Abstract
A number of bacteria bind factor H (FH), the negative regulator of the alternative complement pathway, to avoid complement-mediated killing. Here we show that a gram-negative enteric pathogen, Yersinia enterocolitica serotype O:3, uses two virulence-related outer membrane (OM) proteins to bind FH. With Y. enterocolitica O:3 mutant strains displaying different combinations of surface factors relevant to complement resistance, we demonstrated that the major receptor for FH is the OM protein YadA. Another OM protein, Ail, also contributes to FH binding provided that it is not blocked by distal parts of the lipopolysaccharide (i.e., the O antigen and the outer core hexasaccharide). Importantly, we demonstrated that surface-bound FH was functional; both YadA- and Ail-bound FH displayed cofactor activity for factor I-mediated cleavage of C3b. With truncated recombinant FH constructs, we located the binding site of Ail specifically to short consensus repeats 6 and 7 of FH, while YadA showed a novel type of FH-binding pattern and appears to bind FH throughout the entire FH molecule. We thus conclude that Y. enterocolitica, via YadA and Ail, recruits functionally active FH to its surface. FH binding appears to be an important mechanism of the complement resistance of this pathogen.
The complement system is part of the primary line of human innate immunity. It defends the host against invading microbes by its abilities to opsonize and directly lyse microorganisms and attract phagocytic cells to the site of microbial invasion. Complement resistance (CR) is thus a crucial feature of virulent bacteria. The gram-negative food-borne pathogen Yersinia enterocolitica usually causes enterocolitis, mesenteric lymphadenitis, and, as sequelae, postinfectious reactive arthritis and erythema nodosum (11). Y. enterocolitica expresses the outer membrane (OM) proteins YadA and Ail (7, 10, 12, 42, 56), which confer resistance to both the classical pathway (CP) and the alternative pathway (AP) of complement activation. The latter pathway, initiated spontaneously via binding of activated plasma C3 to the microbial surface, is especially important for pathogen elimination. The nonspecific nature of the AP, although beneficial in terms of antimicrobial defense, could have deleterious consequences for the host if complement activation were not tightly regulated. The main serum proteins responsible for the regulation of AP activation are factor H (FH) and its alternatively spliced variant FH-like protein 1. FH functions as a cofactor in factor I (FI)-mediated cleavage of C3b, inhibits the association of factor B with C3b and participates in the dissociation of the preformed AP C3 convertase C3bBb (28, 41, 59). FH consists of 20 short consensus repeat domains (SCRs) of about 60 amino acid residues each (27).
A number of microorganisms have evolved the capacity to bind FH and exploit its protective properties to avoid complement-mediated killing. The sialic acid-containing capsules of group B streptococci and the lipooligosaccharide of Neisseria gonorrhoeae are thought to mediate FH binding (32, 45). There are also bacterial surface proteins which contribute to FH binding. These include the M protein of Streptococcus pyogenes (20), the β protein of group B streptococcus (4), OspE and CRASP-1 of Borrelia burgdorferi (18, 26), and Por1A of N. gonorrhoeae (44). Also, YadA has been shown to bind FH from human serum (12, 47). Direct binding of purified FH to YadA, however, could not be demonstrated (46).
YadA is encoded by the yadA gene located on the 70-kb virulence plasmid (pYV) (14, 57, 61), and its expression is under the control of the virF/lcrF-encoded transcriptional activator driving YadA expression exclusively at 37°C (31, 51). YadA is a homotrimeric autotransporter protein with about 44-kDa monomers that form a lollipop-shaped structure on the bacterial surface (19, 49, 54, 60). The YadA structure includes an N-terminal head domain, a coiled-coil stalk, and a C-terminal membrane anchor (19). Serum resistance determinants of YadA were ascribed to the stalk and membrane anchor domains (46).
Ail is a 17-kDa protein encoded chromosomally (36, 37). Ail is predicted to form eight OM-spanning amphipathic β strands and four short extracellular loops (35). Ail belongs to a family of structurally related OM proteins (5, 6, 16, 43, 55). Only one of these proteins, Salmonella enterica serovar Typhimurium Rck, shows functional homology, CR, to Ail (17). The CR-conferring regions of Ail are located in the C-terminal end of loop 2 and the N-terminal end of loop 3 (35).
The lipopolysaccharide (LPS) of Y. enterocolitica O:3, required for successful colonization of the gut (3, 52), participates in CR indirectly. Both the LPS O polysaccharide (O antigen [OAg]) and the outer core hexasaccharide (OC) are linked to the inner core. Expression of both OAg and OC is temperature regulated and optimal below 30°C. In vitro, at 37°C, stationary-phase bacteria display reduced levels of both OAg (1, 2) and OC (38) on their surfaces. These LPS compounds most likely sterically block the access of complement components to the OM proteins, such as small-sized Ail (7).
In this report, we have characterized the ability of Y. enterocolitica O:3 to bind the human complement inhibitor FH. We show that Y. enterocolitica O:3 is able to bind purified FH directly, as well as to acquire it from serum. To identify the FH receptor on Y. enterocolitica, we examined the FH-binding properties of Y. enterocolitica O:3 strains expressing all possible combinations of YadA, Ail, OAg, and OC. This study confirmed the FH-binding potential of YadA and additionally revealed that Ail also binds FH. We located the binding sites on FH for Ail at SCRs 6 and 7 and showed that YadA appears to bind to SCRs throughout the entire length of FH. Both YadA- and Ail-bound FH retained the cofactor activity, suggesting that Y. enterocolitica is able to recruit to its surface functionally active FH and that this is an important contribution to the CR of the pathogen.
MATERIALS AND METHODS
Bacteria, plasmids, bacteriophages, and growth conditions.
The bacterial strains, plasmids, and bacteriophages used in this study are listed in Table 1. For adsorption of goat anti-human FH antiserum and for the ability to bind 125I-labeled FH, bacteria were grown to stationary phase overnight in 5 ml of MedECa (7) at 37°C without shaking. For the cofactor assay and examination of the bacterial ability to bind 125I-labeled, I-labeled, nonlabeled, or serum FH, overnight bacterial cultures were diluted 1:20 in fresh medium and incubated for 3 h at 37°C without shaking to obtain bacteria in the exponential phase of growth. When appropriate, antibiotics were added to the growth medium at the following concentrations: kanamycin, 100 μg/ml in agar plates and 20 μg/ml in broth; ampicillin, 50 μg/ml.
TABLE 1.
Bacterial strains, bacteriophages, and plasmids used in this work
| Bacterial strain, bacteriophage, or plasmid | Description | Source or reference |
|---|---|---|
| Y. enterocolitica strains | ||
| 6471/76 (YeO3) | Serotype O:3, patient isolate, wild type; SP YAOCa | 49 |
| YeO3-Ail | Δail::Km-GenBlock, Kmr; SP Y-OC | 7 |
| YeO3-Ail-R | Spontaneous rough derivative of YeO3-Ail, Kmr; SP Y--C | 7 |
| YeO3-Ail-OCR | Spontaneous OC mutant derivative of YeO3-Ail-R, Kmr; SP Y--- | 7 |
| YeO3-trs11 | Δ(wzx-wbcL)::Km-GenBlock, derivative of YeO3, Kmr; SP YAO-2 | 53 |
| YeO3-trs11-R | Spontaneous rough derivative of YeO3-trs11, Kmr; SP YA--2 | 53 |
| YeO3-OC | Δ(wzx-wbcQ), derivative of YeO3; SP YAO-1 | 7 |
| YeO3-OCR | Spontaneous rough derivative of YeO3-OC; SP YA--1 | 7 |
| YeO3-Ail-OC | Δail::Km-GenBlock, derivative of YeO3-OC, Kmr; SP Y-O- | 7 |
| YeO3-R2 | Spontaneous rough derivative of YeO3; SP YA-C | 3 |
| YeO3-028 | ΔyadA::Km-GenBlock, derivative of YeO3, Kmr; SP -AOC2 | 7 |
| YeO3-028-R1 | Spontaneous rough derivative of YeO3-O28, Kmr; SP -A-C1 | 7 |
| YeO3-028-OC | Δ(wzx-wbcQ) derivative of YeO3-028, Kmr; SP -AO-1 | 7 |
| 6471/76-c (YeO3-c) | Virulence plasmid cured derivative of YeO3; SP -AOC1 | 49 |
| YeO3-R1 | Spontaneous rough derivative of YeO3-c; SP -A-C2 | 3 |
| YeO3-c-OC | Δ(wzx-wbcQ) derivative of YeO3-c; SP -AO-3 | 7 |
| YeO3-c-OCR | Spontaneous rough derivative of YeO3-c-OC; SP -A--2 | 7 |
| YeO3-c-trs8 | Δ(wzx-wbcL)::Km-GenBlock, derivative of YeO3-c, Kmr; SP -AO-2 | 53 |
| YeO3-c-trs8R | Spontaneous rough derivative of YeO3-c-trs8, Kmr; SP -A--1 | 53 |
| YeO3-c-Ail | Δail::Km-GenBlock, derivative of YeO3-c, Kmr; SP --OC | 7 |
| YeO3-c-Ail-OC | Spontaneous OC mutant derivative of YeO3-c-Ail, Kmr; SP --O- | 7 |
| YeO3-c-Ail-R | Spontaneous rough derivative of YeO3-c-Ail, Kmr; SP ---C | 7 |
| YeO3-c-Ail-OCR | Spontaneous OC mutant derivative of YeO3-c-Ail-R, Kmr; SP ---- | 7 |
| E. coli JM103 | Sequencing host strain | 34 |
| Plasmids | ||
| pYMS4450 | Promoterless yadA cloned into pL2.1; yadA, Ampr | 50 |
| pL2.1 | pBR322 with the Ptac promoter, Ampr | 58 |
| Bacteriophages | ||
| φR1-37 | OC-specific phage of Y. enterocolitica O:3 | 53 |
| φR8-01 | Infects OAg- and OC-negative Y. enterocolitica O:3 bacteria grown at 22°C | This work |
YadA, Ail, OAg, and OC SPs.
Complement components.
FH and FI (catalog no. 341274 and 341280, respectively) were supplied by Calbiochem (La Jolla, CA). C3b was generated from C3 by factor B, factor D, and Mg2+ as described previously (24). FH was labeled with 125I (NEN, Boston, MA) or NaI (catalog no. 383112; Sigma) by the Iodogen method (48). FH constructs SCR 1-5, SCR 1-6, SCR 1-7, SCR 8-11, SCR 11-15, and SCR 8-20 were produced in the baculovirus expression system as described previously (29).
Antibodies and antisera.
The antibodies used were horseradish peroxidase (HRP)-conjugated rabbit anti-goat immunoglobulin G (catalog no. P449; Dako), HRP-conjugated swine anti-rabbit immunoglobulin G (catalog no. P217; Dako), rabbit anti-human C3c (catalog no. A0062; Dako), rabbit anti-human C3d (catalog no. A0063; Dako), and goat antiserum against human FH (catalog no. A312; Quidel). In immunoblotting (see below), we observed that the latter antiserum contained a high titer of YadA-specific antibodies and these were removed from the antiserum by 1 h of adsorption on ice with YadA-expressing bacteria, YeO3 and YeO3-R2. YadA-specific monoclonal antibodies (MAbs) 2A9, 2G12, and 3G12, raised against Triton X-114-enriched YadA, have been described earlier (50).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
The samples, heated in Laemmli buffer at 95°C for 5 to 10 min, were subjected to electrophoresis in 8 or 10% polyacrylamide gels and transferred onto nitrocellulose membranes. After blocking with 5% skim milk-phosphate-buffered saline (PBS), the membranes were incubated overnight at +4°C with the appropriate primary antibody diluted in blocking solution. After washes with PBS, primary antibodies were detected with HRP-conjugated rabbit anti-mouse (1:2,000), rabbit anti-goat antibodies (1:2,000), and swine anti-rabbit (1:2,000) immunoglobulins. Peroxidase activity was detected with the enhanced chemiluminescence system.
NHS.
Normal human serum (NHS) was obtained from healthy human donors who were devoid of anti-Yersinia antibodies as described previously (7).
Binding of 125I-labeled FH to whole bacteria.
A 125I-labeled FH-binding assay was performed as described previously (18, 21, 22). Briefly, bacteria grown to exponential or stationary phase were washed in Veronal-buffered saline (VBS; 142 mM NaCl, 1.8 mM sodium barbital, 3.3 mM barbituric acid, pH 7.4) and suspended in VBS containing 0.1% gelatin (GVB). Bacteria were adjusted to 1010 CFU/ml in 1/3-diluted GVB and incubated for 30 min at 37°C with 125I-labeled FH (ca. 46,000 cpm) in a total volume of 40 μl of 1/3 GVB. After incubation, the mixtures were centrifuged (10,000 × g, 3 min) through 20% sucrose in 1/3 VBS containing 0.1% gelatin (1/3 GVB). Tubes were frozen at −70°C, the pellets were cut off, and the radioactivity in the pellets and supernatants was measured in a gamma counter. The binding of FH was calculated as a percentage of the total radioactivity input.
Binding of nonlabeled or I-labeled FH to whole bacteria.
Bacteria (3 × 109) grown to the exponential phase were washed three times in PBS (binding assay with nonlabeled FH) or VBS (binding assay with I-labeled FH) and then incubated with 6 μg of nonlabeled or I-labeled FH in a total volume of 50 μl of PBS (nonlabeled FH) or in (i) VBS, (ii) GVB, or (iii) VBS supplemented with 100 μg/ml collagen type I (I-labeled FH). This gives a final FH concentration of 120 μg/ml, i.e., below the physiological concentration (500 μg/ml). The incubation was performed at 37°C for 30 min with vigorous shaking (850 rpm). Ca. 108 PBS- or VBS-washed bacteria were suspended in 15 μl of the washing buffer, and 3 μl of 5× reducing Laemmli buffer was added. After heating, 10 μl of the samples was subjected to immunoblotting.
Analysis of the binding of serum FH to bacteria by immunoblotting and ELISA.
Bacteria were grown to the exponential phase as described above and washed three times in PBS. Bacteria (3 × 109) were incubated in 100 μl of 50% heat-inactivated serum (HIS; 7) for 30 min at 37°C with vigorous shaking (850 rpm), followed by three washes with PBS. Bacteria were resuspended in PBS, and the volume was adjusted on the basis of the optical density at 600 nm (OD600) so that ca. 108 bacteria (in 60 μl) were transferred to enzyme-linked immunosorbent assay (ELISA) plate wells (Nunc PolySorp), which were allowed to dry overnight at 37°C. At the same time, 5 × 107 bacteria (30 μl) were mixed 1:1 with reducing Laemmli buffer and subjected to 8% SDS-PAGE and immunoblotting with polyclonal goat anti-FH antiserum (1:2,000) as the primary antibody. Quantitative analysis of FH bands was performed with the Typhoon 9400 ImageQuant analyzer (Amersham Biosciences, Piscataway, NJ).
The ELISA plate wells were washed three times with PBS, blocked with 100 μl of 5% skim milk in PBS for 2 h at 37°C, washed again three times with PBS, and incubated with preadsorbed polyclonal goat anti-FH antibodies diluted 1:7,500 in 5% skim milk for 1 h at room temperature (RT). After three washes with PBS, wells were incubated with HRP-conjugated rabbit anti-goat antibodies (1:5,000) for 1 h at RT. Wells were washed again three times with PBS, and 1,2-phenylenediamine dihydrochloride substrate (Dako S2045) was added. After a 15-min incubation, the reaction was stopped with 0.5 M H2SO4 and the OD492 was determined. Background OD492 values (obtained with the wells not containing bacteria but treated the same way as the bacterium-containing wells) were subtracted from all of the binding values. The average OD492 background value was 0.050, with a range of 0.045 to 0.060. Experiments were repeated three times in duplicate. In a few instances where the duplicate OD492 values differed from each other by more than twofold, both values were excluded from the data.
Bacteriophage sensitivity assay.
Bacteriophage φR8-01 (Table 1) was isolated from sewage as described previously (53) with strain YeO3-c-trs8R as the host strain. Bacteria were grown to the logarithmic phase in MedECa at RT. A 100-μl aliquot of the bacterial cultures was mixed with 3 ml of melted 50°C soft agar (0.4%). The mixtures were poured onto agar plates, and 10-μl drops of serially diluted bacteriophages φR8-01 and φR1-37 (from 103 to 107 PFU/ml) were spotted onto the soft agar. Plates were incubated overnight at RT, and the sensitivity of the strain to the phages was indicated by the clear lysis zone in the bacterial growth in the soft agar.
Purification of YadA with Triton X-114 (Tx-114).
The extracts Tx-YadA (from Escherichia coli JM103/pYMS4450) and Tx-Ctrl (the vector control extract from E. coli strain JM103/pL2.1) were prepared as described previously (50), with slight modifications. Briefly, bacteria were grown overnight at 37°C in 400 ml of Luria broth supplemented with ampicillin. Bacteria were centrifuged (3,000 × g, 15 min) and incubated on ice for 1 h in 20 ml of lysis buffer (10 mM EDTA, 50 mM glucose, 25 mM Tris-HCl [pH 8.0], 5 mg/ml lysozyme). Tx-114, prepared as described previously, was added to the lysate to a final concentration of 5%. Extraction was carried out by incubating the mixture at 4°C for 24 h with slow rocking. Subsequently, the mixture was incubated overnight at 37°C to separate the water and Tx-114 phases, followed by centrifugation (4,000 × g, 10 min) to clear the phases. The Tx-114 phase was recovered and stored at 4°C.
FH affinity blotting.
Tx-YadA and Tx-Ctrl (see above) were subjected to 8% SDS-PAGE. Proteins were electrotransferred onto nitrocellulose membrane. The fragment of the membrane containing the proteins with molecular masses greater than 160 kDa was cut out and blocked with 5% skim milk. Subsequently, the membrane was incubated with the purified FH (50 μg/ml). Binding of the protein was detected by immunoblotting with goat anti-human FH.
Cofactor assay for C3b inactivation.
The cofactor assay was performed to analyze FI-mediated cleavage of C3b to iC3b. Bacteria (4 × 109) were incubated with FH (final concentration, 30 μg/ml) in 40 μl of 1/3 PBS for 30 min at 37°C with shaking. After washing three times with 1/3 PBS, the bacterial concentration of the suspension was estimated by measuring the OD600. About 109 bacteria were pelleted and resuspended in 30 μl of PBS containing FI (final concentration, 20 μg/ml) and C3b (final concentration, 35 μg/ml). The reaction mixtures were incubated for 45 min at 37°C with shaking. A reaction mixture in which C3b was incubated for 45 min at 37°C with 20 μg/ml FH and FI was used as a positive control. In addition, negative controls comprising C3b incubated without FI, FH, or both were included. After incubation, the samples were centrifuged and supernatants were collected, mixed with reducing Laemmli buffer, and subjected to 10% SDS-PAGE and immunoblotting with rabbit anti-human C3c and rabbit anti-human C3d (1:2,000) as the primary antibodies. To control FH binding to the bacteria, the bacterial pellets were resuspended in 20 μl of 1/3 PBS, mixed with reducing Laemmli buffer, and subjected to 8% SDS-PAGE and immunoblotting with nonadsorbed goat antiserum against human FH (1:2,000).
Binding of truncated recombinant FH constructs to bacteria.
Bacteria were grown to log phase and washed three times with 1/3 VBS. The bacteria (1.8 × 109) were incubated in 200 μl of 1/3 VBS alone or containing SCR 1-5, 1-6, 1-7, 8-11, 11-15, or 8-20 in equimolar (0.3 μM) concentrations for 30 min at 37°C with shaking (850 rpm). Following incubation, bacteria were washed three times with 1/3 VBS and bacterial pellets were suspended in 30 μl of 1/3 VBS and mixed with Laemmli buffer. The samples were subjected to 12% SDS-PAGE, and the truncated FH constructs were detected by immunoblotting with goat antiserum against human FH as described above. We used control blotting to confirm that all of the recombinant FH constructs were recognized by the goat anti-FH antiserum.
Influence of bovine serum albumin (BSA), NaCl, and heparin on FH binding.
Bacteria (4 × 109) grown to mid-logarithmic phase were incubated with purified FH (final concentration of 30 μg/ml) in 1/3 PBS supplemented with BSA (0 to 1,000 μg/ml), heparin (0 to 1,000 μg/ml), or NaCl (50 to 650 mM) for 30 min at 37°C. After incubation, bacteria were washed three times with 1/3 PBS. Bacteria (5 × 108 in 30 μl of 1/3 PBS) were mixed with Laemmli buffer. The samples were subjected to 8% SDS-PAGE and immunoblotting with nonadsorbed goat antiserum against human FH. Quantitative analysis of FH binding was performed with the NIH ImageJ software. The intensities of the FH protein bands were quantitated, and the influence of loading differences was eliminated by relating the values to the intensity of a nonspecifically reacting bacterial protein band in the same lane.
Statistical methods.
Statistical analyses were performed with the two-sample t test; P < 0.05 was considered statistically significant.
RESULTS
Identification of FH receptor(s) on Y. enterocolitica O:3.
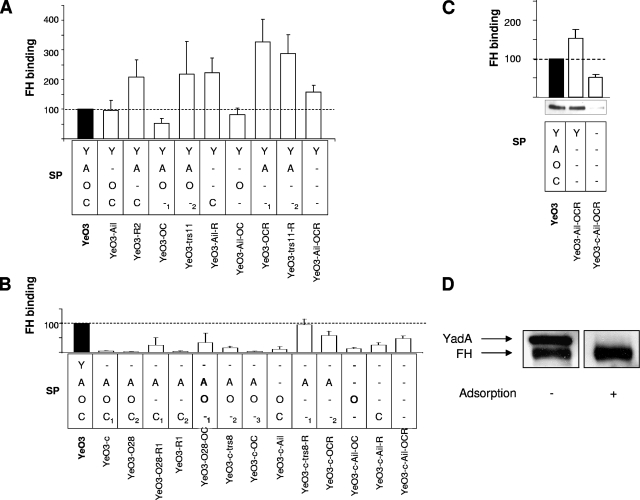
To find a bacterial receptor responsible for FH binding, a set of 23 Y. enterocolitica O:3 strains, expressing YadA, Ail, OAg, and OC in all possible combinations was used (7). In a previous work, we demonstrated that the level of expression of these factors by the mutant strains was not affected by the type or number of mutations introduced (7). For clarity, we have given the strains a letter code for the expressed surface phenotypes (SPs) of the strains (Table 1) as follows: YadA, Y; Ail, A; OAg, O; OC, C. Thus, SP YAOC means that all four factors are expressed and SP Y--C means that YadA and OC are expressed and Ail and OAg are not. Identical SP codes of different strains are differentiated by subscript arabic numerals (Table 1). This set of strains was tested for the ability to bind FH from 50% HIS both by ELISA (Fig. 1) and by immunoblotting (Fig. 1 and data not shown) with goat antiserum against human FH for detection. In immunoblotting, we noticed that the antiserum contained YadA-specific antibodies; however, these were efficiently removed by adsorption with YadA-expressing bacteria (Fig. 1D). Unless otherwise indicated, the preadsorbed antiserum was used for FH detection in all of the experiments.
FIG. 1.
FH binding to Y. enterocolitica O:3. (A and B) FH binding to YadA-positive and -negative strains, respectively, from 50% HIS was quantitated by ELISA. The wild-type strain FH-binding level was set to 100% (black bars), and those of the mutant strains are expressed relative to it. The data were obtained from three independent experiments. Error bars represent standard deviations. The factors expressed by the strains tested are marked as follows: YadA, Y; Ail, A; OAg, O; OC, C; the strains of the same phenotype obtained by different genetic methods are indicated by the subscript numbers 1, 2, and 3 (see Table 1 for details). The average OD492 value for wild-type strain YeO3 was 0.74 ± 0.26 (range, 0.35 to 1.36). (C) Comparison of the ELISA and immunoblotting analyses of FH binding to selected Y. enterocolitica O:3 strains. (D) Presence of cross-reacting anti-YadA antibodies in goat antiserum against human FH (Quidel). Detection of FH bound to wild-type YeO3 bacteria by immunoblotting with the antiserum before and after adsorption with YadA-expressing bacteria is shown.
FH ELISA.
The results of our FH-binding analysis by ELISA are shown in Fig. 1. In general, all strains expressing YadA were able to bind serum FH (Fig. 1A, 53 to 326% of the wild-type level) while much less FH binding to most other strains occurred (Fig. 1B, 2 to 47% of the wild-type level). The difference in FH binding between these two groups was statistically significant (P < 0.0001).
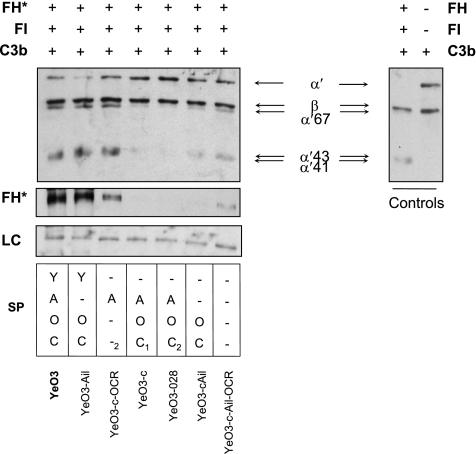
FIG. 2.
The cofactor activity of FH bound to the bacterial surface. In the left and middle panels, bacteria were preincubated with FH (final concentration, 30 μg/ml), washed extensively, and exposed to FI and C3b. Bacterial surface-bound FH is indicated by the asterisk, while in the control panel at the right, reactions were carried out without bacteria. C3b and its cleavage products were detected in the supernatants with a mixture of polyclonal antibodies against C3c and C3d. Inactivation of C3b is demonstrated by the reduced intensity of the C3b α′ chain and the appearance of the α′ chain cleavage fragments of 67, 43, and 41 kDa. FH bound to bacteria was detected in pellets by immunoblotting with polyclonal antiserum against human FH. A nonspecific bacterial protein band recognized by the anti-FH antiserum is shown as a loading control (LC). The factors expressed by the tested strains are marked as in Fig. 1.
As demonstrated in Fig. 1A, expression of YadA in the absence of OAg resulted in increased binding (SP YA-C, SP Y--C, SP Y--1, SP Y--2, and SP Y---), perhaps due to better exposure of YadA on the bacterial surface. Interestingly, most of the YadA-expressing strains bound as much FH as their Ail-lacking counterparts (Fig. 1A, compare SP YAOC versus SP Y-OC, SP YA-C versus SP Y--C, and SP YAO-1 versus SP Y-O-), suggesting that YadA is the major FH-binding OM protein. The contribution of Ail to FH binding when YadA was expressed could only be observed when both OAg and OC were not produced. Thus, SP YA-- strains YeO3-OCR and YeO3-trs11-R, missing both OAg and OC, bound significantly more FH (P < 0.029 and P < 0.001, respectively) than the SP Y--- strain, the Ail-negative derivative YeO3-Ail-OCR (Fig. 1A). It is worth noting that the latter strain, expressing only YadA, was still able to bind more FH than the wild type (P < 0.001). Thus, in the absence of OAg, YadA is likely to be better exposed and this could favor FH binding. On the contrary, lack of OC in YadA- and OAg-expressing SP YAO-1 and SP Y-O- strains (YeO3-OC and YeO3-Ail-OC) did not greatly affect the bacterial capacity to bind FH. Interestingly, SP YAO-2 strain YeO3-trs11, which, as shown earlier (7), expresses less OAg than its phenotypic counterpart SP YAO-1 strain YeO3-OC, bound significantly more FH (P < 0.0046). This further supports the hypothesis that in the absence of the OAg blocking effect, FH binding to better-exposed YadA would be facilitated.
LPS blocking effect.
To support the conclusion of the OAg blocking effect on YadA, we examined whether OAg could block the access of YadA-specific MAbs 2A9, 3G12, and 2G12 to their epitopes. These MAbs recognize epitopes within the neck and the very N-terminal region of the YadA stalk (M. Biedzka-Sarek et al., submitted for publication). The concentrations of the MAbs used were adjusted so that most of the MAb was adsorbed by YadA-expressing bacteria under the adsorption conditions. The MAbs were incubated with the YadA-positive strains expressing or lacking OAg (SP YAOC and SP YA-C), and their YadA-negative counterparts (SP -AOC and SP -A-C) were used as negative controls. Following incubation, bacteria were centrifuged and the remaining amount of MAbs in the supernatants was semiquantitated by dot blotting from 1:2 dilution series. Although differences in the adsorption of MAbs between the YadA-expressing and YadA-negative strains were clear, no difference between the YadA-positive strains expressing or lacking OAg could be observed (data not shown). These results indicated that OAg did not block the MAb epitopes located in the N-terminal end of the stalk; however, the possibility remains that it could block the YadA regions that are involved in FH binding and located closer to the C terminus and thus the OM (Biedzka-Sarek et al., submitted).
Both YadA and Ail bind FH.
The YadA-negative mutants also provided evidence for the involvement of YadA in FH binding (Fig. 1B). Almost all YadA-negative strains bound less FH than the wild-type strain. The exceptions were SP -A--1,2 strains YeO3-c-trs8-R and YeO3-c-OCR, which express Ail in the absence of both OAg and OC (Fig. 1B). These strains bound amounts of FH comparable to that bound by the wild-type strain. These observations suggest that to bind FH, Ail needs to be very well exposed on the OM since neither the removal of OAg in the SP -A-C1,2 strains (YeO3-O28-R and YeO3-R1) nor that of OC in the SP -AO-1-3 strains (YeO3-O28-OC, YeO3-c-trs8, and YeO3-c-OC) was enough to facilitate Ail-mediated FH binding (Fig. 1B). To demonstrate the masking potential of LPS, we compared the sensitivities of different Y. enterocolitica O:3 strains, grown at RT, to bacteriophages φR1-37 and φR8-01 (Table 2). The results obtained supported the LPS masking hypothesis. φR1-37 could reach its OC receptor only in the absence of blocking OAg, and φR8-01 infection took place exclusively when both OAg and OC were not expressed (Table 2). This suggests that the still unknown receptor of φR8-01 does not protrude far from the OM and is efficiently masked by both OC and OAg. While φR1-37 infectivity was not affected by the growth temperature of the host bacteria, bacteriophage φR8-01 produced almost no plaques on bacteria grown at 37°C (data not shown). Thus, the φR8-01 receptor is either strongly downregulated at 37°C or masked by surface structures expressed under these conditions (other that OAg or OC).
TABLE 2.
Characterization of the LPS masking effect by analysis of the phage sensitivities of bacteria grown at 22°C
| Strain | SP (22°C) | φR8-01 | φR1-37 |
|---|---|---|---|
| YeO3 | --OC | − | − |
| YeO3-c | --OC | − | − |
| YeO3-Ail | --OC | − | − |
| YeO3-c-Ail | --OC | − | − |
| YeO3-OC | --O- | − | − |
| YeO3-c-OC | --O- | − | − |
| YeO3-Ail-OC | --O- | − | − |
| YeO3-c-Ail-OC | --O- | − | − |
| YeO3-R2 | ---C | − | + |
| YeO3-R1 | ---C | − | + |
| YeO3-Ail-R | ---C | − | + |
| YeO3-c-Ail-R | ---C | − | + |
| YeO3-OCR | ---- | + | − |
| YeO3-c-OCR | ---- | + | − |
| YeO3-Ail-OCR | ---- | + | − |
| YeO3-c-Ail-OCR | ---- | + | − |
In general, the immunoblotting and ELISA results correlated well (Fig. 1 and data not shown). Only YeO3-c-Ail-OCR, YeO3-trs11, and YeO3-Ail-R showed relatively higher FH binding in ELISA than in immunoblotting. We can only speculate that this may be due to some nonspecific reactivity in ELISA.
Collectively, these results demonstrated that Y. enterocolitica could acquire FH from human serum and that YadA was the main FH receptor on the bacterial surface. In addition to YadA, Ail was able to bind FH but solely when not masked by LPS OAg and OC.
FH bound to Y. enterocolitica is functional.
FH functions in the negative regulation circuit of AP activation. Specifically, it acts as a cofactor in FI-mediated progressive cleavage of the C3b α′ chain into several fragments (67, 43, 41, and 30 kDa). With the cofactor assay, we aimed to examine whether YadA- and Ail-bound FH remains functionally active. Y. enterocolitica bacteria were incubated with purified FH, and after intensive washes, bacteria were incubated with purified FI and C3b. Following incubation, the samples were centrifuged and supernatants were analyzed for C3b cleavage with the mixture of anti-C3c and anti-C3d antibodies that recognizes both the intact C3b α′ chain and its cleavage products while the lysed pellets were examined for FH deposition on bacteria with goat anti-FH antiserum (Fig. 2).
Similar to FH binding from HIS, purified FH could be detected on YadA-expressing bacteria while Ail bound FH only in the absence of LPS (Fig. 1 and 2). Bacteria that expressed Ail in the presence of LPS or were YadA and Ail negative (SP --OC and SP ---) bound negligible amounts of FH. This further indicated that neither OAg nor OC directly binds FH. On the other hand, we could also see slightly more binding of purified FH to YeO3-c-Ail-OCR (SP ---) than to YeO3-c-Ail or YeO3-028 (SP --OC and SP -AOC2, respectively; Fig. 2). This could also mean that the loss of the LPS surface structures might have generated or exposed novel FH-binding specificities such as the LPS inner core or small OM proteins.
The ability of Y. enterocolitica strains to cleave C3b in the presence of FI was reflected in their ability to bind FH. FH bound to the wild-type (SP YAOC), SP Y-OC, and SP -A--2 bacteria retained its cofactor activity, as shown by the cleavage of the C3b α′ chain into 67-, 43-, and 41-kDa fragments (Fig. 2, left panel; note the reduction of the intact α′ band intensity and the simultaneous appearance of the α′ cleavage fragments). No or very modest cofactor activity was observed with SP -AOC1, SP -AOC2, SP --OC, and SP --- bacteria displaying no or scarce FH on their surface.
These results showed that YadA- and Ail-bound FH functions as an FI cofactor for C3b cleavage.
Purified FH binds directly to YadA.
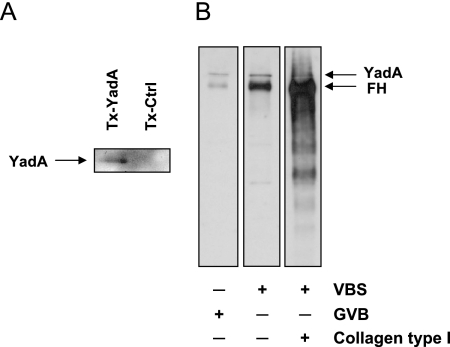
As the analyses described above suggested that Y. enterocolitica binds purified FH via both YadA and Ail, we wanted to further show that the interaction between YadA, the main FH receptor on Y. enterocolitica, and FH is direct. To this end, Tx-114 membrane protein extracts from E. coli expressing YadA (Tx-YadA) and E. coli carrying the empty vector (Tx-Ctrl) were prepared. The extracts were subjected to SDS-PAGE and transferred onto nitrocellulose membrane. The fragment of the membrane containing the proteins with molecular masses greater than 160 kDa was cut out and incubated with purified FH. As shown in Fig. 3A, binding to Tx-YadA, but not to Tx-Ctrl, was detected.
FIG. 3.
Binding of FH to YadA. (A) Binding of purified FH to YadA shown by affinity blotting. Triton X-114 membrane protein extracts from E. coli expressing YadA (Tx-YadA) or from E. coli carrying the empty vector (Tx-Ctrl) were subjected to 8% SDS-PAGE and transferred onto nitrocellulose membrane. The part of the membrane containing the proteins with molecular masses greater than 160 kDa was incubated with purified FH. Binding of FH was detected by immunoblotting with preadsorbed goat anti-human FH and HRP-conjugated rabbit anti-goat antibodies. Binding of FH to the YadA band is indicated by the arrow. (B) Divergent effects of gelatin and collagen on FH binding to YadA. Binding of purified FH to YadA-expressing wild-type Y. enterocolitica bacteria was tested in VBS (gelatin free), GVB (contains 0.1% gelatin), or VBS supplemented with type I collagen (0.01%). After extensive washings, bound FH was identified by immunoblotting with nonadsorbed polyclonal antiserum against human FH. The use of nonadsorbed serum allowed the simultaneous detection of the YadA band (indicated by the arrow), which served as a loading control.
Since demonstration of direct binding of purified FH to Y. enterocolitica with 125I-labeled FH was not successful earlier (46), we also repeated 125I-labeled FH-binding experiments by following a protocol successfully applied to streptococci and Borrelia (18, 21, 22). These experiments, however, failed to demonstrate any FH binding (data not shown). To verify whether 125I labeling affected FH tyrosines involved in the interaction with Y. enterocolitica, the binding experiment was repeated with FH labeled with nonradioactive I. I-labeled FH, however, was acquired by Y. enterocolitica O:3 as efficiently as the nonlabeled regulator (data not shown). We then examined the effect of the buffers commonly used in the 125I-labeled FH-binding assay, such as VBS and GVB. In the latter, gelatin is used to nonspecifically prevent bacterial aggregation. With YadA-expressing Y. enterocolitica O:3, however, the effect was quite the opposite and strong aggregation of bacteria was observed. This resulted in a significant reduction of the binding of nonlabeled FH (Fig. 3B). Gelatin is a denatured form of collagen and has been shown to inhibit 50% of YadA-mediated collagen binding (13). Thus, the lack of 125I-labeled FH binding to Y. enterocolitica O:3 in GVB could result from the blocking of FH-specific sites on YadA by gelatin and/or by the strong gelatin-induced aggregation. Interestingly, when the gelatin in the buffer was replaced with collagen, a significant increase in the amount of Y. enterocolitica O:3-bound FH was observed. Our preliminary studies show that collagen per se is able to bind FH (Fig. 3B). Thus, by binding to collagen, Y. enterocolitica could also indirectly acquire FH, thereby increasing its chances to avoid complement attack. More studies elucidating the role of collagen as a shield protecting Y. enterocolitica against complement attack are warranted.
Locations of YadA- and Ail-binding sites on FH.
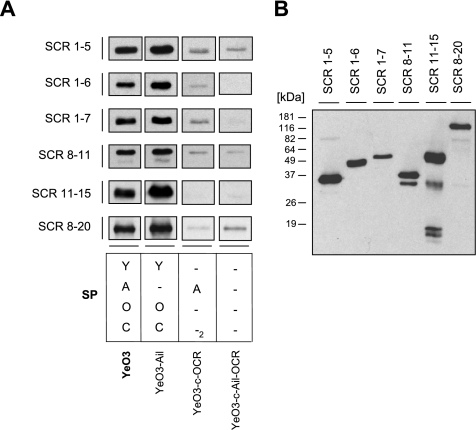
To identify the FH region(s) involved in binding to Y. enterocolitica O:3, we examined the binding of truncated recombinant FH fragments representing SCRs 1-5, 1-6, 1-7, 8-11, 11-15, and 8-20 to wild-type (SP YAOC), SP Y-OC, SP -A--2, and SP --- bacteria.
Bacteria were incubated with truncated fragments of FH and washed, and whole-cell lysates were subjected to SDS-PAGE and immunoblotting with a goat anti-FH antiserum that recognizes all of the FH fragments tested (Fig. 4B). As shown in Fig. 4A, all of the FH fragments bound to wild-type bacteria and the YadA-expressing strain of SP Y-OC. The Ail-expressing strain, SP -A--2, on the other hand, specifically bound only SCRs 1-6 and 1-7. Ail binding to SCRs 1-5, 8-11, and 8-20 was at the level of the negative control SP --- strain and thus was not considered specific (Fig. 4A).
FIG. 4.
(A) Mapping of the binding region on FH for YadA and Ail of Y. enterocolitica O:3. Strain YeO3 (SP YAOC), YeO3-Ail (SP Y-OC), YeO3-c-OCR (SP -A--2), and YeO3-c-Ail-OCR (SP ---) bacteria were incubated with truncated recombinant FH constructs representing SCRs 1-5, 1-6, 1-7, 8-11, 11-15, and 8-20. Following incubation, the bacteria were washed and whole-cell lysates were run in 12% SDS-PAGE and analyzed by immunoblotting with goat anti-FH and HRP-conjugated rabbit anti-goat antibodies. The factors expressed by the strains tested are marked as in Fig. 1. (B) Detection of recombinant FH constructs with goat anti-FH antiserum and HRP-conjugated rabbit anti-goat antibodies.
On the basis of these results, we conclude that the binding site for Ail on FH is located within SCRs 6 and 7 while YadA appears to bind throughout the entire FH.
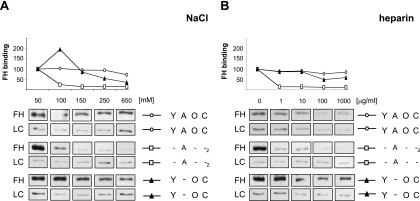
Effects of salt and heparin on FH binding to Y. enterocolitica.
The nature of FH binding to YadA and Ail was examined by using the wild-type strain and strains expressing either Ail (the SP -A--2 strain) or YadA (the SP Y-OC strain). Bacteria were incubated with purified FH in 1/3 PBS or 1/3 PBS supplemented with NaCl to create a salt concentration gradient ranging from 50 to 650 mM. After incubation with FH, the bacteria were washed, whole-cell lysates were run in SDS-PAGE, and the bound FH was detected with goat antiserum against human FH. The results showed that FH binding to Ail by the SP -A--2 strain was maximal at the lowest salt concentration and decreased dramatically with increasing salt concentrations (Fig. 5A). To the contrary, YadA-mediated FH binding was more salt resistant and only a twofold decrease in FH binding to the SP Y-OC strain was observed at an NaCl concentration of 250 mM (Fig. 5A). FH binding to the wild-type bacteria under these conditions was not affected and dropped slightly only at an NaCl concentration of 650 mM (Fig. 5A).
FIG. 5.
Characterization of FH interaction with Y. enterocolitica. The effects of NaCl (A) and heparin (B) on the binding of purified FH to Y. enterocolitica O:3 strains YeO3 (SP YAOC), YeO3-Ail (SP Y-OC), YeO3-c-OCR (SP -A--2), and YeO3-c-Ail-OCR (SP ---) are shown. Bacteria were incubated with FH (final concentration, 30 μg/ml) in 1/3 PBS with various amounts of NaCl (100 to 650 mM) or heparin (0 to 1,000 μg/ml). Following incubation, the bacteria were washed and whole-cell lysates were subjected to 8% SDS-PAGE and immunoblotting with the nonadsorbed goat antiserum against human FH. Quantitative analysis of FH binding was performed with the NIH ImageJ software. The intensities of the FH protein bands were quantitated, and the influence of loading differences was eliminated by relating the values to the intensity of a nonspecifically reacting bacterial protein band in the same lane (loading control [LC]). The FH-binding level of the bacteria incubated in 1/3 PBS alone was set to 100, and the FH-binding level of the bacteria in 1/3 PBS supplemented with NaCl (100 to 650 mM) and heparin (1 to 1,000 μg/ml) was expressed relative to the FH-binding level of the bacteria in 1/3 PBS alone. The factors expressed by the strains tested are marked as in Fig. 1.
Heparin interaction sites for FH have been mapped to SCRs 7, 13, and 20 (8, 9, 40). We tested whether heparin inhibits the binding of FH to YadA- or Ail-expressing bacteria (Fig. 5B). Bacteria were incubated with purified FH in the presence of heparin (0 to 1,000 μg/ml) in 1/3 PBS, and the bound FH was detected by immunoblotting as described above. Heparin efficiently inhibited FH binding to the SP -A--2 strain, such that even at the lowest heparin concentration used, 1 μg/ml, FH binding was almost completely abolished (Fig. 5B). This indicates that the binding sites for heparin and Ail on FH are likely to be identical or at least overlap. This also corroborates our above observation that the Ail-binding site on FH resides on SCRs 6 and 7. The YadA-mediated FH binding determined with the SP Y-OC strain was only affected at a heparin concentration of 100 μg/ml and did not drop further at 1,000 μg/ml, the highest concentration of heparin used. This finding is supported by the ability of YadA to bind all of the truncated FH fragments (Fig. 4A). As wild-type bacteria express both YadA and Ail but the latter is likely to be blocked by the LPS, binding of FH occurs mainly via YadA. Thus, similar to the SP Y-OC strain, FH binding to the wild-type bacteria was slightly reduced only at the highest heparin concentrations used, 100 to 1,000 μg/ml (Fig. 5B).
FH binding to all of the three strains tested was affected only by the highest BSA concentration used (1,000 μg/ml, 25% of control binding; data not shown), indicating that YadA- and Ail-mediated binding to FH is specific.
In conclusion, the FH-Ail interaction appears to depend on ionic interactions between the proteins and Ail shares the binding site on FH with heparin. On the contrary, the YadA-FH interaction is more salt and heparin resistant.
DISCUSSION
The binding of the AP regulator FH to Y. enterocolitica serotype O:3 was characterized in this work. We showed that Y. enterocolitica is able to recruit FH both from serum and in a purified form, suggesting that the Y. enterocolitica-FH interaction is direct and independent of other serum proteins. Relevant to the importance of this interaction in bacterial pathogenesis, we also demonstrated that FH bound to bacteria retained its biological function and acted as a cofactor for FI-mediated cleavage of C3b.
To identify FH receptors on Y. enterocolitica, we examined the FH-binding potential of strains expressing different combinations of the putative serum resistance determinants YadA, Ail, OAg, and OC. In general, FH binding to these strains (Fig. 1A and B) correlated well with their serum sensitivity examined in the previous study (7). We demonstrated that both main serum resistance factors of Y. enterocolitica O:3, YadA and Ail, bind FH. YadA was shown to be the major FH binder, while Ail contributed to FH binding only when not blocked by OAg and OC. Both YadA- and Ail-bound FH displayed a cofactor activity.
China et al. identified YadA as an FH receptor based on an affinity blotting experiment by incubating membrane-bound YadA with NHS, and FH binding to the YadA band was detected with anti-FH antibodies (12). In this work, we demonstrated a direct interaction between YadA and FH with an analogous affinity blotting experiment; however, instead of whole serum, purified FH was used. Binding of FH to the YadA band was detected (Fig. 3), suggesting that the YadA-FH interaction is direct and does not require other complement proteins or serum factors. Earlier attempts to show a direct interaction between YadA and purified radiolabeled FH had failed (46). Here we provided evidence that the failure could result from the presence of gelatin in the assay buffer (Fig. 3B). To further alleviate the biological significance of the YadA-mediated FH binding, we showed that YadA-expressing bacteria could use FH to efficiently promote FI-mediated cleavage of C3b, the key protein of complement activation. This is in accordance with studies showing that more C3b deposits on bacteria lacking YadA (12, 56).
Finally, we showed that YadA bound truncated recombinant FH fragments representing the entire FH and that the binding was not affected by heparin (Fig. 4 and 5). Such interaction with all parts of FH has not been reported earlier for any other microbes. In general, the C terminus of FH has been identified as the favorite site targeted by microbes, e.g., B. burgdorferi, N. gonorrhoeae, and Candida albicans (18, 33, 45). The central part of FH was shown to be a target for Streptococcus pneumoniae (22), while S. pyogenes and B. burgdorferi were shown to engage the N-terminal portion of FH, SCR 7 (25, 26, 39). SCR fragment 1-5 contains the domain responsible for the regulatory functions of FH, i.e., the cofactor and decay-accelerating activities (15, 28, 30). As YadA-bound FH acted as a cofactor for FI-mediated cleavage of C3b, the SCR 1-5 interaction apparently did not interfere with these regulatory functions of FH.
Ail-mediated serum resistance has been evident only when YadA has been deleted from the bacteria or when Ail has been expressed in E. coli (10). The molecular mechanism of Ail-mediated resistance has been unknown. In this study, we have shown that Ail, in addition to YadA, is able to recruit FH when well surface exposed. Heparin efficiently blocked FH binding to Ail, and that supported the finding that the binding domain of Ail on FH is located within SCRs 6 and 7. In addition, we showed that the Ail-FH interaction is ionic in nature and that Ail-bound FH retains an FI cofactor activity (Fig. 3 and 5).
LPS OAg and OC alone are not able to protect Y. enterocolitica O:3 bacteria against complement-mediated killing (7). In this study, we further showed that they do not participate in the recruitment of FH onto the Yersinia surface. Our results clearly showed that FH binds primarily through YadA, regardless of OAg and OC expression. The absence of OAg, however, potentiated the FH-binding capacity of YadA-expressing bacteria. Moreover, removal of both OAg and OC most likely unveils Ail, the second FH receptor that was able to bind FH solely in the absence of these distal LPS parts. The blocking potential of OAg and OC was clearly demonstrated by the phage sensitivity experiments (Table 2). The expression of OAg and OC is reduced in bacteria grown in vitro at 37°C (1, 2, 38), and this is also reflected in phage sensitivity data (23, 53). How bacteria regulate LPS expression during infection in the host is not known. One cannot exclude, however, the possibility that, at a certain stage(s) of infection, Ail, due to suppressed expression of the LPS distal parts, is unveiled and functions as an FH receptor.
Conclusions.
In conclusion, we have demonstrated that YadA and Ail mediate FH binding to Y. enterocolitica O:3 but the latter only when well surface exposed. We located the Ail-binding sites on FH to SCRs 6 and 7 and showed that YadA binds to the entire length of FH. As both YadA- and Ail-bound FH functions as a cofactor for FI, it is very likely that it protects Y. enterocolitica O:3 against AP-mediated killing.
Acknowledgments
This work has been supported by the Academy of Finland (projects 114075, 201358, and 104361 to M.S.) and the European Union Network of Excellence EuroPathoGenomics (contract LSHB-CT-2005-512061). M. Biedzka-Sarek has been supported by the University of Helsinki Graduate School in Biotechnology and Molecular Biology.
We are grateful to Nathalie Friberg and Hanna Tossavainen for the purification of the truncated FH fragments and Heli Mönttinen, Laura Kalin, and Chun-Mei Li for excellent technical assistance.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 14 July 2008.
REFERENCES
- 1.al-Hendy, A., P. Toivanen, and M. Skurnik. 1991. The effect of growth temperature on the biosynthesis of Yersinia enterocolitica O:3 lipopolysaccharide: temperature regulates the transcription of the rfb but not of the rfa region. Microb. Pathog. 1081-86. [DOI] [PubMed] [Google Scholar]
- 2.al-Hendy, A., P. Toivanen, and M. Skurnik. 1991. Expression cloning of Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb. Pathog. 1047-59. [DOI] [PubMed] [Google Scholar]
- 3.al-Hendy, A., P. Toivanen, and M. Skurnik. 1992. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect. Immun. 60870-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Areschoug, T., M. Stalhammar-Carlemalm, I. Karlsson, and G. Lindahl. 2002. Streptococcal beta protein has separate binding sites for human factor H and IgA-Fc. J. Biol. Chem. 27712642-12648. [DOI] [PubMed] [Google Scholar]
- 5.Barondess, J. J., and J. Beckwith. 1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346871-874. [DOI] [PubMed] [Google Scholar]
- 6.Beer, K. B., and V. L. Miller. 1992. Amino acid substitutions in naturally occurring variants of Ail result in altered invasion activity. J. Bacteriol. 1741360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biedzka-Sarek, M., R. Venho, and M. Skurnik. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 732232-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackmore, T. K., J. Hellwage, T. A. Sadlon, N. Higgs, P. F. Zipfel, H. M. Ward, and D. L. Gordon. 1998. Identification of the second heparin-binding domain in human complement factor H. J. Immunol. 1603342-3348. [PubMed] [Google Scholar]
- 9.Blackmore, T. K., T. A. Sadlon, H. M. Ward, D. M. Lublin, and D. L. Gordon. 1996. Identification of a heparin binding domain in the seventh short consensus repeat of complement factor H. J. Immunol. 1575422-5427. [PubMed] [Google Scholar]
- 10.Bliska, J. B., and S. Falkow. 1992. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc. Natl. Acad. Sci. USA 893561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.China, B., M. P. Sory, B. T. N′Guyen, M. De Bruyere, and G. R. Cornelis. 1993. Role of the YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect. Immun. 613129-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emödy, L., J. Heesemann, H. Wolf-Watz, M. Skurnik, G. Kapperud, P. O'Toole, and T. Wadström. 1989. Binding to collagen by Yersinia enterocolitica and Yersinia pseudotuberculosis: evidence for yopA-mediated and chromosomally encoded mechanisms. J. Bacteriol. 1716674-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gemski, P., J. R. Lazere, and T. Casey. 1980. Plasmid associated with pathogenicity and calcium dependency of Yersinia enterocolitica. Infect. Immun. 27682-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon, D. L., R. M. Kaufman, T. K. Blackmore, J. Kwong, and D. M. Lublin. 1995. Identification of complement regulatory domains in human factor H. J. Immunol. 155348-356. [PubMed] [Google Scholar]
- 16.Heffernan, E. J., J. Harwood, J. Fierer, and D. Guiney. 1992. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J. Bacteriol. 17484-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heffernan, E. J., L. Wu, J. Louie, S. Okamoto, J. Fierer, and D. G. Guiney. 1994. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect. Immun. 625183-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellwage, J., T. Meri, T. Heikkilä, A. Alitalo, J. Panelius, P. Lahdenne, I. J. Seppälä, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 2768427-8435. [DOI] [PubMed] [Google Scholar]
- 19.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 195989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horstmann, R. D., H. J. Sievertsen, J. Knobloch, and V. A. Fischetti. 1988. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc. Natl. Acad. Sci. USA 851657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarva, H., J. Hellwage, T. S. Jokiranta, M. J. Lehtinen, P. F. Zipfel, and S. Meri. 2004. The group B streptococcal beta and pneumococcal Hic proteins are structurally related immune evasion molecules that bind the complement inhibitor factor H in an analogous fashion. J. Immunol. 1723111-3118. [DOI] [PubMed] [Google Scholar]
- 22.Jarva, H., R. Janulczyk, J. Hellwage, P. F. Zipfel, L. Bjorck, and S. Meri. 2002. Streptococcus pneumoniae evades complement attack and opsonophagocytosis by expressing the pspC locus-encoded Hic protein that binds to short consensus repeats 8-11 of factor H. J. Immunol. 1681886-1894. [DOI] [PubMed] [Google Scholar]
- 23.Kawaoka, Y., K. Otsuki, and M. Tsubokura. 1983. Growth temperature-dependent variation in the bacteriophage-inactivating capacity and antigenicity of Yersinia enterocolitica lipopolysaccharide. J. Gen. Microbiol. 1292739-2747. [DOI] [PubMed] [Google Scholar]
- 24.Koistinen, V., S. Wessberg, and J. Leikola. 1989. Common binding region of complement factors B, H and CR1 on C3b revealed by monoclonal anti-C3d. Complement Inflamm. 6270-280. [DOI] [PubMed] [Google Scholar]
- 25.Kotarsky, H., J. Hellwage, E. Johnsson, C. Skerka, H. G. Svensson, G. Lindahl, U. Sjöbring, and P. F. Zipfel. 1998. Identification of a domain in human factor H and factor H-like protein-1 required for the interaction with streptococcal M proteins. J. Immunol. 1603349-3354. [PubMed] [Google Scholar]
- 26.Kraiczy, P., J. Hellwage, C. Skerka, H. Becker, M. Kirschfink, M. M. Simon, V. Brade, P. F. Zipfel, and R. Wallich. 2004. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 2792421-2429. [DOI] [PubMed] [Google Scholar]
- 27.Kristensen, T., R. A. Wetsel, and B. F. Tack. 1986. Structural analysis of human complement protein H: homology with C4b binding protein, beta 2-glycoprotein I, and the Ba fragment of B2. J. Immunol. 1363407-3411. [PubMed] [Google Scholar]
- 28.Kühn, S., C. Skerka, and P. F. Zipfel. 1995. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H1. J. Immunol. 1555663-5670. [PubMed] [Google Scholar]
- 29.Kühn, S., and P. F. Zipfel. 1995. The baculovirus expression vector pBSV-8His directs secretion of histidine-tagged proteins. Gene 162225-229. [DOI] [PubMed] [Google Scholar]
- 30.Kühn, S., and P. F. Zipfel. 1996. Mapping of the domains required for decay acceleration activity of the human factor H-like protein 1 and factor H. Eur. J. Immunol. 262383-2387. [DOI] [PubMed] [Google Scholar]
- 31.Lambert de Rouvroit, C., C. Sluiters, and G. R. Cornelis. 1992. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6395-409. [PubMed] [Google Scholar]
- 32.Marques, M. B., D. L. Kasper, M. K. Pangburn, and M. R. Wessels. 1992. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infect. Immun. 603986-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meri, T., A. Hartmann, D. Lenk, R. Eck, R. Wurzner, J. Hellwage, S. Meri, and P. F. Zipfel. 2002. The yeast Candida albicans binds complement regulators factor H and FHL-1. Infect. Immun. 705185-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Messing, J., R. Crea, and P. H. Seeburg. 1981. A system for shotgun DNA sequencing. Nucleic Acids Res. 9309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, V. L., K. B. Beer, G. Heusipp, B. M. Young, and M. R. Wachtel. 2001. Identification of regions of Ail required for the invasion and serum resistance phenotypes. Mol. Microbiol. 411053-1062. [DOI] [PubMed] [Google Scholar]
- 36.Miller, V. L., J. B. Bliska, and S. Falkow. 1990. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J. Bacteriol. 1721062-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 561242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muszynski, A. 2004. Characterization of lipopolysaccharides from mutants of Yersinia enterocolitica O:3 cultivated at different temperatures. Ph.D. thesis. University of Silesia, Katowice, Poland.
- 39.Pandiripally, V., L. Wei, C. Skerka, P. F. Zipfel, and D. Cue. 2003. Recruitment of complement factor H-like protein 1 promotes intracellular invasion by group A streptococci. Infect. Immun. 717119-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pangburn, M. K., M. A. L. Atkinson, and S. Meri. 1991. Localization of the heparin-binding site on complement factor H. J. Biol. Chem. 26616847-16853. [PubMed] [Google Scholar]
- 41.Pangburn, M. K., R. D. Schreiber, and H. J. Müller-Eberhard. 1977. Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein β1H for cleavage of C3b and C4b in solution. J. Exp. Med. 146257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilz, D., T. Vocke, J. Heesemann, and V. Brade. 1992. Mechanism of YadA-mediated serum resistance of Yersinia enterocolitica serotype O3. Infect. Immun. 60189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pulkkinen, W. S., and S. I. Miller. 1991. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J. Bacteriol. 17386-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ram, S., D. P. McQuillen, S. Gulati, C. Elkins, M. K. Pangburn, and P. A. Rice. 1998. Binding of complement factor H to loop 5 of porin protein 1A: a molecular mechanism of serum resistance of nonsialylated Neisseria gonorrhoeae. J. Exp. Med. 188671-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ram, S., A. K. Sharma, S. D. Simpson, S. Gulati, D. P. Mcquillen, M. K. Pangburn, and P. A. Rice. 1998. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J. Exp. Med. 187743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 1853735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roggenkamp, A., K. Rückdeschel, L. Leitritz, R. Schmitt, and J. Heesemann. 1996. Deletion of amino acids 29 to 81 in adhesion protein YadA of Yersinia enterocolitica serotype O:8 results in selective abrogation of adherence to neutrophils. Infect. Immun. 642506-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salacinski, P. R., C. McLean, J. E. Sykes, V. V. Clement-Jones, and P. J. Lowry. 1981. Iodination of proteins, glycoproteins, and peptides using a solid-phase oxidizing agent, 1,3,4,6-tetrachloro-3α,6α-diphenyl glycoluril (Iodogen). Anal. Biochem. 117136-146. [DOI] [PubMed] [Google Scholar]
- 49.Skurnik, M. 1984. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Appl. Bacteriol. 56355-363. [DOI] [PubMed] [Google Scholar]
- 50.Skurnik, M., Y. El Tahir, M. Saarinen, S. Jalkanen, and P. Toivanen. 1994. YadA mediates specific binding of enteropathogenic Yersinia enterocolitica to human intestinal submucosa. Infect. Immun. 621252-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skurnik, M., and P. Toivanen. 1992. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Bacteriol. 1742047-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skurnik, M., R. Venho, J.-A. Bengoechea, and I. Moriyón. 1999. The lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3 is required for virulence and plays a role in outer membrane integrity. Mol. Microbiol. 311443-1462. [DOI] [PubMed] [Google Scholar]
- 53.Skurnik, M., R. Venho, P. Toivanen, and A. al-Hendy. 1995. A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol. Microbiol. 17575-594. [DOI] [PubMed] [Google Scholar]
- 54.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3517-529. [DOI] [PubMed] [Google Scholar]
- 55.Stoorvogel, J., M. J. van Bussel, J. Tommassen, and J. A. van de Klundert. 1991. Molecular characterization of an Enterobacter cloacae outer membrane protein (OmpX). J. Bacteriol. 173156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tertti, R., E. Eerola, O.-P. Lehtonen, T. Ståhlberg, M. Viander, and A. Toivanen. 1987. Virulence-plasmid is associated with the inhibition of opsonization in Yersinia enterocolitica and Yersinia pseudotuberculosis. Clin. Exp. Immunol. 68266-274. [PMC free article] [PubMed] [Google Scholar]
- 57.Vesikari, T., T. Nurmi, M. Mäki, M. Skurnik, C. Sundqvist, K. Granfors, and P. Grönroos. 1981. Plasmids in Yersinia enterocolitica serotypes O:3 and O:9: correlation with epithelial cell adherence in vitro. Infect. Immun. 33870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viitanen, A.-M., P. Toivanen, and M. Skurnik. 1990. The lcrE gene is part of an operon in the lcr region of Yersinia enterocolitica O:3. J. Bacteriol. 1723152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whaley, K., and S. Ruddy. 1976. Modulation of the alternative complement pathways by β1H globulin. J. Exp. Med. 1441147-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaleska, M., K. Lounatmaa, M. Nurminen, E. Wahlström, and P. H. Mäkelä. 1985. A novel virulence-associated cell surface structure composed of 47-kd protein subunits in Yersinia enterocolitica. EMBO J. 41013-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zink, D. L., J. C. Feeley, J. G. Wells, C. Vanderzant, J. C. Vickery, W. D. Roofs, and G. A. O'Donovan. 1980. Plasmid-mediated tissue invasiveness in Yersinia enterocolitica. Nature 283224-226. [DOI] [PubMed] [Google Scholar]