Abstract
Interactions between salivary agglutinin and the adhesin P1 of Streptococcus mutans contribute to bacterial aggregation and mediate sucrose-independent adherence to tooth surfaces. We have examined biofilm formation by S. mutans UA159, and derivative strains carrying mutations affecting the localization or expression of P1, in the presence of fluid-phase or adsorbed saliva or salivary agglutinin preparations. Whole saliva- and salivary agglutinin-induced aggregation of S. mutans was adversely affected by the loss of P1 and sortase (SrtA) but not by the loss of trigger factor (RopA). Fluid-phase salivary agglutinin and, to a lesser extent, immobilized agglutinin inhibited biofilm development by S. mutans in the absence of sucrose, and whole saliva was more effective at decreasing biofilm formation than salivary agglutinin. Inhibition of biofilm development by salivary agglutinin was differently influenced by particular mutations, with the P1-deficient strain displaying a greater inhibition of biofilm development than the SrtA- or RopA-deficient strains. As expected, biofilm-forming capacities of all strains in the presence of salivary preparations were markedly enhanced in the presence of sucrose, although biofilm formation by the mutants was less efficient than that by the parental strain. Aeration strongly inhibited biofilm development, and the presence of salivary components did not restore biofilm formation in aerated conditions. The results disclose a potent ability of salivary constituents to moderate biofilm formation by S. mutans through P1-dependent and P1-independent pathways.
Streptococcus mutans, a primary etiologic agent of human dental caries, is particularly effective at forming biofilms on the hard tissues of the human oral cavity. Adherence of S. mutans to dental surfaces is the first step in the formation of biofilms by this organism and is mediated by sucrose-dependent and sucrose-independent mechanisms (8, 16). S. mutans expresses several surface adhesins that can bind to salivary pellicles formed on the teeth (23), whereas sucrose-dependent adherence is mediated by glucan binding proteins and water-insoluble glucans produced from sucrose by glucosyltransferase (GTF) enzymes (16).
Salivary agglutinin (also known as gp340) is a high-molecular-weight glycoprotein in human saliva that mediates the adhesion and aggregation of S. mutans (4, 14, 18) via the cell wall-associated adhesin P1 (a member of the AgI/II family of cell surface proteins), encoded by spaP (14). P1 interaction with fluid phase salivary agglutinin mediates aggregation of S. mutans (4, 10, 20), whereas adsorption of salivary agglutinin to solid surfaces provides a site for initial adhesion of the organism (15). Thus, salivary agglutinin is believed to facilitate bacterial clearance from the oral cavity or to promote colonization, depending on whether it is in solution or adsorbed on a surface (14, 20).
The expression or biological activity of S. mutans P1 can be influenced by multiple gene products, including luxS, encoding the synthesis for autoinducer 2 (AI-2) (30), ropA, encoding a peptidyl-prolyl isomerase (7), and srtA, encoding the sortase enzyme that covalently couples LPXTG-containing proteins, including P1, to the cell wall (13, 17). While it is not exactly clear how AI-2 levels impact spaP transcription, the loss of RopA appears to alter the maturation of P1 in a way that specifically diminishes its ability to interact with solid-phase agglutinin without affecting its ability to bind to fluid-phase agglutinin preparations (7). The loss of sortase, on the other hand, does not allow cells to anchor P1 to the peptidoglycan, causing P1 to be secreted into the culture supernatant fluid (17). Strains of S. mutans carrying mutations in ropA or srtA display aberrant biofilm formation in an in vitro biofilm assay when saliva is absent and glucose is the primary carbohydrate source (19, 24, 32).
A large body of literature is accumulating that is helping to define the genetic regulatory circuits underlying biofilm maturation by S. mutans and related oral bacteria. This progress has largely been facilitated by the examination of the effects of specific mutants on biofilm formation and architecture and on homeostatic mechanisms of adherent populations. Appropriately, these studies have been conducted in the absence of saliva or its constituents to allow for the disclosure of bacterial factors required for establishing and maintaining the intermolecular interactions needed to create a biofilm or to maintain homeostasis in the presence of stresses induced by mass transport limitations (5). However, evidence is emerging that interaction of S. mutans and other oral streptococci with salivary components not only alters the adhesive capacity of the organisms but may also induce changes in gene expression that could potentially influence biofilm maturation (25). Notably, while differences in the way in which P1 interacts with fluid-phase or surface-bound agglutinin have been reported (4, 20), their effects on biofilm maturation by S. mutans remain largely unexplored. Here, we utilized an established in vitro model to explore the effects of salivary components on the abilities of S. mutans to form biofilms and used strains carrying specific mutations that are known to affect P1 biogenesis to begin to probe the basis for the observed effects.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The streptococcal strains used for aggregation and biofilm assays were S. mutans UA159 serotype c and three mutant derivatives constructed by insertion of an antibiotic resistance determinant gene (erythromycin for ropA, tetracycline for spaP, and kanamycin for srtA) with the concomitant deletion of each gene. The RopA-deficient strain was constructed from S. mutans UA159 by insertion of an erythromycin resistance determinant and concurrent deletion of the ropA gene (32). To create a deletion of the srtA gene, 5′ and 3′ flanking regions of the gene were amplified from chromosomal DNA from S. mutans UA159, ligated together using BamHI sites designed in the primer set, and cloned into pGEM-T Easy Vector (Promega, Madison, WI). The primers used were srtA-A (TGATGCCATGCTCTTCTTTA) and srtA-BamHI-B (TCCTAGATTGGATCCCTTTTTTCAT) for the 5′ flanking region and srtA-BamHI-C (GCAGTCCTAGGATCCAATATCATTT) and srtA-D (CCTTCTGAACAGAATCGTTGC) for the 3′ flanking region. The plasmid was digested with BamHI, and a kanamycin cassette (NPKm), released from pALH124 (2) with the same enzyme, was inserted for the generation of a nonpolar mutation. The desired mutagenic plasmid was selected after PCR amplification using vector-originated m13 primers, isolated, and used to transform S. mutans UA159. Transformants were selected on brain heart infusion (BHI) agar containing kanamycin (1,000 μg/ml), and the double-crossover mutant of srtA was confirmed by PCR and sequencing. The P1-deficient derivatives of S. mutans UA159 and NG8 were constructed by insertion of a tetracycline resistance determinant and concurrent deletion of the spaP gene (6).
All strains were maintained on BHI medium. When needed, tetracycline, erythromycin, and kanamycin were added to the medium at 10, 10, and 1,000 μg/ml, respectively. For biofilm formation, S. mutans and its derivatives were grown in a semidefined biofilm medium (BM) with either 20 mM glucose (BM-glucose) or sucrose (BM-sucrose) as the carbohydrate source, as previously described (21).
Saliva collection.
Unstimulated whole saliva (UWS) was collected from healthy volunteers with good oral health into a chilled, sterile tube via a spitting method. The volunteers had refrained from eating, drinking, and brushing for at least 2 h prior to collection. The saliva was centrifuged at 4,000 × g for 10 min to remove cellular debris. The resulting supernatants were pooled and stored at −20°C for 1 to 2 weeks for agglutinin preparations. For aggregation and biofilm assays, the clarified saliva was used after filter sterilization through a 0.22-μm Acrodisc filter (Pall Corporation, Ann Arbor, MI). After filtration, the protein concentration of UWS was approximately 0.90 to 1.20 mg ml−1, as determined by the bicinchoninic acid protein assay (Pierce, Rockford, IL), with bovine serum albumin as a standard. For all the following experiments, we tested saliva from a total of six different donors. In all cases, the effects of the individual salivary preparations on the phenotypes that were evaluated were the same. Thus, for the data reported below, a pool of saliva from a total of six different donors was utilized.
Agglutinin preparation.
Salivary agglutinin was prepared from S. mutans NG8 and its P1-deficient strain by the affinity purification method of Brady et al. (4). Salivary agglutinin preparations (SAP) were also obtained from a spaP mutant of UA159 using the same protocol. The protein concentration of the agglutinin preparations from S. mutans NG8 (SAP) and those prepared using P1-deficient mutant strains (mock agglutinin preparation [MAP]) were approximately 0.02 to 0.10 mg ml−1. There were no significant differences in protein concentrations between the preparations (data not shown).
Electrophoresis.
UWS, SAP, and MAP were subjected to electrophoresis through a 3 to 8% Tris-acetate gradient gel (Invitrogen, Carlsbad, CA). Following electrophoresis, the gels were stained with a glycoprotein detection kit (Sigma-Aldrich, St. Louis, MO) and a silver-staining kit (Bio-Rad Laboratories, Hercules, CA) according to the suppliers' instructions. The molecular weights were determined by comparing the mobility of each of the unknown components with high-molecular-weight markers (Invitrogen). Western blotting was performed to detect gp340 using a commercially available antibody (Affinity BioReagents, Boulder, CO), as recommended by the supplier.
Aggregation assay.
Aggregation experiments were performed as previously described (22). Briefly, S. mutans UA159 and its derivatives were grown overnight in BHI broth. The bacteria were harvested by centrifugation at 4,000 × g for 10 min, washed twice with phosphate-buffered saline (PBS), and resuspended in PBS to an optical density at 600 nm (OD600) of approximately 0.6. Nine hundred microliters of bacterial suspension, 5 μl of 0.1 M CaCl2, and 100 μl of SAP, MAP, UWS, or PBS were mixed in a test tube, vortexed, and transferred to cuvettes. After equilibrating the cuvettes for 5 min at room temperature, the OD600 of the samples was recorded at 10-min intervals at 37°C for 120 min in a spectrophotometer equipped with a temperature-controlled multicuvette positioner. Percent aggregation (percent decrease in OD600) was calculated as (OD600 at 0 min − OD600 at 120 min)/(OD600 at 0 min) × 100. All assays were performed at least five times.
Biofilm formation assays.
Biofilm assays were done as described previously using polystyrene 96-well (flat-bottom) cell culture clusters (Costar 3595; Corning Inc., Corning, NY) (32). Briefly, overnight cultures of S. mutans UA159 and its derivatives were transferred to prewarmed BHI and grown at 37°C in a 5% CO2, aerobic atmosphere to the mid-exponential phase (OD600 = 0.5). The cultures were then diluted 1:100 in prewarmed BM. Biofilm formation assays were performed in the following two different ways: salivary preparations were added to each well with the cell suspensions (fluid-phase salivary preparations), or wells were first coated with salivary preparations before being inoculated with cell suspensions (surface-bound salivary preparations). For the experiments with surface-bound salivary preparations, each well was conditioned with 100 μl of SAP, MAP, UWS, or PBS. The plates were incubated at 37°C for 2 h with gentle shaking and then washed three times with PBS. Immediately after air drying for 30 min, 200 μl of the cell suspensions was inoculated into the wells. For the experiments with fluid-phase salivary preparations, 200 μl of the cell suspension was inoculated into the wells concurrently with 20 μl of SAP, MAP, UWS, or PBS. After inoculation, all plates were incubated at 37°C in a 5% CO2 atmosphere for 24 h. The culture medium was then decanted, and the plates were gently washed twice with 200 μl of sterile distilled water to remove planktonic and loosely bound cells. The adherent bacteria were stained with 50 μl of 0.1% crystal violet for 15 min. After rinsing twice with 200 μl of water, the bound dye was extracted from the stained cells using 200 μl of 99% ethanol. Biofilm formation was then quantified by measuring the absorbance of the solution at 575 nm in a spectrophotometer.
To analyze the effects of oxygen on biofilm formation by S. mutans, the cell culture clusters containing cell suspensions and salivary preparations were incubated at 37°C on a rotary shaker at 150 rpm in an aerobic atmosphere, as previously described (1). All biofilm experiments were repeated five times, with quantification assays performed from quadruplicate wells each time.
Confocal microscopy.
S. mutans UA159 biofilms for use in confocal microscopy were generated in eight-well Lab-Tek Chamber Permanox slides (Nagle Nunc International, Rochester, NY). Overnight cultures were transferred to fresh BHI broth and allowed to grow to the late exponential phase (OD600 = 0.5). The cultures were then diluted 1:100 in BM-glucose or BM-sucrose in the presence of adsorbed or fluid-phase salivary preparations as described above. Following 24 h of growth, each well was washed twice with 400 μl of Tris-buffered saline (TBS) (pH 7.0), stained for 20 min in the dark with 300 μl of TBS containing 10 μM SYTO 13 (Invitrogen), and washed once with 400 μl of TBS. The chamber walls were gently removed, 120 μl of TBS was deposited on each biofilm, and the chamber slides were covered with a coverslip that was secured with Superglue. Biofilms were examined by a Leica inverted fluorescent microscope with a Yokogawa spinning disk confocal system (Yokogawa Corporation, Newnan, GA). Images were obtained using a 60× oil objective. Three independent biofilm experiments were performed, and at least three image stacks per experiment were collected. Simulated xyz three-dimensional images were generated by using the ImageJ software package (National Institutes of Health, Montgomery, MD). The unit volume of cells (μm3 per μm2) was analyzed from the image stacks using Measure Stack (OptiNav, Bellevue, WA).
Cell surface hydrophobicity of S. mutans strains.
Bacterial cells from late-exponential-phase cultures were washed twice and suspended in PUM buffer (17.4 g of K2HPO4·3H2O, 7.26 g of KH2PO4, 1.8 g of urea, 0.2 g of MgSO4·7H2O, and distilled water to 1,000 ml, pH 7.1) to an OD600 of 0.8, as previously described (27). Then, 0.6 ml of hexadecane (Sigma-Aldrich) was added to 1.2 ml of the cell suspensions. The mixtures were vortexed for 60 s, and the aqueous phase was allowed to settle for 15 min. The cell density of the aqueous phase was then measured at 600 nm. The percentage of cells partitioned to hexadecane was calculated as (OD600 before adsorption − OD600 after adsorption)/(OD600 before adsorption) × 100.
Statistical analysis.
Bacterial aggregation assays were analyzed using a two-way analysis of variance (ANOVA) with respect to bacterial strain and salivary preparations. A two-way or three-way ANOVA was used to analyze biofilm formation and the interaction effects caused by different strains, types of salivary preparations, and pretreatment of the plates. Cell surface hydrophobicity was analyzed by a one-way ANOVA. Multiple comparisons were performed using the Bonferroni correction at a level of P < 0.05.
RESULTS AND DISCUSSION
Various salivary components adhere to and aggregate S. mutans.
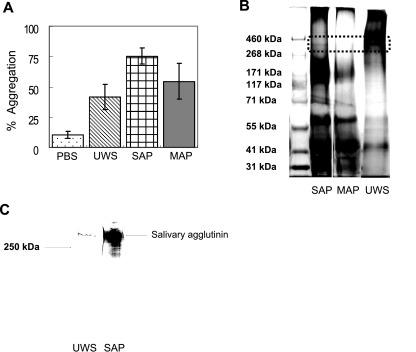
When comparing aggregation activities of SAP (from the wild-type NG8) with those of MAP (the NG8-derived spaP mutant) or UWS, SAP aggregated S. mutans UA159 more effectively than MAP or UWS (Fig. 1A). MAP and UWS showed moderate aggregation activities between SAP and PBS (SAP > MAP > UWS > PBS; P < 0.01). Self-aggregation of S. mutans UA159 was minimal in the absence of salivary preparations (<15% after 120 min in PBS).
FIG. 1.
(A) Bacterial aggregation profiles of S. mutans UA159. SAP and MAP were prepared from S. mutans NG8, and 100 μl of UWS, SAP, or MAP was added to 900 μl of bacterial suspensions. Aggregation was calculated as the percent reduction in OD after 120 min relative to that of the initial cell suspension. Aggregation data were repeated at least five times, and error bars represent standard deviations. There were statistically significant differences in bacterial aggregation capacity among salivary preparations (SAP > MAP > UWS > PBS; P < 0.01). (B) Tris-acetate gel (3 to 8%) electrophoresis of high-molecular-weight markers (lane 1), SAP (lane 2), MAP (lane 3), and UWS (lane 4). Following electrophoresis, proteins were stained with both a glycoprotein detection kit and a silver-staining kit. The dotted rectangle indicates the position of salivary agglutinin migration, which stains as a band of approximately 340 kDa in UWS and SAP but is absent in MAP. (C) Western blot analysis of UWS (lane 1) and SAP (lane 2) using an antibody to gp340 (Affinity BioReagents, Golden, CO).
Interestingly, MAP demonstrated intermediate aggregating activities that were stronger than those of UWS, suggesting that salivary components other than agglutinins can take part in the aggregation of S. mutans. The simplest explanation for MAP showing enhanced aggregating properties over UWS is that the adsorption of MAP to S. mutans enriches for proteins that are able to bind to the surface of this organism. Alternatively, MAP may be more efficient at aggregating S. mutans because UWS contains factors that inhibit agglutination, perhaps by the formation of complexes with aggregation-promoting proteins under the assay conditions used. Electrophoretic analysis of the different salivary preparations demonstrated that a 340-kDa protein, corresponding in molecular mass to that of salivary agglutinin, was present in UWS and SAP but was not detected in MAP (Fig. 1B). In addition, salivary agglutinin was significantly enriched in SAP compared with that in UWS, as assessed by Western blotting with a commercially available antibody to gp340 (Fig. 1C). However, there were no apparent differences in the protein profiles of salivary components in SAP and MAP except for the 340-kDa band. Clearly though, various salivary components in addition to agglutinin selectively adhered to S. mutans strains, which could explain why MAP aggregated S. mutans without containing substantial amounts of gp340. These findings are consistent with previous studies that showed that high-molecular-weight mucin, acidic proline-rich proteins (11, 12), 55- and 60-kDa salivary glycoproteins (3), and a 440-kDa parotid glycoprotein (9) could serve as ligands for S. mutans.
Defects in the expression or localization of P1 affect the interaction of S. mutans with salivary agglutinin in the fluid phase.
We analyzed the interaction of P1 with various salivary preparations using strains of S. mutans lacking P1, trigger factor (RopA), or sortase (SrtA). The ropA mutant strain produces normal amounts of P1 and can interact normally with fluid-phase agglutinin, but the ability to bind to surface-bound agglutinin preparations is reduced by about 50% (7). Strains lacking sortase express P1 at normal levels but fail to properly anchor the protein to the cell surface, resulting in the secretion of P1 into the supernatant fluid (13, 17).
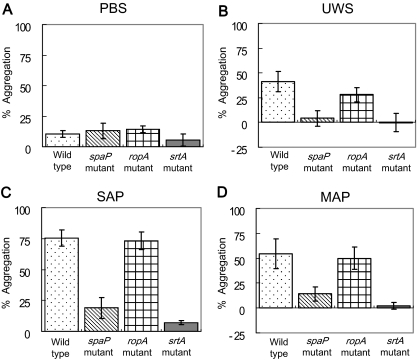
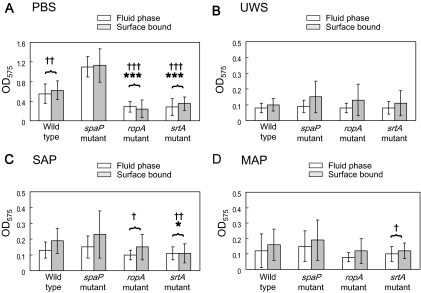
There were significant differences in the degree of bacterial aggregation between the wild-type and mutant strains. Generally, the wild-type and RopA-defective strains aggregated better in the presence of salivary preparations than did the srtA and spaP mutants (the wild-type and ropA mutant > spaP mutant > srtA mutant; P < 0.001) (Fig. 2), emphasizing the importance of the production and proper localization of P1 for interaction with salivary agglutinin. Notably, aggregation of the srtA mutant was substantially weaker than that of the P1 mutant, which is likely explained by the fact that the loss of sortase results in improper localization of not only P1 but other surface proteins, including WapA and WapE (19), which may affect bacterial aggregation. The finding that there was no difference in bacterial aggregation between the wild-type and RopA-deficient strains is consistent with the finding that RopA-deficient strains do not show aberrant interaction with fluid-phase agglutinin but do differ in their interaction with adsorbed agglutinin (7). The basis for this finding is believed to be that the loss of RopA causes abnormal folding of some or all P1 molecules on the surface of the cell. However, effects on the saliva-bacteria interactions of ropA inactivation arising from misfolding or maturation of other surface proteins cannot be excluded at this time.
FIG. 2.
Bacterial aggregation by wild-type Streptococcus mutans UA159 (dots) and its spaP (diagonal lines), ropA (hatching), and srtA (solid) mutants in the presence of PBS (A), UWS (B), SAP (C), and MAP (D). Salivary preparations and aggregation assays were conducted as described in the methods section and in the legend for Fig. 1. Aggregation data were repeated at least five times, and the data were analyzed using a two-way ANOVA with respect to bacterial strain and salivary preparations. The error bars represent standard deviations. The wild-type and RopA-deficient strains aggregated more effectively than the P1- and SrtA-deficient strains (P < 0.001). SAP aggregated S. mutans strains better than UWS and MAP (P < 0.001). In addition, a significant interaction was detected between strains and salivary preparations. Salivary preparations aggregated the parental strain and the ropA mutant better than the spaP and srtA mutants (P < 0.001).
Salivary preparations influence biofilm development by S. mutans in the absence of sucrose.
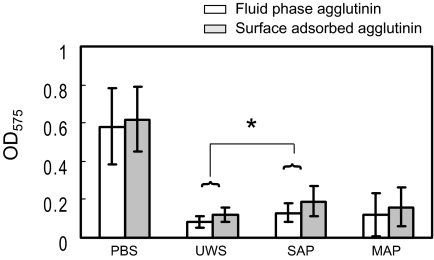
When glucose was used as a carbohydrate source, both fluid-phase and adsorbed salivary preparations significantly decreased biofilm development by S. mutans compared to that by PBS controls (Fig. 3). Pretreatment of the wells with salivary preparations resulted in slightly less inhibition of biofilm formation than was observed when the salivary preparations were present in the fluid phase (P < 0.05). Compared to PBS, the UWS, SAP, and MAP samples inhibited biofilm formation by approximately 85%, 75%, and 80%, respectively, in the fluid phase, whereas these preparations inhibited biofilm formation by approximately 80%, 70%, and 75%, respectively, when used to pretreat microtiter wells. The inhibition of biofilm development was not due to alterations in bacterial growth by salivary preparations, because there was no difference in bacterial growth rates or final yields of cells grown in the presence or absence of salivary preparations (data not shown). These findings indicate that both fluid-phase and adsorbed salivary preparations have specific capacities to impair biofilm development.
FIG. 3.
Comparisons of biofilm development by Streptococcus mutans UA159 when fluid-phase (white bars) and surface-phase (gray bars) salivary preparations were utilized. The cultures were grown in BM medium supplemented with 20 mM glucose. Fluid-phase salivary preparations were added directly to each well with the cell suspensions, whereas wells were coated with salivary preparations before inoculation with cell suspensions in the case of adsorbed salivary preparations. Biofilm formation was assayed on polystyrene microtiter plates after staining with crystal violet. Data shown are obtained from at least five independent experiments performed in quadruplicate. The error bars represent standard deviations. Biofilm development was inhibited more by fluid-phase salivary preparations than by surface-bound salivary preparations (P < 0.05), except for PBS, which showed no difference. Among salivary preparations, SAP allowed biofilm formation better than UWS (*, P < 0.05), while MAP allowed biofilm development that was intermediate to SAP and UWS.
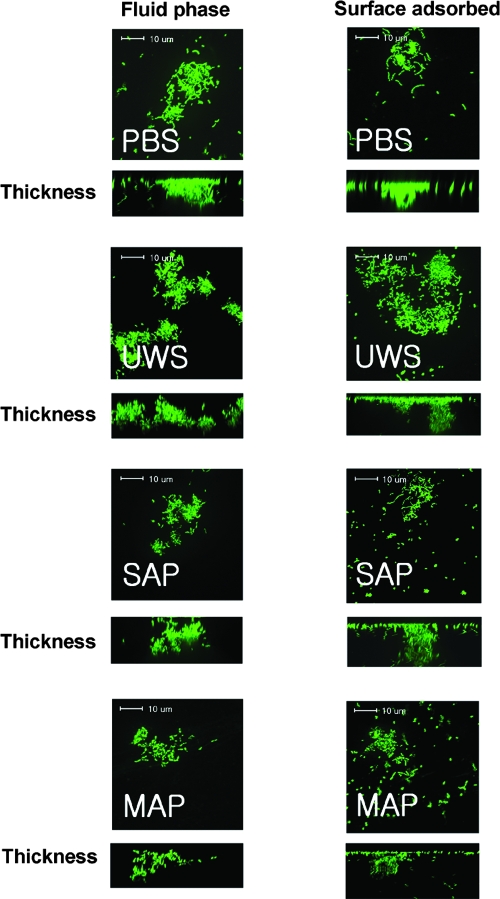
Confocal images of S. mutans biofilms formed in plastic slides showed results similar to those of the microtiter plate model. In the absence of sucrose, S. mutans biofilms showed short, scattered chains and microcolonies on the surfaces in the presence of adsorbed or fluid-phase salivary preparations compared to the longer chains and adherent clusters formed in the absence of salivary preparations (Fig. 4). Although some cell aggregates were found more frequently in the presence of SAP or MAP than in the presence of UWS, there were no significant differences in microscopic characteristics among salivary preparations. The mean volumes of biomass in the presence of fluid-phase PBS, UWS, SAP, and MAP were 2.33 ± 1.11, 0.17 ± 0.12, 0.24 ± 0.06, and 0.25 ± 0.06 μm3 per μm2, respectively. In the case of adsorbed salivary preparations, the mean volume of each biomass was 2.08 ± 0.76, 0.28 ± 0.08, 0.37 ± 0.16, and 0.32 ± 0.13 μm3 per μm2, respectively. There were no apparent differences in microscopic characteristics of the biofilms when comparing adsorbed or fluid-phase samples prepared with the same salivary preparations or with PBS (e.g., SAP biofilms appeared similar regardless of whether the preparation was used to coat the slides or was present during biofilm formation).
FIG. 4.
Confocal microscopic images of S. mutans UA159 glucose biofilms. Biofilms of S. mutans UA159 were grown in BM supplemented with 20 mM glucose in the presence of fluid-phase PBS, UWS, salivary agglutinin preparations (SAG), and mock agglutinin preparations (MAG) and stained with SYTO 13. Data presented are representative of three independent experiments.
The negative effects of fluid-phase salivary components on biofilm formation are probably exerted through multiple routes. First, occupation by salivary components of surface adhesins that are important for interactions with polystyrene, or other modifications that reduce the affinity of the organisms for substratum, would diminish the avidity of the adherence of the biofilms to the plastic. This hypothesis was confirmed by directly measuring adhesion, which revealed that fluid-phase salivary preparations significantly inhibited the initial adhesion of S. mutans UA159 to polystyrene by about 90% compared to the PBS control (UWS < SAP) (data not shown). Second, salivary constituents binding to bacterial surface structures that are needed for interbacterial interactions that support biofilm formation could also block maturation of the biofilms. It is also possible that the salivary preparations alter gene expression patterns in S. mutans in a way that renders the final biofilm less stable than cells cultured in the absence of salivary components (25). The finding that the chain length of the bacteria is altered in the presence of salivary preparations (Fig. 4) adds some credence to the idea that saliva induces changes in the expression or presentation of surface proteins, perhaps via the AtlA autolytic pathway and the Vic signal transduction complex (1). Both of these pathways are critical for biofilm formation, influence surface characteristics, and affect chain length in S. mutans.
It is also of interest that biofilm development was inhibited by surface-adsorbed salivary components, albeit less so than by fluid-phase components. Generally, saliva coating should promote bacterial adherence by providing binding receptors, but it may also diminish retention of the biofilms by decreasing the surface free energy of the underlying materials (26, 31). Such surface modification could reduce the strength of attachment of the biofilm to the substratum, resulting in decreases of the amounts of adherent biomass remaining after washing and staining. Notably, differences in biofilm development are not associated with differences in the protein concentration of the different preparations, because there was no significant difference in biofilm development among the salivary preparations when the final protein concentration of UWS was adjusted to the same concentrations of SAP or MAP (data not shown). The fact that biofilm development in the presence of SAP is more efficient than in the presence of MAP suggests that the initial stages of adherence and biofilm formation by S. mutans may be stimulated by salivary agglutinin, as well as by other salivary proteins, such as high-molecular-weight mucin and acidic proline-rich proteins (11, 12), but that other factors in saliva are responsible for the inhibition in biofilm maturation. Our adhesion data also showed that adsorbed salivary preparations significantly inhibited the initial adhesion of S. mutans UA159 to polystyrene wells (UWS < SAP). In contrast to biofilm development, however, the inhibition of initial adhesion by adsorbed salivary preparations was greater than that by fluid-phase preparations. These data may indicate that the effects on the biofilm maturation of fluid-phase and surface-adsorbed salivary preparations are exerted through different pathways, although additional research will be needed to confirm or refute this idea.
Effects of sucrose or aeration on biofilm formation by S. mutans in the presence of salivary preparations.
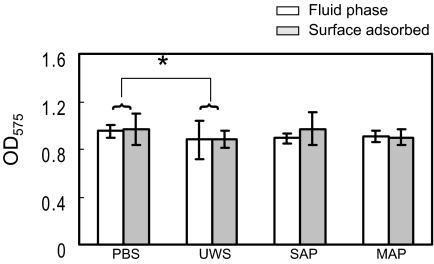
As expected, biofilm formation in BM-sucrose showed a significantly different pattern compared with biofilms formed in BM-glucose (Fig. 4 and 5). Biofilm-forming capacities that were inhibited by salivary preparations were markedly restored when sucrose was used as a carbohydrate source. In addition, there was no significant difference in biofilm development when the effects of fluid-phase and adsorbed salivary preparations on biofilm development were compared in sucrose-containing medium. This result is not entirely surprising given that the production of, and binding to, water-insoluble glucans very effectively promotes vigorous biofilm formation by S. mutans independently of saliva-P1 interactions (28, 29). However, fluid-phase and surface-bound UWS decreased biofilm development compared to PBS (PBS > UWS; P < 0.05) when sucrose was the growth carbohydrate (Fig. 5). In addition, there was a tendency for SAP to allow better biofilm formation than UWS when the salivary preparations were adsorbed, although this difference was not statistically significant. Thus, enrichment for salivary agglutinin or depletion of other constituents from UWS in the SAP samples modulates the biofilm-forming capacity in the presence of sucrose. Saliva, particularly adsorbed saliva, has been shown to enhance glucan synthetic activities of GTFs, to alter glucan end product structure, and to enhance attachment of S. mutans (28). The ability of surface-bound SAP to support biofilm formation in the presence of sucrose better than UWS may be due to the enrichment in SAP for factors that promote surface deposition and stimulate the activities of GTF enzymes. In microscopic images, biofilms in BM-sucrose showed thick cell aggregates around scattered microcolonies (Fig. 6). There were some variations in the volumes of cell biomass due to the existence of cell aggregates, and the volumes of biomass in BM-sucrose ranged from 0.4 to 1.2 μm3 per μm2. There were no significant differences in cellular volumes as a function of the different salivary preparations.
FIG. 5.
Comparison of biofilm development by Streptococcus mutans UA159 when fluid-phase (white bars) and surface-phase (gray bars) salivary preparations were utilized. The cultures were grown in BM medium supplemented with 20 mM sucrose. Salivary treatment and biofilm quantification were as described in Materials and Methods and in the legend for Fig. 3. Data were obtained from at least five independent experiments performed in quadruplicate. Error bars represent standard deviations. There were no significant differences in biofilm development between fluid-phase and adsorbed salivary preparations. Significant differences in biofilm development were detected only between PBS and UWS. *, P < 0.05; two-way ANOVA.
FIG. 6.
Confocal microscopic images of S. mutans UA159 sucrose biofilms. Biofilms of S. mutans UA159 were formed in BM supplemented with 20 mM sucrose in the presence of fluid-phase PBS, UWS, salivary agglutinin preparations (SAG), and mock agglutinin preparations (MAG), and stained with SYTO 13. There was no significant difference in microscopic images between adsorbed and fluid-phase salivary preparations. Data presented here are representative of three independent experiments.
When cultures were aerated, the capacity to form biofilms was decreased by over 80% compared to that of the strains grown in a 5% CO2 atmosphere. This finding is consistent with a previous study from our laboratory that showed strong inhibition of biofilm maturation by S. mutans by oxygen (1). This inhibition was shown to be due, at least in part, to changes in the cell surface composition of S. mutans involving the Vic signal transduction system and the AtlA autolysin (1). In addition, the amount of biofilm formed was at the low end of detection when fluid-phase and adsorbed salivary preparations were utilized (data not shown). Therefore, the presence of salivary preparations was not able to reverse the inhibitory effects of oxygen on biofilm formation, regardless of the growth carbohydrate.
Biofilm formation of S. mutans can be significantly influenced by mutations that affect P1.
The first P1-deficient strain of S. mutans UA159 was engineered identically to the well-characterized derivative of S. mutans NG8 that has been studied extensively from the perspective of agglutinin interactions. However, the UA159 spaP mutant also contains a deletion of the approximately one-third of the bsp gene located 5′ to spaP. To ensure that the characteristics of the P1-deficient strain were attributable to the loss of only spaP, otherwise-isogenic derivatives of UA159 with deleted spaP and bsp genes were constructed by insertion of an antibiotic resistance determinant gene, as described for the srtA mutant. In all cases, the regions used to integrate the gene were amplified by PCR and sequenced to ensure that no mutations had been introduced into flanking regions. When the same aggregation and biofilm assays were performed, both spaP mutants yielded similar results and the deletion of bsp did not significantly influence the characteristics of aggregation and biofilm development by S. mutans UA159. The only notable difference between the spaP mutant and the spaP strain lacking part of bsp was that the former was about 30% less efficient at forming biofilms than the latter, and about 20% more efficient than the wild-type strain in BM-glucose when salivary preparations were absent. However, there were no significant differences in biofilm development among the P1-deficient strains and wild-type strains in BM-glucose in the presence of salivary preparations. In the present study, only the data generated from the spaP mutant containing a deletion of the bsp region are presented because of its similarity to the already well-characterized derivative of S. mutans NG8.
Similar to that by the wild-type strain, biofilm formation by S. mutans mutant strains was significantly inhibited by salivary preparations, but there were some notable differences in the behavior of particular strains (Fig. 7). When cultivated with glucose as the primary carbohydrate, the strains harboring deletions of the ropA or srtA gene displayed significantly decreased biofilm development compared to that of the parental strain, irrespective of the salivary preparation used (wild type > ropA and srtA mutants) (Fig. 7). However, the differences in biofilm development between these mutants and the parental strain were less obvious in the presence of SAP or MAP (Fig. 7C and D). Surprisingly, the P1-deficient strain did not show decreased biofilm development in agglutinin- or saliva-coated wells compared to the parental strain (Fig. 7B and C). Although P1 is clearly important for the initial adherence of S. mutans to saliva-coated materials (6, 15), these results indicate that it is not required for biofilm maturation. In fact, the P1-defective strains formed biofilms more efficiently than the parental strain in the absence of salivary preparations (Fig. 7A), suggesting that P1 may directly or indirectly diminish the ability of the organisms to form the stable intercellular interactions required for biofilm formation in the absence of sucrose. Such inhibition could occur from remodeling of the cell surface due to the loss of P1 or through direct inhibitory effects on intercellular adhesive interactions by P1. Notably, adsorbed salivary preparations decreased biofilm formation by the P1 mutant to an extent similar to that of the parental strain, perhaps supporting the notion that P1 somehow interferes with intercellular interactions (Fig. 7B to D).
FIG. 7.
Results of biofilm formation of Streptococcus mutans UA159 and its derivatives in BM supplemented with 20 mM glucose, with respect to PBS (A), UWS (B), SAP (C), and MAP (D). Biofilm formation assays were performed in the following two different ways: salivary preparations were added to each well with the cell suspensions (fluid-phase salivary preparations) (white bars), or wells were first coated with salivary preparations before inoculation with cell suspensions (surface-phase salivary preparations) (gray bars). Biofilm assays and data analysis were as described in Materials and Methods and in the legend for Fig. 6. Biofilm development was inhibited more by fluid-phase salivary preparations than by surface-bound salivary preparations (P < 0.05). The differences in biofilm development between the P1-defective strain and other strains are marked by a cross, and those between the wild type and other strains are marked by an asterisk. †, P < 0.05; ††, P < 0.01; †††, P < 0.001; *, P < 0.05; ***, P < 0.001. See the text for more details.
Recently, Pecharki et al. (25) reported differences in biofilm formation between S. mutans LT11 and its spaP mutant in the presence of saliva. Notably, they found that biofilm formation by P1 mutants of S. mutans and S. intermedius was inhibited to a greater degree than that by the wild-type strain in the presence of saliva, although saliva significantly decreased biofilm formation by all strains. In addition, there was no significant difference in biofilm formation between the strains in the absence of saliva. The differences between the findings of Pecharki et al. (25) and those presented herein are likely attributable to experimental design and culture conditions. Specifically, biofilms of S. mutans LT11 were formed in tryptone soy broth with 25% stimulated saliva for 48 h. Tryptone soy broth is substantially richer than the semidefined BM used here, which could impact cell surface characteristics and interaction of the bacteria with salivary molecules, alter adhesion to polystyrene, accelerate bacterial growth, and lead to differences in the final pH of the biofilms, all of which could lead to alterations in biofilm maturation or stability. Nonetheless, both studies support that the biofilm-forming behavior of abundant and pathogenic oral streptococci is dramatically altered in the presence of saliva.
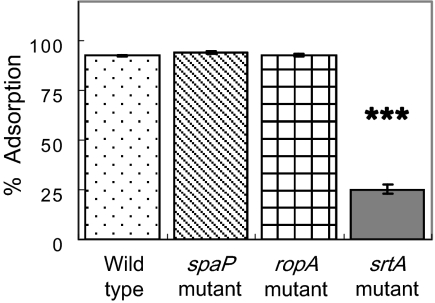
The greater effect of the losses of the srtA and ropA genes on biofilm formation compared to the effect of the loss of spaP is particularly interesting. A deficiency of srtA is known to elicit defects in the localization of other surface proteins containing LPXTG anchor motifs, including those encoded by fruA, wapA, and wapE, which may contribute to altered biofilm formation (19). Thus, a general defect in proper cell surface biogenesis probably contributes to the biofilm defect of the SrtA-deficient strain. Also, diminished surface hydrophobicity of the srtA mutant (Fig. 8) may make it difficult for the cells to remain attached by the relatively weak forces that mediate adhesion to the polystyrene wells. The effects of the loss of RopA on biofilm formation have been established (32), but the underlying basis for this defect is not known. From the data presented here, it seems unlikely that the effects of RopA on biofilm maturation are exerted through P1, given that the loss of P1 actually enhances biofilm formation. Strains lacking RopA do form longer chains than the parental strain (32), which could affect the affinity of the cell population for the substratum. More likely, though, is that defects in the proper folding of surface or membrane proteins associated with the loss of the enzymatic activity of RopA (peptidyl-prolyl isomerase) contribute to the observed phenotype.
FIG. 8.
Cell surface hydrophobicity of Streptococcus mutans UA159 strains. The bacterial suspensions (1.2 ml) were mixed with 0.6 ml of hexadecane. Adsorption was calculated as the percent loss in OD of the aqueous phase relative to that of the initial cell suspension. Error bars represent standard deviations. ***, P < 0.001; one-way ANOVA.
Collectively, these findings may have a high degree of biological relevance. Both the biofilm formation and initial adherence of S. mutans were significantly inhibited by salivary preparations. Thus, saliva may play a significant role in suppressing S. mutans levels in vivo while favoring the establishment and persistence of commensals. Given the complexity of the bacterial cell surface and of the salivary proteome, dissecting the molecular basis for these findings will require more experimentation. However, such studies can provide the needed information for the design of strategies to more effectively promote the establishment and persistence of diverse biofilm communities with the capacity to suppress the emergence of a pathogenic microflora.
Sucrose and aeration partially mask defects of P1 expression or localization.
The differences between biofilm formation by the wild-type strain and the mutants were greatly reduced when the cells were grown in sucrose-containing medium. In BM-sucrose, SrtA-defective and RopA-defective strains displayed markedly better biofilm formation than in BM-glucose, although biofilm formation was not restored to the level of the wild-type or P1-deficient strains (wild type and spaP mutant > srtA mutant) (data not shown). In addition, there were no significant differences in biofilm formation between the wild-type and P1-deficient strains. Salivary preparations did not significantly influence the level of biofilm formation among the different mutant strains when sucrose was present (wild type and spaP mutant > srtA mutant). Collectively, these results provide evidence that the loss of RopA or SrtA, but not P1, may impact the expression, localization, or folding of GTF or glucan binding proteins in a way that adversely affects biofilm formation. Consistent with this hypothesis, Wen et al. (32) showed that the loss of RopA resulted in downregulation of the expression of the GTFs that synthesize water-insoluble (GtfB) and water-soluble (GtfD) glucans.
Under aerobic conditions, the capacity to form biofilms was significantly inhibited regardless of strains and salivary preparations (data not shown). In fact, the level of biofilm formation was too low to draw any firm conclusions about the role of the gene products examined in this study other than that the loss of the gene products does not reverse the inhibitory effects of oxygen, contrary to what was noted in the case of the AtlA autolysin and the VicK sensor kinase component of a two-component signal transduction system (1). Thus, it is unlikely that AtlA or VicK alters biofilm formation through the RopA or SrtA pathway.
Summary.
More effective strategies are needed to prevent and treat the oral diseases that are elicited by biofilms on the hard and soft tissues of the oral cavity. Despite major advances in dissecting the genetics and virulence attributes of oral pathogens, our current understanding of the formation and persistence of oral biofilms, and in particular the role of saliva in governing the composition and biological activity of biofilms, is at best rudimentary. This study reveals that salivary components have a profound impact not only on bacterial adherence but also on the ability of an important oral pathogen to efficiently form biofilms. Likewise, we have shown that the major adhesin of S. mutans, P1, is dispensable for biofilm maturation and that factors that require RopA and SrtA for proper biogenesis play substantial roles in biofilm formation in the presence or absence of saliva and sucrose. Further detailed analysis of the effects of the loss of SrtA and RopA on the secretome of S. mutans and the analysis of the interaction of srtA and ropA mutant strains with specific salivary constituents will shed light on the underlying mechanisms by which saliva moderates the biofilm-forming capacity of S. mutans. It is also noteworthy that complex interactions of a variety of species of oral bacteria with dramatically different adhesive and physiologic capacities drive the formation of natural biofilms on surfaces in the human oral cavity. Exploration of the effects of salivary molecules on multispecies consortia will be necessary to gain a better understanding of the molecular mechanisms governing the development of normal and pathogenic oral biofilms.
Acknowledgments
We thank Scott Grieshaber for his assistance with the confocal microscopy and Paula Crowley for valuable discussions and editorial assistance.
This work was supported by grants DE13239, DE08007, and DE13882 from the NIDCR.
Editor: A. Camilli
Footnotes
Published ahead of print on 14 July 2008.
REFERENCES
- 1.Ahn, S. J., and R. A. Burne. 2007. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J. Bacteriol. 1896293-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 741631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babu, J. P., E. H. Beachey, D. L. Hasty, and W. A. Simpson. 1986. Isolation and characterization of a 60-kilodalton salivary glycoprotein with agglutinating activity against strains of Streptococcus mutans. Infect. Immun. 51405-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady, L. J., D. A. Piacentini, P. J. Crowley, P. C. F. Oyston, and A. S. Bleiweis. 1992. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect. Immun. 601008-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burne, R. A., R. G. Quivey, Jr., and R. E. Marquis. 1999. Physiologic homeostasis and stress responses in oral biofilms. Methods Enzymol. 310441-460. [DOI] [PubMed] [Google Scholar]
- 6.Crowley, P. J., L. J. Brady, S. M. Michalek, and A. S. Bleiweis. 1999. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect. Immun. 671201-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowley, P. J., T. S. Seifert, R. Isoda, M. van Tilburg, M. W. Oli, R. Robinette, W. P. McArthur, A. S. Bleiweis, and L. J. Brady. 2008. Requirements for surface expression and function of adhesin P1 from Streptococcus mutans. Infect. Immun. 762456-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cvitkovitch, D. G., Y. H. Li, and R. P. Ellen. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Investig. 1121626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ericson, T., A. Carler, and Z. Dagerskog. 1976. Salivary aggregation factors, p. 151-152. In H. M. Stiles, W. J. Loesche, and T. C. O'Brien (ed.), Microbial aspects of dental caries, vol. 2. Information Retrieval Inc., Washington, DC. [Google Scholar]
- 10.Ericson, T., and J. Rundegren. 1983. Characterization of a salivary agglutinin reacting with a serotype c strain of Streptococcus mutans. Eur. J. Biochem. 133255-261. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons, R. J., L. Cohen, and D. I. Hay. 1986. Strains of Streptococcus mutans and Streptococcus sobrinus attach to different pellicle receptors. Infect. Immun. 52555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbons, R. J., and D. I. Hay. 1989. Adsorbed salivary acidic proline-rich proteins contribute to the adhesion of Streptococcus mutans JBP to apatitic surfaces. J. Dent. Res. 681303-1307. [DOI] [PubMed] [Google Scholar]
- 13.Igarashi, T., E. Asaga, and N. Goto. 2003. The sortase of Streptococcus mutans mediates cell wall anchoring of a surface protein antigen. Oral Microbiol. Immunol. 18266-269. [DOI] [PubMed] [Google Scholar]
- 14.Jenkinson, H. F., and D. R. Demuth. 1997. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 23183-190. [DOI] [PubMed] [Google Scholar]
- 15.Kishimoto, E., D. I. Hay, and R. J. Gibbons. 1989. A human salivary protein which promotes adhesion of Streptococcus mutans serotype c strains to hydroxyapatite. Infect. Immun. 573702-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koga, T., H. Asakawa, N. Okahashi, and S. Hamada. 1986. Sucrose-dependent cell adherence and cariogenicity of serotype c Streptococcus mutans. J. Gen. Microbiol. 1322873-2883. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. F., and T. L. Boran. 2003. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect. Immun. 71676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, S. F., A. Progulske-Fox, G. W. Erdos, D. A. Piacentini, G. Y. Ayakawa, P. J. Crowley, and A. S. Bleiweis. 1989. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II). Infect. Immun. 573306-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lévesque, C. M., E. Voronejskaia, Y.-C. C. Huang, R. W. Mair, R. P. Ellen, and D. G. Cvitkovitch. 2005. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect. Immun. 733773-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loimaranta, V., N. S. Jakubovics, J. Hytonen, J. Finne, H. F. Jenkinson, and N. Stromberg. 2005. Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect. Immun. 732245-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 1821374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magnusson, I., and T. Ericson. 1976. Effect of salivary agglutinins of reactions between hydroxyapatite and a serotype c strain of Streptococcus mutans. Caries Res. 10273-286. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell, T. J. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. Microbiol. 1219-230. [DOI] [PubMed] [Google Scholar]
- 24.Nobbs, A. H., R. M. Vajna, J. R. Johnson, Y. Zhang, S. L. Erlandsen, M. W. Oli, J. Kreth, L. J. Brady, and M. C. Herzberg. 2007. Consequences of a sortase A mutation in Streptococcus gordonii. Microbiology 1534088-4097. [DOI] [PubMed] [Google Scholar]
- 25.Pecharki, D., F. C. Petersen, S. Assev, and A. A. Scheie. 2005. Involvement of antigen I/II surface proteins in Streptococcus mutans and Streptococcus intermedius biofilm formation. Oral Microbiol. Immunol. 20366-371. [DOI] [PubMed] [Google Scholar]
- 26.Quirynen, M., and C. M. Bollen. 1995. The influence of surface roughness and surface-free energy on supra- and subgingival plaque formation in man. A review of the literature. J. Clin. Periodontol. 221-14. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg, M. 2006. Microbial adhesion to hydrocarbons: twenty-five years of doing MATH. FEMS Microbiol. Lett. 262129-134. [DOI] [PubMed] [Google Scholar]
- 28.Schilling, K. M., and W. H. Bowen. 1988. The activity of glucosyltransferase adsorbed onto saliva-coated hydroxyapatite. J. Dent. Res. 672-8. [DOI] [PubMed] [Google Scholar]
- 29.Schilling, K. M., and W. H. Bowen. 1992. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect. Immun. 60284-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sztajer, H., A. Lemme, R. Vilchez, S. Schulz, R. Geffers, C. Y. Y. Yip, C. M. Levesque, D. G. Cvitkovitch, and I. Wagner-Döbler. 2008. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 190401-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weerkamp, A. H., H. C. van der Mei, and H. J. Busscher. 1985. The surface free energy of oral streptococci after being coated with saliva and its relation to adhesion in the mouth. J. Dent. Res. 641204-1210. [DOI] [PubMed] [Google Scholar]
- 32.Wen, Z. T., P. Suntharaligham, D. G. Cvitkovitch, and R. A. Burne. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect. Immun. 73219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]