(a) in which protein was identifiedb in culture under osmotic stress | |||
|---|---|---|---|---|---|---|
| LB | Low-iron medium | Urine | MM | |||
| PMI0047† | Putative secreted 5′-nucleotidase | E, S | E | LB | ||
| PMI0103* | Conserved hypothetical protein | S | ||||
| PMI0631 | Dps | S | (S) | (LB, MM) | ||
| PMI0776* | PqiA | S | ||||
| PMI0874 | Conserved hypothetical protein | E (S) | (S) | |||
| PMI1150 | ZnuB | E (S) | (S) | |||
| PMI1449 | UspG1 | S (E) | (S) | |||
| PMI1451 | UspG2 | S (E) | (S) | (LB) | ||
| PMI1488* | Hns | S | (LB) | |||
| PMI1506 | MipA | E | ||||
| PMI1905* | Conserved hypothetical protein | E | ||||
| PMI3062 | AtpA | E (S) | (S) | (LB) | ||
| PMI3210 | GlpK | (E, S) | LB | |||
| PMI3377 | RplI | E (S) | ||||
†, assessed for contribution to virulence (Table 4); *, corresponding protein is not on gel shown in Fig. 2 but was identified from a subsequent gel.
Bold letters correspond to proteins identified directly by mass spectrometry. Letters in parentheses correspond to proteins identified by the comparison of electrophoretic mobilities to those of identified proteins on other gels. E, exponential phase; S, stationary phase; MM, minimal medium.
Selection of potential vaccine candidates.
For the present study, five representative antigens were chosen for analysis. We chose at least one antigen per category listed in Table 2, with the exception of lipoproteins, motility proteins, and toxins. We did not pursue these three classes of proteins for specific reasons. Lipoproteins are generally on the inner leaflet of the outer membrane and may not, therefore, be exposed to the surface. There are two copies of the gene encoding flagellin that can recombine to form antigenically distinct flagella (8, 9, 44). Since the flagella formed by this recombination event are antigenically distinct, a vaccine employing FlaA as a protective antigen may not be able to target bacterial cells expressing recombinant flagellin. One of the toxins found in this screen, Pta (Proteus toxic agglutinin), recently identified as an autotransporter that is both a cytotoxin and an agglutinin (2), is being assessed as a vaccine candidate in a separate study (P. Alamuri and H. Mobley, unpublished data). We specifically chose FadL and RafY for further study because structures of similar proteins have been resolved and reveal the presence of extracellular loops (19, 53, 60). If these proteins in P. mirabilis have similar structures, these extracellular loops may be accessible to the immune system during infection. We also chose to study two putative outer membrane iron receptors, PMI0842 and PMI2596. The urinary tract is an iron-limited environment, and another uropathogen (UPEC) upregulates a number of antigenic iron acquisition outer membrane proteins during growth in urine (4, 22, 55). Additionally, the use of iron receptors as protective antigens against E. coli, Haemophilus influenzae, and Haemophilus ducreyi infections demonstrates that iron receptors can be used successfully in vaccines (1, 50, 62). We also investigated the role of one of the antigens not predicted to be in the outer membrane, PMI0047. Although this protein is not an ideal vaccine candidate (since it is not predicted to be surface exposed), it was recognized by sera from infected mice. Thus, it is produced during urinary tract infection of mice and may have a role in pathogenesis.
Two outer membrane antigens involved in iron acquisition contribute to P. mirabilis virulence in the urinary tract.
We reasoned that if a protein target is both antigenic and actively required for colonization, it is more likely to serve successfully as a protective antigen for vaccination. For example, if a surface protein is being used as a protective antigen, any bacterium that attempts to escape the host immune response by downregulating this protein will be less likely to persist in the host if the protein is required for colonization. To determine the contributions to virulence, we investigated the roles of five representative antigens (described above) in the colonization of the murine urinary tract. Isogenic insertional mutations in the genes encoding each of the potential vaccine candidates were constructed. Genes were disrupted by the insertion of an intron carrying a kanamycin resistance gene by the method of Pearson and Mobley (45). All mutations were confirmed by PCR (data not shown). The growth of each mutant in LB was tested independently; none of the mutants had growth rates significantly different from that of the wild-type strain (data not shown).
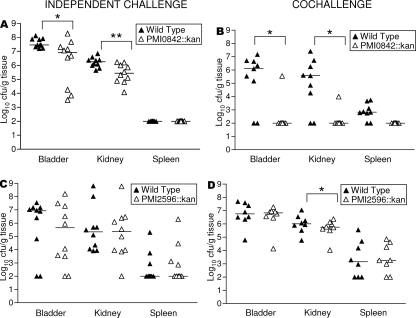
Mutants were tested in the murine model of ascending urinary tract infection by cochallenge with wild-type P. mirabilis. Mutant and wild-type bacteria were mixed in a 1:1 ratio. This suspension was used to inoculate mice transurethrally at a total inoculum of 107 CFU per mouse. After 7 days, urine was collected and mice were euthanized. Urine, bladders, kidneys, and spleens (as an indicator of systemic infection) were quantitatively cultured to determine levels of colonization by both wild-type and mutant strains. Based on data from the cochallenge experiments (Table 4), PMI0047 (secreted 5′ nucleotidase), RafY, and FadL do not appear to contribute to the colonization of the urinary tract. Mutant bacteria with interruptions in genes encoding these proteins colonized the urinary tract in numbers similar to those of wild-type bacteria. Two outer membrane iron receptors did, however, contribute to the colonization of the urinary tract by P. mirabilis (Fig. 3). The strain PMI0842::Kan was outcompeted by the wild-type strain at statistically significant levels in both the bladder and kidneys (P < 0.05 for both tissues) (Fig. 3B). Also in the cochallenge experiments, strain PMI2596::Kan bacteria were recovered from kidneys in significantly lower numbers than wild-type bacteria (P < 0.05) (Fig. 3D). As a control, in vitro cocultures were performed, in which the mutant and wild-type strains were mixed in a 1:1 ratio and used to inoculate the culture medium. Neither mutant was outcompeted by the wild-type strain (data not shown). Thus, the mutants retained wild-type growth rates in vitro and likely were not outcompeted in vivo due to an in vitro growth defect.
TABLE 4.
Assessment of virulence contributions of mutant forms of selected antigens in cochallenge studies
| Antigen | Straina | No. of samplesb | Amt of bacteriac (ratio of mutant to WT bacteria)d in:
|
|||
|---|---|---|---|---|---|---|
| Urine (CFU/ml) | Bladder tissue (CFU/g) | Kidney tissue (CFU/g) | Spleen tissue (CFU/g) | |||
| Secreted 5′-nucleotidase | PMI0047::Kan | 10 | 1.21 × 106 (20.07) | 5.62 × 104 (0.02) | 1.06 × 104 (0.41) | 1.00 × 102 (1.00) |
| WT | 6.03 × 104 | 3.54 × 106 | 2.56 × 104 | 1.00 × 102 | ||
| RafY | PMI0288::Kan | 9 | 6.20 × 103 (0.25) | 3.73 × 104 (0.54) | 1.48 × 104 (0.47) | 1.00 × 102 (1.00) |
| WT | 2.45 × 104 | 6.85 × 104 | 3.16 × 104 | 1.00 × 102 | ||
| TonB-dependent | PMI0842::Kane | 9 | 1.00 × 102 (<0.01) | 1.00 × 102* (<0.01) | 1.00 × 102* (<0.01) | 1.00 × 102 (0.15) |
| receptor | WT | 8.11 × 106 | 1.27 × 106 | 3.74 × 105 | 6.70 × 102 | |
| FadL | PMI1810::Kan | 9 | 2.96 × 105 (0.18) | 3.78 × 105 (0.11) | 1.08 × 105 (0.53) | 1.00 × 102 (1.00) |
| WT | 1.61 × 106 | 3.53 × 106 | 2.03 × 105 | 1.00 × 102 | ||
| TonB-dependent | PMI2596::Kane | 8 | 6.73 × 105 (0.08) | 6.76 × 106 (1.19) | 5.74 × 105* (0.56) | 1.79 × 103 (1.25) |
| receptor | WT | 8.97 × 106 | 5.67 × 106 | 1.03 × 106 | 1.43 × 103 | |
WT, wild type. Mice were cochallenged with the indicated mutant strain and the wild-type strain.
In each experiment, 10 mice were cochallenged; the numbers shown correspond to the numbers of live mice remaining at the ends of the experiments. Urine collection from each mouse was not always possible; therefore, data for urine are based on a lower number of samples, as follows: PMI0047::Kan-WT challenge, 7; PMI0288::Kan-WT challenge, 4; PMI0842::Kan-WT challenge, 4; PMI1810::Kan-WT challenge, 5; and PMI2596::Kan-WT challenge, 5.
The limit of detection was 1.00 × 102 CFU per ml of urine or per g of tissue. Any sample with levels below this limit was considered to contain this amount for statistical analyses. The colonization data shown are the median values for a given group of mice. *, statistically significant compared to wild-type colonization data for the same organ after cochallenge (P < 0.05).
Ratio of the medians of amounts of mutant and wild-type bacteria. A ratio of 1 means that wild-type and mutant bacteria were present at equal levels, a ratio of less than 1 indicates more wild-type than mutant bacteria, and a ratio greater than 1 indicates the presence of more mutant than wild-type cells in the sample.
Data are shown in Fig. 3.
FIG. 3.
Putative iron acquisition outer membrane proteins contribute to P. mirabilis HI4320 virulence in the murine model of urinary tract infection. Each triangle represents an individual mouse. Bars represent medians. Points at 100 CFU/g of tissue represent samples at or below the limit of detection. (A) PMI0842::Kan and wild-type strain independent challenges. (B) PMI0842::Kan and wild-type strain cochallenge. (C) PMI2596::Kan and wild-type strain independent challenges. (D) PMI2596::Kan and wild-type strain cochallenge. *, P < 0.05; **, P < 0.01.
Due to the striking phenotypes of these mutants (particularly that of PMI0842::Kan) in cochallenges, each mutant was tested for virulence in independent challenges. While cochallenges are more sensitive than independent challenges in detecting differences between mutant and wild-type strains because the strains are put into direct competition for niches and nutrients, they cannot conclusively reveal that a putative virulence factor is required for infection. Thus, mice were transurethrally inoculated with either the wild type or the mutant strain at a dose of 107 CFU per mouse. After 7 days, urine, bladders, kidneys, and spleens were quantitatively cultured to determine the levels of bacterial colonization. Independent challenge data show that PMI0842 is required for the maximal colonization of the urinary tract (Fig. 3A). Although the PMI0842::Kan mutant was able to colonize mice, the mutant bacteria were found in significantly lower numbers than wild-type bacteria in both the bladders and kidneys of infected mice (P, <0.05 and <0.01, respectively). In contrast, although PMI2596::Kan was outcompeted in cochallenge experiments (Fig. 3D), in independent challenge experiments the mutant bacteria colonized mice in numbers similar to those of wild-type bacteria (Fig. 3C).
DISCUSSION
This is the first study to identify outer membrane antigens of P. mirabilis by using an immunoproteomics approach. Here, we identified 37 antigens that reacted with sera from mice experimentally infected in the urinary tract with P. mirabilis. Based on their annotation or homology to other proteins, 23 of these proteins are predicted to reside in the outer membrane. These proteins are attractive as potential vaccine candidates since they are expressed in vivo, are antigenic, and are exposed on the surface of the bacterium (and thus to the immune system). Five representative antigens were selected for further study: PMI0047, RafY, PMI0842, FadL, and PMI2596. Two of these proteins, both putative outer membrane iron receptors (PMI0842 and PMI2596), contribute to the fitness of P. mirabilis in the urinary tract. From this study, PMI0842 and PMI2596 emerge as potential vaccine candidates, since each protein both contributes to pathogenesis and is antigenic. PMI0047, RafY, and FadL do not appear to contribute directly to P. mirabilis HI4320 virulence in the urinary tract, as bacteria with insertionally interrupted copies of the corresponding genes colonized the urinary tracts of mice in numbers similar to those of wild-type bacteria.
This study identified five predicted porins as immunogenic: OmpA, OmpF, OmpW, PMI1017, and RafY. OmpA, OmpF, and OmpW are major outer membrane proteins. PMI1017 appears to be a member of the OprD porin family based on homology to other family members. RafY is predicted to belong to the glycoporin family, a family of proteins that typically allow the passage of sugars across the bacterial outer membrane. Other well-known glycoporins include LamB (maltoporin) and ScrY (sucrose porin). In E. coli, RafY allows growth on raffinose but has been shown previously to be a general diffusion pore rather than a carbohydrate-specific one (5). The role of RafY in P. mirabilis has not yet been determined. The structures of LamB from E. coli and ScrY from Salmonella enterica serovar Typhimurium have been resolved (19, 53). Both proteins contain extracellular loops which are surface exposed. If the protein structure of P. mirabilis RafY is similar to those of other glycoporins, it may also have surface-exposed extracellular loops and therefore be an attractive candidate for inclusion in vaccine studies.
Four lipoproteins were identified: Pal, DapX, PMI1842 (homologous to YfgL), and PMI1165 (homologous to YeaY). Lipoproteins are typically found on the inner leaflet of the outer membrane. Two of these lipoproteins (DapX, which belongs to the NlpB family, and YfgL) are involved in a complex with outer membrane protein YaeT, which was also identified as an antigen in this study. Although the complex is conserved in gram-negative bacteria, DapX/NlpB and YfgL are not essential for the growth of E. coli (11, 16, 49, 63). Peptidoglycan-associated lipoprotein (Pal) is released from gram-negative bacteria during sepsis and mediates inflammation and death in mice (25). Pal is part of the Tol-Pal system, which is conserved among gram-negative organisms (56). This system has been implicated previously in the maintenance of cell envelope integrity (37) and cell division machinery (21). Pal has been identified as an antigen in many other bacterial pathogens, including Legionella pneumophila and Campylobacter jejuni (13, 17). A homologous lipoprotein (P6) in H. influenzae is being assessed as a protective antigen (10, 15, 26, 30). Although we initially did not choose to investigate Pal as a vaccine candidate since we are aiming specifically for surface-exposed proteins, these findings from other bacteria suggest that it may warrant further study.
Two flagellar proteins were identified: the major flagellin subunit and the flagellar hook protein (FlgE). P. mirabilis carries two copies of the flagellin gene, flaA and flaB (9). Normally, flaA is expressed and flaB is silent, but the two genes can undergo recombination, resulting in the formation of antigenically distinct flagella (8, 44). The recombination of the two alleles occurs infrequently in wild-type cells (38). Such an event must have occurred under our culture conditions, since mass spectrometry data yielded results for both FlaA and FlaB proteins.
We identified five iron-related outer membrane proteins as antigenic. This finding is not surprising considering the important role iron acquisition plays in bacterial pathogens. Indeed, we chose to examine outer membranes from bacteria cultured under iron limitation conditions because the urinary tract is an iron-limited environment. Iron acquisition systems are upregulated in UPEC during infection in the murine model (55). UPEC upregulates a number of outer membrane iron receptors when cultured in human urine, and many of these have been identified as antigenic (4, 22). Additionally, P. mirabilis iron-related outer membrane receptors have been identified previously as antigenic both in mice and in human patients (48, 54). One of these proteins, also identified as an antigen in the present study, was recently identified as HmuR2 (PMI1426) and characterized as a heme receptor that contributes to virulence in the urinary tract (36).
Two outer membrane antigens predicted to be iron receptors (PMI0842 and PMI2596) contribute to the fitness of P. mirabilis HI4320 during experimental urinary tract infection. PMI0842 was previously identified as a virulence factor by signature-tagged mutagenesis (12) and was recently shown to be upregulated in response to iron limitation (S. D. Himpsl et al., unpublished data). Our cochallenge results confirm the previous finding from signature-tagged mutagenesis. Additionally, our independent challenge data show that PMI0842 is required to achieve maximum colonization of the urinary tract. This result is surprising due to the fact that iron receptors are typically redundant in function; a defect in one system rarely results in attenuation during independent challenges since other iron uptake systems remain functional, as highlighted by Torres and colleagues (59). The role of PMI0842 in iron uptake in P. mirabilis remains to be determined.
This study is also the first to show a role for PMI2596 in P. mirabilis HI4320 virulence in the urinary tract. Although the independent challenge data show that this protein is not required for virulence, cochallenge data highlight its contribution to colonization in the kidneys. The PMI2596 gene is located roughly 2 kb upstream of the nrp operon, which comprises nonribosomal peptide synthesis genes (20). NrpG was previously identified by signature-tagged mutagenesis as a virulence factor in P. mirabilis (12). Interestingly, a strain lacking a functional copy of NrpG was outcompeted by wild-type P. mirabilis HI4320 in the kidneys but not the bladders of mice in a cochallenge experiment (12), the same pattern we observed with PMI2596::Kan. It is currently unknown if the PMI2596 gene is functionally related to the nrp operon.
Although we were able to identify 37 antigens, we do not believe this study generated a complete list of the outer membrane antigens expressed by P. mirabilis in vivo. The methods used to identify these proteins imposed some limitations. First, we were able to identify only proteins that were abundant enough to be detected by Coomassie blue staining. Also, since we ran the protein samples on a denaturing gel prior to screening with sera, we will have likely missed any conformational epitopes. Finally, we did not detect any fimbrial proteins under any culture condition tested. This result is surprising, especially considering the large number of fimbrial operons contained in the genome (46). Specifically, we expected to find proteins that make up MR/P fimbriae, since these fimbriae are expressed (although not optimally) under the conditions tested (7) and the sera used in this experiment have previously detected MR/P fimbriae (27). It is likely that fimbriae were sheared from bacterial cells during fractionation.
This study also identified 14 antigenic proteins that do not appear to be surface exposed. These non-surface-exposed antigens include two apparent DNA binding proteins, a ribosomal protein, various enzymes, and three conserved hypothetical proteins. Since the goal of this study was to identify surface-exposed antigens, these proteins were not investigated further for vaccine studies. Even though these proteins may not be ideal vaccine candidates, they were recognized by sera from infected mice, meaning that they are expressed by P. mirabilis during urinary tract infection and may potentially play a role in pathogenesis. PMI0047 does not contribute to the fitness of P. mirabilis during experimental infection, since bacteria with an interrupted copy of the PMI0047 gene were able to colonize mice in numbers similar to those of the wild-type bacteria. Other proteins on this list, however, may play a role in pathogenesis and are currently under investigation (G. Nielubowicz and H. Mobley, unpublished data).
The sera used to identify antigens in this experiment are from our previous study in which groups of mice were either infected transurethrally with P. mirabilis (vaccinated with live organisms) or sham vaccinated with phosphate-buffered saline (27). Both groups of mice were then transurethrally challenged with 106 CFU of P. mirabilis HI4320 to determine whether having a previous infection protected mice from subsequent infection. Vaccinated mice were colonized in the urine, bladder, and kidneys but had significantly lower colonization levels in the kidneys than the sham-vaccinated mice (P = 0.016). Therefore, in the study described here, we identified antigens (which we hope will be protective) by using sera from mice that were not completely protected. It may seem counterintuitive to use sera from nonprotected mice to search for protective antigens, but it is important to point out that these mice had strong immunoglobulin responses to MR/P fimbriae (27). MR/P fimbriae or protein subunits (MrpH and MrpA) have previously been used successfully in a protective vaccine in the murine model (32, 33, 47, 51, 52). Even though these antigens can be protective and were present during infection (as evidenced by the strong immunoglobin response that was detected), their presence was not able to protect the mice fully from subsequent infection. Higher levels of antibody to specific proteins induced by subunit vaccinations may, however, be effective in protecting the urinary tract from colonization. The correlates of protection in the urinary tract are not yet well understood. Multiple studies have shown that the production of IgG and IgM in serum does not necessarily correlate with protection (29, 51, 52). However, data on the production of local IgA and protection are not as clear. Some studies have shown no correlation (51, 52), while others have suggested one (27, 33). It is known that antigen-specific responses occur in the urinary tract and accelerate the clearance of at least one uropathogen, UPEC (58).
Future work will determine whether these proteins are conserved among P. mirabilis strains. We are also interested in assessing whether these antigens could provide protection against other bacteria which are commonly found in polymicrobial cUTIs, such as Providencia and Morganella species. Although the corresponding genome sequences have not been published, work is currently under way in our laboratory to determine whether homologs of the genes encoding antigenic P. mirabilis proteins are present in these species (E. Flannery and H. Mobley, unpublished data). We will also determine the extent of protection offered by immunization with selected newly identified antigens in the murine model of urinary tract infection. P. mirabilis antigenic proteins identified in this study are being overexpressed in and purified from E. coli. Mice will be immunized with antigen cross-linked to cholera toxin, which is known to be an effective mucosal adjuvant (18) and has been used previously with P. mirabilis MrpH for an effective vaccine (33). By taking this approach, we hope to develop an effective vaccine against this agent of cUTIs.
Acknowledgments
We thank Virginia Lockatell and David Johnson for providing mouse sera and Mark Shirtliff and Sandy Jacobsen for their assistance in mass spectrometry interpretation. Proteomics data were provided by the Michigan Proteome Consortium (http://www.proteomeconsortium.org), which is supported in part by funds from the Michigan Life Sciences Corridor (State of Michigan MEDC grant no. GR239).
Funding was provided by the Molecular Mechanisms of Microbial Pathogenesis training grant T32 AI07528 (G.R.N.) and Public Health Service grants AI43363 and AI059722 from the National Institutes of Health (H.L.T.M.).
Editor: R. P. Morrison
Footnotes
Published ahead of print on 14 July 2008.
REFERENCES
- 1.Afonina, G., I. Leduc, I. Nepluev, C. Jeter, P. Routh, G. Almond, P. E. Orndorff, M. Hobbs, and C. Elkins. 2006. Immunization with the Haemophilus ducreyi hemoglobin receptor HgbA protects against infection in the swine model of chancroid. Infect. Immun. 742224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alamuri, P., and H. L. Mobley. 2008. A novel autotransporter of uropathogenic Proteus mirabilis is both a cytotoxin and an agglutinin. Mol. Microbiol. 68997-1017. [DOI] [PubMed] [Google Scholar]
- 3.Allocati, N., B. Favaloro, M. Masulli, M. F. Alexeyev, and C. Di Ilio. 2003. Proteus mirabilis glutathione S-transferase B1-1 is involved in protective mechanisms against oxidative and chemical stresses. Biochem. J. 373305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alteri, C. J., and H. L. Mobley. 2007. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect. Immun. 752679-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen, C., D. Krones, C. Ulmke, K. Schmid, and R. Benz. 1998. The porin RafY encoded by the raffinose plasmid pRSD2 of Escherichia coli forms a general diffusion pore and not a carbohydrate-specific porin. Eur. J. Biochem. 254679-684. [DOI] [PubMed] [Google Scholar]
- 6.Bacheller, C. D., and J. M. Bernstein. 1997. Urinary tract infections. Med. Clin. N. Am. 81719-730. [DOI] [PubMed] [Google Scholar]
- 7.Bahrani, F. K., and H. L. Mobley. 1994. Proteus mirabilis MR/P fimbrial operon: genetic organization, nucleotide sequence, and conditions for expression. J. Bacteriol. 1763412-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belas, R. 1994. Expression of multiple flagellin-encoding genes of Proteus mirabilis. J. Bacteriol. 1767169-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belas, R., and D. Flaherty. 1994. Sequence and genetic analysis of multiple flagellin-encoding genes from Proteus mirabilis. Gene 14833-41. [DOI] [PubMed] [Google Scholar]
- 10.Bertot, G. M., P. D. Becker, C. A. Guzman, and S. Grinstein. 2004. Intranasal vaccination with recombinant P6 protein and adamantylamide dipeptide as mucosal adjuvant confers efficient protection against otitis media and lung infection by nontypeable Haemophilus influenzae. J. Infect. Dis. 1891304-1312. [DOI] [PubMed] [Google Scholar]
- 11.Bouvier, J., A. P. Pugsley, and P. Stragier. 1991. A gene for a new lipoprotein in the dapA-purC interval of the Escherichia coli chromosome. J. Bacteriol. 1735523-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burall, L. S., J. M. Harro, X. Li, C. V. Lockatell, S. D. Himpsl, J. R. Hebel, D. E. Johnson, and H. L. Mobley. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 722922-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnens, A., U. Stucki, J. Nicolet, and J. Frey. 1995. Identification and characterization of an immunogenic outer membrane protein of Campylobacter jejuni. J. Clin. Microbiol. 332826-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davanloo, P., A. H. Rosenberg, J. J. Dunn, and F. W. Studier. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 812035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeMaria, T. F., D. M. Murwin, and E. R. Leake. 1996. Immunization with outer membrane protein P6 from nontypeable Haemophilus influenzae induces bactericidal antibody and affords protection in the chinchilla model of otitis media. Infect. Immun. 645187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggert, U. S., N. Ruiz, B. V. Falcone, A. A. Branstrom, R. C. Goldman, T. J. Silhavy, and D. Kahne. 2001. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science 294361-364. [DOI] [PubMed] [Google Scholar]
- 17.Engleberg, N. C., D. C. Howe, J. E. Rogers, J. Arroyo, and B. I. Eisenstein. 1991. Characterization of a Legionella pneumophila gene encoding a lipoprotein antigen. Mol. Microbiol. 52021-2029. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson, K., and J. Holmgren. 2002. Recent advances in mucosal vaccines and adjuvants. Curr. Opin. Immunol. 14666-672. [DOI] [PubMed] [Google Scholar]
- 19.Forst, D., W. Welte, T. Wacker, and K. Diederichs. 1998. Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose. Nat. Struct. Biol. 537-46. [DOI] [PubMed] [Google Scholar]
- 20.Gaisser, S., and C. Hughes. 1997. A locus coding for putative non-ribosomal peptide/polyketide synthase functions is mutated in a swarming-defective Proteus mirabilis strain. Mol. Gen. Genet. 253415-427. [DOI] [PubMed] [Google Scholar]
- 21.Gerding, M. A., Y. Ogata, N. D. Pecora, H. Niki, and P. A. de Boer. 2007. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 631008-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagan, E. C., and H. L. Mobley. 2007. Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect. Immun. 753941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagberg, L., I. Engberg, R. Freter, J. Lam, S. Olling, and C. Svanborg Eden. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajishengallis, G., S. K. Hollingshead, T. Koga, and M. W. Russell. 1995. Mucosal immunization with a bacterial protein antigen genetically coupled to cholera toxin A2/B subunits. J. Immunol. 1544322-4332. [PubMed] [Google Scholar]
- 25.Hellman, J., J. D. Roberts, Jr., M. M. Tehan, J. E. Allaire, and H. S. Warren. 2002. Bacterial peptidoglycan-associated lipoprotein is released into the bloodstream in gram-negative sepsis and causes inflammation and death in mice. J. Biol. Chem. 27714274-14280. [DOI] [PubMed] [Google Scholar]
- 26.Hotomi, M., T. Saito, and N. Yamanaka. 1998. Specific mucosal immunity and enhanced nasopharyngeal clearance of nontypeable Haemophilus influenzae after intranasal immunization with outer membrane protein P6 and cholera toxin. Vaccine 161950-1956. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, D. E., F. K. Bahrani, C. V. Lockatell, C. B. Drachenberg, J. R. Hebel, R. Belas, J. W. Warren, and H. L. Mobley. 1999. Serum immunoglobulin response and protection from homologous challenge by Proteus mirabilis in a mouse model of ascending urinary tract infection. Infect. Immun. 676683-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, D. E., C. V. Lockatell, M. Hall-Craigs, H. L. Mobley, and J. W. Warren. 1987. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. J. Urol. 138632-635. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, D. E., C. V. Lockatell, R. G. Russell, J. R. Hebel, M. D. Island, A. Stapleton, W. E. Stamm, and J. W. Warren. 1998. Comparison of Escherichia coli strains recovered from human cystitis and pyelonephritis infections in transurethrally challenged mice. Infect. Immun. 663059-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyd, J. M., M. L. Dunkley, and A. W. Cripps. 1995. Enhanced respiratory clearance of nontypeable Haemophilus influenzae following mucosal immunization with P6 in a rat model. Infect. Immun. 632931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legnani-Fajardo, C., P. Zunino, G. Algorta, and H. F. Laborde. 1991. Antigenic and immunogenic activity of flagella and fimbriae preparations from uropathogenic Proteus mirabilis. Can. J. Microbiol. 37325-328. [DOI] [PubMed] [Google Scholar]
- 32.Li, X., J. L. Erbe, C. V. Lockatell, D. E. Johnson, M. G. Jobling, R. K. Holmes, and H. L. Mobley. 2004. Use of translational fusion of the MrpH fimbrial adhesin-binding domain with the cholera toxin A2 domain, coexpressed with the cholera toxin B subunit, as an intranasal vaccine to prevent experimental urinary tract infection by Proteus mirabilis. Infect. Immun. 727306-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, X., C. V. Lockatell, D. E. Johnson, M. C. Lane, J. W. Warren, and H. L. Mobley. 2004. Development of an intranasal vaccine to prevent urinary tract infection by Proteus mirabilis. Infect. Immun. 7266-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, X., and H. L. Mobley. 2002. Vaccines for Proteus mirabilis in urinary tract infection. Int. J. Antimicrob. Agents 19461-465. [DOI] [PubMed] [Google Scholar]
- 35.Li, X., H. Zhao, C. V. Lockatell, C. B. Drachenberg, D. E. Johnson, and H. L. Mobley. 2002. Visualization of Proteus mirabilis within the matrix of urease-induced bladder stones during experimental urinary tract infection. Infect. Immun. 70389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lima, A., P. Zunino, B. D'Alessandro, and C. Piccini. 2007. An iron-regulated outer-membrane protein of Proteus mirabilis is a haem receptor that plays an important role in urinary tract infection and in in vivo growth. J. Med. Microbiol. 561600-1607. [DOI] [PubMed] [Google Scholar]
- 37.Lloubes, R., E. Cascales, A. Walburger, E. Bouveret, C. Lazdunski, A. Bernadac, and L. Journet. 2001. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152523-529. [DOI] [PubMed] [Google Scholar]
- 38.Manos, J., and R. Belas. 2004. Transcription of Proteus mirabilis flaAB. Microbiology 1502857-2863. [DOI] [PubMed] [Google Scholar]
- 39.Moayeri, N., C. M. Collins, and P. O'Hanley. 1991. Efficacy of a Proteus mirabilis outer membrane protein vaccine in preventing experimental Proteus pyelonephritis in a BALB/c mouse model. Infect. Immun. 593778-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mobley, H. L., and G. R. Chippendale. 1990. Hemagglutinin, urease, and hemolysin production by Proteus mirabilis from clinical sources. J. Infect. Dis. 161525-530. [DOI] [PubMed] [Google Scholar]
- 41.Mobley, H. L., and J. W. Warren. 1987. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J. Clin. Microbiol. 252216-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 2672871-2881. [DOI] [PubMed] [Google Scholar]
- 43.Molloy, M. P., B. R. Herbert, K. L. Williams, and A. A. Gooley. 1999. Extraction of Escherichia coli proteins with organic solvents prior to two-dimensional electrophoresis. Electrophoresis 20701-704. [DOI] [PubMed] [Google Scholar]
- 44.Murphy, C. A., and R. Belas. 1999. Genomic rearrangements in the flagellin genes of Proteus mirabilis. Mol. Microbiol. 31679-690. [DOI] [PubMed] [Google Scholar]
- 45.Pearson, M. M., and H. L. Mobley. 2007. The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. J. Med. Microbiol. 561277-1283. [DOI] [PubMed] [Google Scholar]
- 46.Pearson, M. M., M. Sebaihia, C. Churcher, M. A. Quail, A. S. Seshasayee, Z. Abdellah, C. Arrosmith, B. Atkin, T. Chillingworth, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, D. Walker, S. Whithead, N. R. Thomson, P. N. Rather, J. Parkhill, and H. L. Mobley. 2008. The complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 1904027-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pellegrino, R., U. Galvalisi, P. Scavone, V. Sosa, and P. Zunino. 2003. Evaluation of Proteus mirabilis structural fimbrial proteins as antigens against urinary tract infections. FEMS Immunol. Med. Microbiol. 36103-110. [DOI] [PubMed] [Google Scholar]
- 48.Piccini, C. D., F. M. Barbe, and C. L. Legnani-Fajardo. 1998. Identification of iron-regulated outer membrane proteins in uropathogenic Proteus mirabilis and its relationship with heme uptake. FEMS Microbiol. Lett. 166243-248. [DOI] [PubMed] [Google Scholar]
- 49.Ruiz, N., B. Falcone, D. Kahne, and T. J. Silhavy. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121307-317. [DOI] [PubMed] [Google Scholar]
- 50.Russo, T. A., C. D. McFadden, U. B. Carlino-MacDonald, J. M. Beanan, R. Olson, and G. E. Wilding. 2003. The siderophore receptor IroN of extraintestinal pathogenic Escherichia coli is a potential vaccine candidate. Infect. Immun. 717164-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scavone, P., A. Miyoshi, A. Rial, A. Chabalgoity, P. Langella, V. Azevedo, and P. Zunino. 2007. Intranasal immunisation with recombinant Lactococcus lactis displaying either anchored or secreted forms of Proteus mirabilis MrpA fimbrial protein confers specific immune response and induces a significant reduction of kidney bacterial colonisation in mice. Microbes Infect. 9821-828. [DOI] [PubMed] [Google Scholar]
- 52.Scavone, P., V. Sosa, R. Pellegrino, U. Galvalisi, and P. Zunino. 2004. Mucosal vaccination of mice with recombinant Proteus mirabilis structural fimbrial proteins. Microbes Infect. 6853-860. [DOI] [PubMed] [Google Scholar]
- 53.Schirmer, T., T. A. Keller, Y. F. Wang, and J. P. Rosenbusch. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science 267512-514. [DOI] [PubMed] [Google Scholar]
- 54.Shand, G. H., H. Anwar, J. Kadurugamuwa, M. R. Brown, S. H. Silverman, and J. Melling. 1985. In vivo evidence that bacteria in urinary tract infection grow under iron-restricted conditions. Infect. Immun. 4835-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 726373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sturgis, J. N. 2001. Organisation and evolution of the tol-pal gene cluster. J. Mol. Microbiol. Biotechnol. 3113-122. [PubMed] [Google Scholar]
- 57.Tambyah, P. A., and D. G. Maki. 2000. Catheter-associated urinary tract infection is rarely symptomatic: a prospective study of 1,497 catheterized patients. Arch. Intern. Med. 160678-682. [DOI] [PubMed] [Google Scholar]
- 58.Thumbikat, P., C. Waltenbaugh, A. J. Schaeffer, and D. J. Klumpp. 2006. Antigen-specific responses accelerate bacterial clearance in the bladder. J. Immunol. 1763080-3086. [DOI] [PubMed] [Google Scholar]
- 59.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 696179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van den Berg, B., P. N. Black, W. M. Clemons, Jr., and T. A. Rapoport. 2004. Crystal structure of the long-chain fatty acid transporter FadL. Science 3041506-1509. [DOI] [PubMed] [Google Scholar]
- 61.Warren, J. W., J. H. Tenney, J. M. Hoopes, H. L. Muncie, and W. C. Anthony. 1982. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J. Infect. Dis. 146719-723. [DOI] [PubMed] [Google Scholar]
- 62.Webb, D. C., and A. W. Cripps. 1999. Immunization with recombinant transferrin binding protein B enhances clearance of nontypeable Haemophilus influenzae from the rat lung. Infect. Immun. 672138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, T., J. Malinverni, N. Ruiz, S. Kim, T. J. Silhavy, and D. Kahne. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121235-245. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, H., X. Li, D. E. Johnson, I. Blomfield, and H. L. Mobley. 1997. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Mol. Microbiol. 231009-1019. [DOI] [PubMed] [Google Scholar]