Abstract
Klebsiella pneumoniae is recognized as an important gram-negative opportunistic pathogen. The ability of bacteria to adhere to host structures is considered essential for the development of infections; however, few studies have examined the influence of adhesion factors on K. pneumoniae virulence. In this study, we cloned and characterized the type 1 fimbria gene cluster of a clinical K. pneumoniae isolate. Although this cluster was not identical to the Escherichia coli type 1 fimbria gene cluster, an overall high degree of structural resemblance was demonstrated. Unique to the K. pneumoniae fim gene cluster is the fimK gene, whose product contains an EAL domain, suggesting that it has a role in regulation of fimbrial expression. Like expression of type 1 fimbriae in E. coli, expression of type 1 fimbriae in K. pneumoniae was found to be phase variable, and an invertible DNA element (fim switch) was characterized. An isogenic type 1 fimbria mutant was constructed and used to evaluate the influence of type 1 fimbriae in different infection models. Type 1 fimbriae did not influence the ability of K. pneumoniae to colonize the intestine or infect the lungs, but they were determined to be a significant virulence factor in K. pneumoniae urinary tract infection. By use of a PCR-based assay, the orientation of the fim switch during colonization and infection was investigated and was found to be all “off” in the intestine and lungs but all “on” in the urinary tract. Our results suggest that during colonization and infection, there is pronounced selective pressure in different host environments for selection of either the type 1 fimbriated or nonfimbriated phenotype of K. pneumoniae.
The gram-negative enterobacterium Klebsiella pneumoniae is a common cause of urinary tract infections and respiratory tract infections in immunocompromised individuals (51). Frequently, the bacteria disseminate from the initial focus of infection to the bloodstream. Thus, K. pneumoniae is the second most common agent of gram-negative sepsis (71). Epidemiological studies have revealed that the first step in the majority of K. pneumoniae infections is colonization of the patient's gastrointestinal tract (44). So far, K. pneumoniae has generally been considered an opportunistic pathogen, primarily causing nosocomial infections in immunocompromised patients. However, in recent years an increasing number of severe K. pneumoniae infections in otherwise healthy individuals have been reported, especially in Southeast Asia (26, 28, 34, 70). These infections are often community acquired and primarily present as pyogenic liver abscesses frequently complicated by devastating dissemination of the infection to the eyes and central nervous system. The increasing occurrence of infections in otherwise healthy individuals stresses the necessity of elucidating the pathogenic mechanism of K. pneumoniae. Several virulence factors have been suggested for K. pneumoniae. Most clinical isolates produce vast amounts of capsular polysaccharide covering the bacterial surface. The capsule protects the bacteria against opsonization and phagocytosis, and the significance of the capsule in K. pneumoniae virulence has been demonstrated in several studies (6, 32, 60, 64, 65). Also, lipopolysaccharide and iron acquisition systems have been identified as significant virulence factors in in vivo studies using relevant infection models (33, 43, 46, 58, 64).
Adhesion of bacteria to host mucosal surfaces is generally considered an essential step in the development of infection. In gram-negative bacteria adhesion is often mediated by fimbriae, which are filamentous organelles expressed on the surface of the bacterial cell. Most clinical K. pneumoniae isolates are able to produce the two fimbrial adhesins, type 1 fimbriae and type 3 fimbriae. Type 3 fimbriae mediate adhesion to several cell types in vitro (21, 67); however, the receptor for the fimbriae has not been identified yet. Although type 3 fimbriae have been shown to play an essential role in biofilm formation in vitro (9, 24, 31), the influence of the fimbriae on virulence has not been evaluated in vivo yet. Type 1 fimbriae mediate adhesion to mannose-containing structures on host cells and extracellular matrix and are present in many species of Enterobacteriaceae. However, there are significant genetic, serological, and functional differences between type 1 fimbria variants in the different species (4, 10, 17, 39). In Escherichia coli the expression of type 1 fimbriae exhibits phase variation mediated by an invertible DNA element, the fim switch, which contains the promoter for the fimA gene encoding the major fimbrial subunit protein FimA (1). Two recombinases encoded by the fimB and fimE genes mediate inversion of the switch, resulting in a fimbriated state when the switch is in the “on” position and a nonfimbriated state when the switch is in the “off” position (27). Several studies have demonstrated that type 1 fimbriae play an important role in E. coli urinary tract infection (5, 30, 45, 62), whereas the influence of the fimbriae in other types of infections has been only sparsely investigated. In K. pneumoniae the influence of type 1 fimbriae on virulence is unclear as, to our knowledge, no infection studies using well-defined isogenic fimbria mutants have been described previously. In this study, we characterized the K. pneumoniae type 1 fimbria gene cluster and evaluated the influence of the fimbriae in three relevant K. pneumoniae infection models. Furthermore, the in vivo expression of type 1 fimbriae during colonization and infection was investigated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
K. pneumoniae C3091 is a streptomycin-resistant human urinary tract infection isolate (47, 64). Construction of a C3091 isogenic type 1 fimbria mutant is described below. K. pneumoniae strains C2918, C4712, and C747 are clinical isolates described previously (66).
For cloning, E. coli strain EPI100 (Epicentre) or nonfimbriated E. coli strain HB101 was used. Bacteria were cultured at 37°C on solid Luria-Bertani (LB) medium or in liquid LB medium. When appropriate, media were supplemented with antibiotics at the following concentrations: apramycin, 30 μg/ml; chloramphenicol, 12.5 μg/ml; kanamycin, 25 μg/ml; and streptomycin, 100 μg/ml.
Hemagglutination assay.
Expression of type 1 fimbriae was detected by determining the ability of bacterial cells to agglutinate a freshly prepared solution of 5% washed guinea pig erythrocytes. The assays were performed on glass slides in the presence and absence of 5% mannose.
DNA manipulation.
Isolation of chromosomal DNA was carried out by use of a Geneelute bacterial genomic DNA kit (Sigma), and plasmid DNA isolation was carried out by use of a Qiaprep Spin miniprep kit (Qiagen) used according to the manufacturer's instructions. Sequencing was conducted commercially by MWG-Biotech AG (Germany). Restriction endonucleases were used as recommended by the manufacturer (New England Biolabs).
Fosmid library construction and screening for the fimK gene.
A fosmid library of chromosomal DNA from C3091 was constructed by use of an EpiFOS fosmid library production kit as recommended by the manufacturer (Epicentre). Briefly, genomic DNA was manually sheared to obtain a mean fragment size of 40 kb by passing it through a small-bore pipette tip. After end repair, fragments of the correct size were purified by agarose gel electrophoresis, which was followed by ligation into fosmid vector pEpiFos-5 encoding resistance to chloramphenicol. The ligated DNA was packaged into lambda phage and transfected into E. coli EPI100 cells.
A total of 1,152 fosmid clones were isolated and screened for presence of the fimK gene by colony blot hybridization on Hybond-N+ membranes (Amersham) under stringent conditions according to the manufacturer's directions. A digoxigenin-labeled nucleotide probe for fimK was prepared by use of a PCR digoxigenin synthesis kit (Roche), as described by the manufacturer, using primers FimK-F and FimK-R (Table 1) designed using the previously described fimK nucleotide sequence (GenBank accession no. L23111).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ → 3′) |
|---|---|
| FimK-F | GCGGCTGGCTGGGGTGAAAAAG |
| FimK-R | AGTCGACGCGCCGGAAAGATAACG |
| Kn1 | GTGTAGGCTGGAGCTGCTTC |
| Kn2 | ATGGGAATTAGCCATGGTCC |
| UpfimB-F | TGCGGGTATCATCAAGAGG |
| UpfimB-R | GAAGCAGCTCCAGCCTACACCAGCAACGGGTGGGTAGT |
| DwfimK-F | GGACCATGGCTAATTCCCATCTGGTGCTGGAATATGCTGAAACT |
| DwfimK-R | CGATAACACCCGCGAATACGAC |
| K1 | CAGTCATAGCCGAATAGCCT |
| K2 | CGGTGCCCTGAATGAACTGC |
| Upfim | AGCCCAAGCCTTCATCATCA |
| Dwfim | CACCGGCGAACACCTCAGCATTAT |
| CAS168 | GGGACAGATACGCGTTTGAT |
| CAS169 | GGCCTAACTGAACGGTTTGA |
Construction of an isogenic type 1 fimbria mutant.
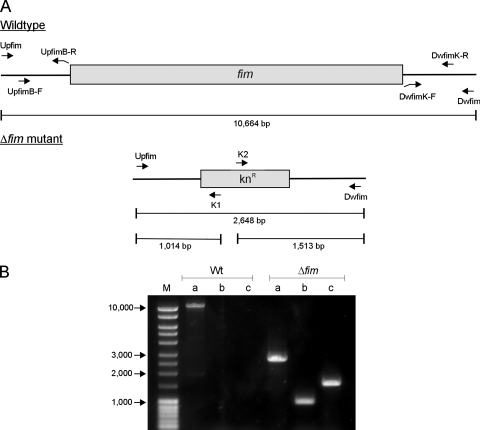
The fim gene cluster in C3091 was deleted by allelic exchange with a kanamycin resistance-encoding cassette flanked by regions homologous to the regions up- and downstream of the fim gene cluster (Fig. 1). All primers used are listed in Table 1. The cassette was generated by using a modification of a three-step PCR procedure described previously (8). In the first step, the kanamycin resistance-encoding gene (kan) was amplified from pKD4 by use of primers Kn1 and Kn2 (7). Second, from C3091 chromosomal DNA a 496-bp region and a 481-bp region flanking the fim gene cluster were PCR amplified by use of primers UpfimB-F and UpfimB-R and primers DwfimK-F and DwfimK-R, respectively. At their 5′ ends, pUpfimB-R and pDwfimK-F contained 20-bp regions homologous to the extremities of the kan gene. In the third step, the flanking regions were added on each side of the kan gene by mixing 100 ng of each fragment, followed by PCR amplification using primers UpfimB-F and DwfimK-R. The PCR product was purified and electroporated into C3091 harboring the thermosensitive plasmid pKOBEGApra encoding the lambda Red recombinase. The C3091Δfim mutant was selected by growth on LB medium plates containing kanamycin at 37°C, and loss of the pKOBEGApra plasmid was verified by the inability of the mutant to grow on LB agar plates containing apramycin. Correct allelic exchange was verified by PCR analysis using combinations of primers inside the kan gene (primers K1 and K2) (7) and primers flanking the deleted fim gene cluster (primers Upfim and Dwfim) (Fig. 1). To verify that the mutant was not able to express type 1 fimbriae, the Δfim mutant and the wild-type strain were analyzed for the ability to agglutinate guinea pig erythrocytes. As expected, the wild-type strain exhibited mannose-sensitive agglutination, whereas the Δfim mutant did not agglutinate guinea pig erythrocytes.
FIG. 1.
(A) Genetic organization of the fim region in wild-type strain C3091 and the constructed Δfim mutant. Primers used for mutant construction and for verification of correct allelic exchange are indicated. (B) Verification of correct allelic exchange in the Δfim mutant by PCR analysis using primers flanking the type 1 fimbria gene cluster in combination with primers specific for the kanamycin resistance-encoding cassette. Lane M contained DNA molecular size markers. Lanes a, primers Upfim and Dwfim; lanes b, primers Upfim and K1; lanes c, primers K2 and Dwfim. Wt, wild type.
Mouse model of gastrointestinal colonization.
Six- to eight-week-old female CFW1 mice (Harlan) were used for intestinal colonization experiments as described previously (35). Briefly, mice were individually caged and provided with drinking water containing 5 g of streptomycin sulfate per liter. After 24 h, 100-μl bacterial suspensions containing approximately 1 ×105 CFU were given orally to the mice by pipette. During the colonization experiment, cages were changed daily and the mice received water containing streptomycin continuously. At the indicated times, 0.5 g of feces was collected and homogenized in 5 ml of 0.9% NaCl, and dilutions were plated on selective media.
Mouse lung infection model.
An intranasal infection model described previously was also used (12, 55). Six- to eight-week-old female NMRi mice (Harlan) were anesthetized. The mice were hooked on a string by the front teeth, and 50 μl of a bacterial suspension containing approximately 5 × 107 CFU was dripped onto the nares. The mice readily aspirated the solution and were left hooked on the string for 10 min before they were returned to their cages. The mice were sacrificed 1 or 2 days after inoculation. For recovery of bacteria, lungs, spleens, and livers were collected in 1 ml of 0.9% NaCl and homogenized, and serial dilutions were plated on selective media.
Mouse model of ascending urinary tract infection.
Six- to eight-week-old female CFW1 mice (Harlan) were used for urinary tract infection. The model used has been described in detail previously (23). Three days before inoculation and throughout the experiment, 5% glucose was added to the drinking water as this has been shown to promote urinary tract infection in mice (25). The mice were anesthetized and inoculated transurethrally with 50 μl of a bacterial suspension containing approximately 5 × 108 CFU by using plastic catheters. A catheter was carefully pushed horizontally through the urethral orifice until it reached the top of the bladder, and the bacterial suspension was slowly injected into the bladder. The catheter was immediately removed, and the mice were subjected to no further manipulations until they were sacrificed. The mice were sacrificed 1 or 3 days after inoculation. For recovery of bacteria, bladders and kidneys were collected in 1 ml of 0.9% NaCl and homogenized, and serial dilutions were plated on selective media.
Fimbrial switch orientation assay.
A modification of a previously described method was used to determine the orientation of the K. pneumoniae fim switch (64). All culture and organ samples were boiled for 5 min in phosphate-buffered saline immediately after collection and then kept at −20°C until they were used. After thawing, the culture and urine samples were boiled for 5 min and centrifuged at 12,000 × g for 15 min, and 10 μl of each supernatant was diluted 1:10 with a 10% suspension of Chelex 100 resin (Bio-Rad). After the suspension was boiled for 5 min, it was centrifuged at 12,000 × g for 5 min, and 2 μl of the supernatant was used as the template for PCR. The bladder samples were homogenized manually and then prepared for PCR as described above. The colon, cecum, and lung samples were homogenized manually, and DNA was purified for PCR amplification by use of a QIAamp DNA mini kit (Qiagen) as described by the manufacturer.
Primers CAS168 and CAS169 (Table 1) were used to amplify an 817-bp region containing the fim switch with the Expand high-fidelity PCR system (Roche). The PCR cycle conditions were as follows: one cycle of 94°C for 2 min; 30 cycles of 94°C for 15 s, 52°C for 1 min, and 72°C for 1 min; and one cycle of 72°C for 7 min. The PCR products were cut with HinfI and separated on a 1.2% agarose gel.
Transmission electron microscopy.
Negative staining was carried out as follows. A Formvar-coated carbon-reinforced copper grid was placed, film side down, on a droplet of a culture suspension for 2 min. The excess liquid was then removed with a piece of filter paper, and the grid was stained for 30 s on droplets of 2% ammonium molybdate (pH 7.4) or for 15 s on droplets of 1.25% phosphotungstic acid (pH 6.5). Electron microscopy was carried out with a Philips 201C electron microscope at 60 kV.
Data analysis and statistical methods.
Nucleotide sequences were analyzed by use of the Lasergene v. 5.03 software (DNAStar, Inc.). The competitive index was calculated by dividing the ratio of wild-type bacteria to mutant bacteria recovered from infected organs by the ratio of wild-type bacteria to mutant bacteria in the inoculum. The Wilcoxon rank sum test was used to analyze the data from the infection studies with zero as the comparator for the log10 of the competitive index values. P values less than 0.05 were considered statistically significant.
Nucleotide sequence accession number.
The nucleotide sequence of the type 1 fimbria gene cluster of K. pneumoniae C3091 described in this paper has been deposited in the GenBank database under accession number EU044788.
RESULTS
Cloning and characterization of the K. pneumoniae type 1 fimbria gene cluster.
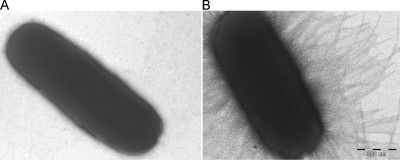
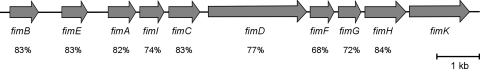
The fim gene cluster encoding K. pneumoniae type 1 fimbriae was isolated by use of a fosmid library of chromosomal K. pneumoniae C3091 DNA expressed in E. coli strain EPI100 and probed for the fimK gene. One hybridization-positive clone was selected from the screening. From the selected clone a 12-kb region containing the entire fim gene cluster was subcloned into cloning vector pUC18, which yielded pCAS624. To validate that the cloned fragment allowed phenotypic expression of K. pneumoniae type 1 fimbriae, pCAS624 was transformed into the nonfimbriated E. coli strain HB101 (52). Expression of type 1 fimbriae was verified on the basis of the ability of HB101(pCAS624) to agglutinate guinea pig erythrocytes in a mannose-sensitive manner. Furthermore, transmission electron microscopy confirmed that HB101(pCAS624) expressed fimbriae on the surface of the cell, whereas the HB101 parent strain, as expected, was nonfimbriated (Fig. 2). By using sequencing, the K. pneumoniae fim gene cluster was found to exhibit an overall high level of structural resemblance to the type 1 fimbria gene cluster of E. coli (Fig. 3). Thus, homologues of all genes in the E. coli fim cluster could be identified in K. pneumoniae. For the individual gene products the levels of amino acid identity ranged from 72 to 84% compared to the E. coli homologues. A major difference between the fim gene clusters of the two species was the presence of the fimK gene directly downstream of fimH in K. pneumoniae. This gene has previously been found to influence expression of type 1 fimbriae in a K. pneumoniae strain and is therefore considered part of the K. pneumoniae fim cluster (18). Sequence analysis indicated that the closest homologue of the fimK product is a hypothetical protein in Citrobacter koseri with 54% amino acid sequence similarity, whereas no homologues in species other than K. pneumoniae could be identified.
FIG. 2.
Transmission electron micrographs of (A) E. coli HB101 and (B) E. coli HB101 transformed with pCAS624 encoding K. pneumoniae type 1 fimbriae.
FIG. 3.
Genetic organization of the type 1 fimbria gene cluster (fim) in K. pneumoniae C3091. The levels of amino acid identity to the E. coli homologues are indicated for the individual genes. The fimK gene is unique to the K. pneumoniae fim gene cluster.
Identification and characterization of the K. pneumoniae fim switch.
To establish whether type 1 fimbria expression in K. pneumoniae C3091 is regulated by an invertible DNA element containing the fimA promoter, as it is in E. coli, sequence analysis of the region upstream of the fimA gene was performed. Indeed, a 314-bp region flanked by inverted repeat nucleotide sequences could be identified directly upstream of the fimA gene (Fig. 4). The sequence of the 9-bp inverted repeats (5′TTGGGGCCA) in K. pneumoniae C3091 was found to be identical to the sequence in E. coli; however, the overall nucleotide similarity of the fim phase switch regions in the two species was only 41%.
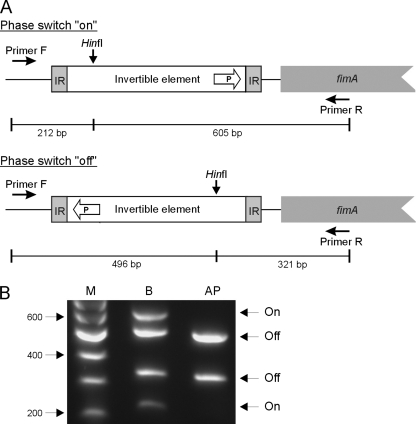
FIG. 4.
Characterization of the K. pneumoniae fim phase switch. (A) Type 1 fimbria expression in K. pneumoniae is regulated by an invertible DNA element (phase switch) containing the promoter (P) of the major fimbrial subunit-encoding gene, fimA. The orientation of the phase switch, “on” or “off,” can be determined by PCR amplification of the switch region, followed by digestion with HinfI. Due to the asymmetric location of the HinfI cleavage site within the invertible element, different fragments are obtained depending on the orientation of the phase switch. IR, inverted repeat. (B) Detection of the fim phase switch orientation in cultures grown in LB broth (lane B) or on LB agar plates (lane AP). Lane M contained DNA molecular size markers.
Agglutination experiments revealed that C3091 does not express type 1 fimbriae when it is cultivated on LB agar plates, in contrast to the expression when it is cultivated in LB broth with shaking. To verify that the identified fim phase switch is invertible, the switch region was PCR amplified from C3091 cultures cultivated either in LB broth or on LB agar plates. Due to the asymmetric location of the cleavage site of restriction endonuclease HinfI inside the invertible element, DNA fragments that were different sizes were obtained after HinfI cleavage depending on the orientation of the fim phase switch (Fig. 4A). HinfI cleavage of the PCR-amplified switch region obtained from the broth-grown culture resulted in four fragments that were of expected sizes, corresponding to a mixture of cells with the fim phase switch in the “on” or “off” position, confirming that the identified fim phase switch is indeed invertible (Fig. 4B). In agreement with the agglutination results, only DNA fragments corresponding to the fim phase switch in the “off” position were detected when the switch assay was performed with an agar plate-grown culture; thus, cultivation on solid media switches the invertible element toward the “off” position, thereby downregulating expression of type 1 fimbriae.
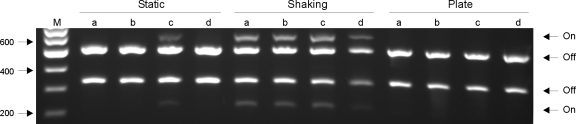
To further investigate the influence of growth conditions on type 1 fimbria phase switching, the orientations of the switch in exponentially growing and late-stationary-phase cultures were compared. Furthermore, the influence of shaking during growth was investigated. In the cultures grown with shaking, the phase switch was found to be in the “off” orientation only during exponential growth, whereas both orientations were detected in the late stationary phase (Table 2). In contrast, in the statically grown cultures only the “off” orientation was detected for the exponentially growing and late-stationary-phase cultures. To investigate whether prolonged growth in static media could turn the switch to the “on” orientation, the switch assay was used with cultures that had been passaged daily four times and with cultures grown for 96 h. Again, whereas the switch in both orientations was detected in all cultures grown with shaking, only the “off” orientation was detected in all statically grown cultures (Table 2). To investigate whether type 1 fimbria phase switching varies between K. pneumoniae strains, the switch orientations under different in vitro culture conditions were also assessed for three other clinical strains, strains C2918, C4712, and C747. The switch orientation in strains C2918 and C747 was found to be identical to that in C3091 in static and shaking overnight broth cultures, as well as in cultures grown on agar plates; i.e., the “on” orientation was detectable only in broth cultures grown with shaking (Fig. 5). Only strain C4712 differed from C3091 as in this strain the “on” orientation was also detectable in statically grown broth cultures (Fig. 5). In cultures grown for 96 h and cultures that had been passaged daily four times, the switch orientation in C2918 and C747 was identical to that in C3091 (Table 2). Again, C4712 differed from the three other strains because the “on” orientation was also detectable in statically grown broth cultures (results not shown).
TABLE 2.
Orientation of the fim switch in strain C3091 under different in vitro culture conditions
| Culture conditions | Switch orientation
|
|||
|---|---|---|---|---|
| Exponential phase | Stationary phase | Passaged four times | Grown for 96 h | |
| Static broth | Off | Off | Off | Off |
| Shaking broth | Off | On and off | On and off | On and off |
| Agar plate | NDa | Off | Off | Off |
ND, not determined.
FIG. 5.
Orientation of the fim phase switch in four K. pneumoniae strains after overnight culture in either static broth or shaking broth or on agar plates. Lane M contained DNA molecular size markers. Lanes a, strain C3091; lanes b, strain C2918; lanes c, strain C4712; lanes d, strain 747.
Type 1 fimbriae do not affect K. pneumoniae gastrointestinal colonization.
To investigate the role of type 1 fimbriae in K. pneumoniae virulence, the fim gene cluster in C3091 was deleted by allelic replacement of the entire gene cluster with a kanamycin resistance-encoding cassette as described in Materials and Methods and shown in Fig. 1.
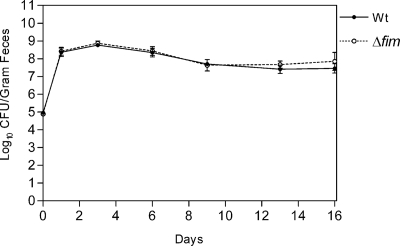
The majority of K. pneumoniae infections are preceded by colonization of the patient's gastrointestinal tract. To evaluate the influence of type 1 fimbriae on K. pneumoniae colonization, a mixture containing equal numbers of the wild-type strain and the Δfim mutant was fed to three mice, and the bacterial counts in the feces were monitored for 16 days. Both the wild-type strain and the Δfim mutant were able to successfully colonize the intestine. Thus, for all three mice, each inoculated with approximately 1 ×105 bacteria, the bacterial count in feces on day 1 after inoculation was approximately 8.4 ×108 bacteria per g of feces, and the number of the Δfim mutant bacteria was found to be similar to the number of the wild-type strain bacteria (Fig. 6). Thus, type 1 fimbriae did not influence the ability of K. pneumoniae to become established in the intestinal tract. Furthermore, the ability of the Δfim mutant to colonize the intestine long term was similar to the ability of the wild-type strain as the two strains cocolonized at a level of approximately 108 CFU per g of feces during the 16-day colonization period.
FIG. 6.
Colonization of the intestine by wild-type strain C3091 and the type 1 fimbria mutant fed simultaneously to three mice. Means and standard errors of the means are shown. The symbol for day 0 indicates the size of the inoculum. Wt, wild type.
Role of type 1 fimbriae in K. pneumoniae lung infection.
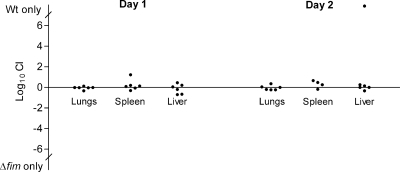
To assess the role of type 1 fimbriae in K. pneumoniae lung infection, two groups of six mice were intranasally inoculated with equal numbers of the Δfim mutant and wild-type strain. The two groups of mice were sacrificed 1 and 2 days after inoculation. In addition to the lungs, spleens and livers were also collected to establish if dissemination of the infection to the bloodstream had occurred. On day 1 after inoculation, the lungs of all mice were profoundly infected, and the bacterial counts ranged from 4 × 107 to 2 × 108 CFU per lung. Furthermore, K. pneumoniae could be detected in the spleens and livers of all mice, and the median levels were 2.3 × 103 and 2.1 × 103 CFU per organ, respectively. Expression of type 1 fimbriae did not influence the ability of K. pneumoniae to infect the lungs or the ability of the bacteria to disseminate from the lungs to the bloodstream and infect the spleen and liver as similar numbers of the Δfim mutant and the wild-type strain were detected in the infected organs (Fig. 7). On day 2 after inoculation, the bacterial counts in infected lungs remained high, ranging from 7 × 106 to 2 × 108 CFU per lung, and, as on day 1, similar numbers of the Δfim mutant and the wild-type strain were detected. The median bacterial counts in infected spleens and livers 2 days after inoculation were lower, 3.8 × 102 and 1.8 × 102 CFU per organ, respectively. Except for one liver sample, similar numbers of the Δfim mutant and wild-type strain were present in the infected organs (Fig. 7). The one liver sample in which only the wild-type strain was detected was the sample with the lowest bacterial count in the experiment, and it is likely a coincidence that only the wild type was detected in this sample.
FIG. 7.
Comparison of the abilities of wild-type strain C3091 and the type 1 fimbria mutant to infect the lungs and disseminate to the spleen and liver. Mice were intranasally inoculated with equal numbers of the wild type and the type 1 fimbria mutant. The competitive index (CI) was determined by dividing the ratio of wild-type bacteria to mutant bacteria recovered from infected organs by the ratio of wild-type bacteria to mutant bacteria in the inoculum. Wt, wild type.
Type 1 fimbriae are an important virulence factor in K. pneumoniae urinary tract infection.
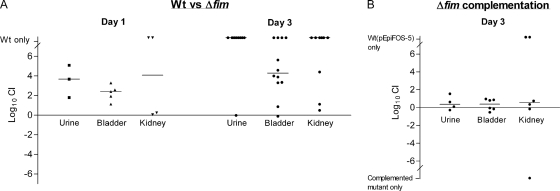
A well-established mouse model was used to evaluate the role of type 1 fimbriae in urinary tract infection. A group of 17 mice were inoculated with a mixture containing equal numbers of the Δfim mutant and the wild-type strain. Five mice were sacrificed on day 1 after inoculation, and the bladders of all of them were found to be infected, with a median bacterial count of 6.8 × 106 CFU per bladder. The wild-type strain had significantly outcompeted the mutant in the bladders by a median of 2.4 log10 (P = 0.03), and in two of four infected kidneys only the wild-type strain was detected (Fig. 8A). The remaining 12 mice were sacrificed 3 days after inoculation. The bladders of all mice were found to be infected, and the median bacterial count was 8.3 × 105 CFU per bladder. Compared to day 1, even more pronounced attenuation of the Δfim mutant was detected as the wild-type was found to outcompete the mutant by a median of 4.3 log10 (P = 0.0005) and in 4 of the 12 infected bladders only the wild-type strain was detected. Type 1 fimbriae were also significantly involved in the ability of K. pneumoniae to ascend to and infect the kidneys, as in the majority of infected kidneys (7/10) only the wild-type strain was detected (P = 0.001).
FIG. 8.
Comparison of the urovirulence of wild-type strain C3091 and the urovirulence of the type 1 fimbria mutant. Mice were inoculated with equal numbers of the wild type and the type 1 fimbria mutant. The competitive index (CI) was determined by dividing the ratio of wild-type bacteria to mutant bacteria recovered from infected organs by the ratio of wild-type bacteria to mutant bacteria in the inoculum. (B) Comparison of the urovirulence of the type 1 fimbria mutant complemented with plasmid pCAS704 containing the fim gene cluster and the urovirulence of the wild-type strain transformed with the cloning vector. Outliers indicate that only the wild-type strain containing pEpiFOS-5 or the complemented mutant was detected in the infected tissue. Wt, wild type.
To verify that the attenuated urovirulence of the mutant was due to abolition of type 1 fimbria expression and not to polar effects of the mutation, C3091Δfim was transformed with pCAS704 containing the fim gene cluster cloned into the pEpiFOS-5 vector. Using the single-copy pEpiFOS-5 fosmid vector for cloning ensured that only one copy of the fim gene cluster was present in the complemented mutant, as in the wild type. In this way altered expression of type 1 fimbriae in the complemented mutant due to the presence of multiple copies of the fim gene cluster was avoided. Indeed, similar expression of type 1 fimbriae in the complemented mutant and the wild-type strain was observed, as evaluated by agglutination of guinea pig erythrocytes with serial dilutions of the wild-type and with serial dilutions of the complemented mutant. Equal numbers of the wild-type strain transformed with pEpiFOS-5 and the complemented mutant were inoculated into six mice that were sacrificed 3 days after inoculation. The complemented mutant exhibited urovirulence that overall was similar to that of the wild-type strain (Fig. 8B). Similar bacterial counts were obtained on agar plates with and without chloramphenicol, showing that loss of the fosmids did not occur during the infection period. Thus, the significant influence of type 1 fimbriae on K. pneumoniae C3091 urovirulence was confirmed.
Expression of type 1 fimbriae during infection.
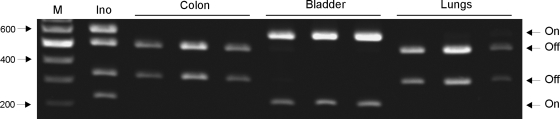
To assess the expression of type 1 fimbriae during K. pneumoniae colonization and infection, the fim switch region was PCR amplified from infected organs, and a bacterial suspension was used to inoculate mice. HinfI cleavage of the PCR-amplified switch region from the inoculum suspension resulted in four fragments; i.e., a mixture of cells expressing and not expressing type 1 fimbriae was present in the suspension used to infect the mice (Fig. 9). In contrast, for colon samples from three mice colonized with K. pneumoniae C3091 only fragments corresponding to the fim switch in the “off” position were detected (Fig. 9). The same results were obtained when the switch assay was used with cecum samples from the colonized mice (results not shown). Thus, the results revealed that during intestinal colonization the fim switch is switched to the “off” position downregulating expression of type 1 fimbriae. Downregulation of type 1 fimbria expression was also observed in the lungs of each of three mice intranasally inoculated with K. pneumoniae C3091. In contrast, type 1 fimbria expression was found to be highly upregulated in infected bladders as only fragments corresponding to the switch in the “on” position were detected for bladder samples from all mice (Fig. 9). The assay was also performed with urine samples from infected mice, and similar to in the bladder results, type 1 fimbria expression was found to be profoundly upregulated as only the “on” position of the fim switch was detected (results not shown).
FIG. 9.
Detection of the fim phase switch orientation in the inoculum suspension (Ino) and K. pneumoniae C3091 cells infecting the colons, bladders, and lungs of mice. Lane M contained DNA molecular size markers.
To investigate whether the phase switch is permanently locked in either the “off” or “on” position in K. pneumoniae cells from colonized or infected tissues, the switch assay was used with bacterial cells isolated from infected organs. Phase switching to the “on” and “off” positions occurred in all cases after overnight growth in shaking broth, indicating that selection of phase-locked mutants did not occur during colonization and infection (results not shown).
DISCUSSION
Even though K. pneumoniae is recognized as one of the most important gram-negative opportunistic pathogens, detailed knowledge of the mechanism by which this bacterium causes infections is limited. Capsule and lipopolysaccharide have been shown to protect the bacterial cell against the host immune system. In contrast, although the ability of bacteria to adhere to host structures is generally considered essential in the development of bacterial infections, knowledge of the influence of specific adhesion factors on K. pneumoniae virulence is sparse.
In the present study, we isolated and characterized the K. pneumoniae type 1 fimbria gene cluster and evaluated the influence of type 1 fimbria expression on virulence by use of relevant in vivo infection models. Type 1 fimbriae are widespread among the enterobacterial species; however, most type 1 fimbria expression and functionality studies have been performed with the E. coli variant. Clegg and coworkers previously isolated and described the K. pneumoniae type 1 fimbria gene cluster and found an overall genetic organization resembling that of the type 1 fimbria gene cluster of E. coli (3, 4, 18, 52). However, only nucleotide sequences of the genes encoding the major fimbrial subunit (fimA), the mannose-binding adhesin (fimH), and the minor components (fimF and fimG) have been published (10, 18). Here, we report the sequence of the complete type 1 fimbria gene cluster of K. pneumoniae clinical isolate C3091. The gene cluster was found to contain homologues of all nine genes included in the type 1 fimbria gene cluster of E. coli. Due to the similarity of the genetic organizations of the fim clusters in the two species, it is likely that type 1 fimbrial structure and biosynthesis in K. pneumoniae resemble type 1 fimbrial structure and biosynthesis in E. coli. Thus, in addition to the fimA gene encoding the major fimbrial subunit and the fimF, fimG, and fimH genes encoding minor structural subunits, homologues of the fimC and fimD genes encoding the E. coli fimbrial chaperone and usher protein, respectively, were identified. Also, a homologue of the E. coli fimI gene was present in the K. pneumoniae fim gene cluster. The function of the fimI gene product is unknown, but this product has been found to be essential for type 1 fimbria biosynthesis in E. coli (69). The amino acid sequence similarity of the individual gene products to the E. coli homologues varied from 72 to 84%. Thus, although the fim gene clusters of E. coli and K. pneumoniae are closely related, they are not identical. This is in agreement with the fact that type 1 fimbriae of E. coli and K. pneumoniae are serologically distinct and the fact that significant differences in the sugar-binding specificities and the capacities to bind to epithelial cells have been demonstrated for the two species (10, 39).
Unique to the K. pneumoniae fim gene cluster is the fimK gene located directly downstream of fimH. The fimK gene product has previously been shown to be involved in type 1 fimbria expression as deletion of fimK results in a scantily fimbriated phenotype (18). Except for a gene encoding a hypothetical protein in C. koseri with 54% amino acid similarity to the protein encoded by fimK, no homologous genes could be identified in species other than K. pneumoniae. Most interestingly, the FimK protein contains a conserved helix-turn-helix DNA-binding domain and a conserved EAL domain. Proteins containing an EAL domain have been shown to mediate degradation of the signaling molecule cyclic di-GMP and to be involved in regulation of a number of cell surface structures, including flagella, surface polysaccharides, and fimbriae, in different species (for a review, see reference 53). Hence, it is obvious to speculate that the fimK gene product is involved in the regulation of fimbrial expression in K. pneumoniae.
Whereas the role of fimK needs to be assessed in future studies, an invertible DNA element mediating phase switching of type 1 fimbria expression was identified and characterized in the present study. The K. pneumoniae fim switch was verified to be invertible by comparing the orientations of the switch in cultures grown in liquid and solid media. A similar fim switch assay has been used previously for E. coli, and the sensitivity of the assay was found to be 2%; i.e., a lack of detectable bands in one orientation indicates that less than 2% of the bacterial population has the fim switch in that orientation (36). Using liquid-grown cultures, both orientations of the phase switch could be detected by the PCR switch assay, whereas growth on solid media turned the switch to the “off” orientation. It is well known that growth conditions affect expression of type 1 fimbriae in E. coli, and in agreement with our results, it has previously been shown that the majority of clinical E. coli isolates downregulate type 1 fimbria expression when they are grown on solid media (22). In contrast, the type 1 fimbria expression profile for K. pneumoniae C3091 in liquid media was distinctly different from the profile previously described for E. coli. Growth in static media is generally thought to induce type 1 fimbriae. However, we found that in C3091 type 1 fimbriae are downregulated in static cultures but upregulated in shaking cultures. To investigate if this was a strain-specific phenomenon, three other clinical K. pneumoniae isolates were subjected to the fim switch assay. In two of the additional isolates the switch orientation was identical to that of C3091, indicating that shaking generally promotes expression of type 1 fimbriae in K. pneumoniae. The “on” orientation was detectable in only one strain when the strain was grown statically. The fact that type 1 fimbria expression is generally lower in K. pneumoniae under growth conditions promoting fimbrial expression in E. coli suggests that there are significant differences in the regulation of type 1 fimbria expression in the two species. In E. coli the orientation of the phase switch is regulated by the recombinases FimB and FimE; FimE turns the switch from the “on” position to the “off” position, while FimB can turn the switch in either direction (27, 40). Homologues of the E. coli FimB- and FimE-encoding genes were identified in the K. pneumoniae gene cluster and may be expected to be functionally equivalent to the E. coli variants. Regulation of type 1 fimbria expression in E. coli is very complex, and several regulatory factors that act by altering the expression of fimB and fimE have been described (11, 16, 61). Furthermore, cross-regulation of the expression of type 1 fimbriae and the expression of other surface structures, including flagella and P fimbriae, has been demonstrated (19, 29). Very little is known about the factors regulating fimbrial expression in K. pneumoniae, but cross-regulation with specific surface structures (e.g., type 3 fimbriae and capsule) is expected and might result in the different type 1 fimbria expression pattern compared to E. coli. Furthermore, whether there is a relationship between strain origin and specific regulation patterns of type 1 fimbria expression remains to be investigated. Thus, it could be hypothesized that in general, uropathogenic strains are more prone to express type 1 fimbriae than strains isolated from the gastrointestinal or respiratory tract.
Several studies have shown that most clinical K. pneumoniae isolates are able to express type 1 fimbriae (20, 37, 48-50, 66, 68), but so far the influence of fimbriae on virulence has not been evaluated by use of well-defined mutants. By using scanning electron microscopy, Fader and Davis found that the bladders and kidneys of rats inoculated with a fimbriated K. pneumoniae strain exhibited more signs of infection than the bladders and kidneys of rats infected with a nonfimbriated variant of the same strain (13, 14). Likewise, Maayan et al. found a higher number of bacteria in the urine from groups of mice inoculated with fimbriated cells than in the urine from groups of mice inoculated with nonfimbriated variants (38). In all the studies described above the workers used spontaneous nonfimbriated variants rather than defined genetic mutants, and the possibility that the variants differed in characteristics other than fimbrial expression cannot be excluded. In the present study a well-defined type 1 fimbria mutant was constructed and evaluated using three relevant mouse models of K. pneumoniae infection. We found that type 1 fimbriae did not influence the ability of C3091 to become established or its ability to colonize the intestine of mice long term. Furthermore, in the K. pneumoniae population colonizing the intestine only the “off” orientation of the fim switch was detectable. In agreement with our results, it has previously been reported that type 1 fimbriae do not enhance the intestine-colonizing ability of E. coli, presumably because expression of type 1 fimbriae reduces the ability of the bacteria to penetrate the intestinal mucus layer (41, 42).
The roles of type 1 fimbriae in K. pneumoniae lung infection and urinary tract infection were found to be profoundly different. The type 1 fimbria mutant was as virulent as the wild-type strain in the lung infection model, and the bacterial population infecting the lungs was found to downregulate fimbrial expression. Furthermore, the ability to express type 1 fimbriae did not influence the level of dissemination of the bacteria from the lungs to the bloodstream. To our knowledge, this is the first study in which the role of type 1 fimbriae in lung infection was investigated.
In contrast to the results for the lungs, type 1 fimbriae were found to be an important virulence factor in K. pneumoniae urinary tract infection. Thus, the urovirulence of the type 1 fimbria mutant was profoundly attenuated, and fimbrial expression was found to be highly upregulated in the bacterial population in urine and infected bladders. The urovirulence of the mutant could be restored by complementation with a plasmid containing the fimbria gene cluster. Thus, Koch's molecular postulates were fulfilled for type 1 fimbriae as a virulence factor in K. pneumoniae, as they have previously been in E. coli (5).
We and others have previously investigated the orientation of the fim switch in E. coli during experimental urinary tract infection (36, 63); however, this is the first study in which the orientations of the fim switch in different infection models were compared. The finding that the fim switch in the different infection scenarios was detected only in the “off” or “on” orientation is intriguing. This indicates that specific environmental signals in the different host environments promote switching to the “on” or “off” position or that the specific host environments provoke strong selection for either fimbriated or nonfimbriated bacteria. Alternatively, mutants with the phase switch locked in either the “on” or “off” position could arise during the infection period. However, this was not the case as phase switching did occur in bacterial cells isolated from infected organs when the cells were cultivated in vitro. It could be speculated that in the urinary tract nonfimbriated bacteria are eliminated by the voiding of urine. However, if phase switching to the “off” position occurred in a substantial part of the bacterial population, both orientations of the phase switch would be expected to be detectable in the urine samples. As only the “on” orientation was detectable in the urine samples, it is likely that environmental signals in the urinary tract promote turning of the switch to the “on” position. In the lungs, expression of type 1 fimbriae may be a disadvantage for the bacteria as type 1 fimbriae have been shown to promote adhesion to phagocytic cells in vitro and in vivo (2, 59). Thus, at phagocyte-rich sites, such as the lungs and bloodstream, selection for the nonfimbriated phenotype is likely to occur as fimbriated cells are rapidly eliminated by phagocytosis. The capsule protects the bacteria against phagocytosis, and it is intriguing that several studies have shown that there is an inverse relationship between capsule expression and the ability to adhere in K. pneumoniae (15, 56, 65). Thus, it could be speculated that cross-regulation of capsule and fimbrial expression may occur in vivo in such a way that capsule expression is upregulated and fimbrial expression is downregulated at phagocyte-rich infection sites and vice versa at infection sites where adhesion is critical (e.g., in the urinary tract). Indeed, a recent study demonstrated that type 1 fimbria-mediated adhesion triggers downregulation of capsule expression in E. coli in vitro (57). The orchestrated expression of different virulence factors in K. pneumoniae infections is an intriguing theme for future studies of this important pathogen.
ADDENDUM
After submission of the manuscript, a study was published which confirmed that fimK is involved in regulation of type 1 fimbria expression (54).
Acknowledgments
We are most grateful to Jean-Marc Ghigo, Unité de Génétique des Biofilms, Institut Pasteur, France, for providing the pKOBEGApra plasmid. We thank Christina Kofoed Johnsen, The Electron Microscopy Unit, Statens Serum Institut, Denmark, for performing the electron microscopy experiments.
C. Struve was partially supported by Danish Research Agency grant 2052-03-0013. M. Bojer was supported by Danish Research Agency grant 22-04-0725.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 16 June 2008.
REFERENCES
- 1.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 825724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhard, W., A. Gbarah, and N. Sharon. 1992. Lectinophagocytosis of type 1 fimbriated (mannose-specific) Escherichia coli in the mouse peritoneum. J. Leukoc. Biol. 52343-348. [DOI] [PubMed] [Google Scholar]
- 3.Clegg, S., S. Hull, R. Hull, and J. Pruckler. 1985. Construction and comparison of recombinant plasmids encoding type 1 fimbriae of members of the family Enterobacteriaceae. Infect. Immun. 48275-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clegg, S., B. K. Purcell, and J. Pruckler. 1987. Characterization of genes encoding type 1 fimbriae of Klebsiella pneumoniae, Salmonella typhimurium, and Serratia marcescens. Infect. Immun. 55281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connell, H., W. Agace, P. Klemm, M. Schembri, S. Marild, and C. Svanborg. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 939827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortes, G., N. Borrell, B. de Astorza, C. Gomez, J. Sauleda, and S. Alberti. 2002. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect. Immun. 702583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derbise, A., B. Lesic, D. Dacheux, J. M. Ghigo, and E. Carniel. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38113-116. [DOI] [PubMed] [Google Scholar]
- 9.Di Martino, P., N. Cafferini, B. Joly, and A. Darfeuille-Michaud. 2003. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 1549-16. [DOI] [PubMed] [Google Scholar]
- 10.Duncan, M. J., E. L. Mann, M. S. Cohen, I. Ofek, N. Sharon, and S. N. Abraham. 2005. The distinct binding specificities exhibited by enterobacterial type 1 fimbriae are determined by their fimbrial shafts. J. Biol. Chem. 28037707-37716. [DOI] [PubMed] [Google Scholar]
- 11.El Labany, S., B. K. Sohanpal, M. Lahooti, R. Akerman, and I. C. Blomfield. 2003. Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol. Microbiol. 491109-1118. [DOI] [PubMed] [Google Scholar]
- 12.Erlendsdottir, H., J. D. Knudsen, I. Odenholt, O. Cars, F. Espersen, N. Frimodt-Moller, K. Fuursted, K. G. Kristinsson, and S. Gudmundsson. 2001. Penicillin pharmacodynamics in four experimental pneumococcal infection models. Antimicrob. Agents Chemother. 451078-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fader, R. C., and C. P. Davis. 1980. Effect of piliation on Klebsiella pneumoniae infection in rat bladders. Infect. Immun. 30554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fader, R. C., and C. P. Davis. 1982. Klebsiella pneumoniae induced experimental pyelitis: the effect of piliation on infectivity. J. Urol. 128197-201. [DOI] [PubMed] [Google Scholar]
- 15.Favre-Bonte, S., B. Joly, and C. Forestier. 1999. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect. Immun. 67554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 1756186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlach, G. F., S. Clegg, N. J. Ness, D. L. Swenson, B. L. Allen, and W. A. Nichols. 1989. Expression of type 1 fimbriae and mannose-sensitive hemagglutinin by recombinant plasmids. Infect. Immun. 57764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerlach, G. F., S. Clegg, and B. L. Allen. 1989. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J. Bacteriol. 1711262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holden, N. J., M. Totsika, E. Mahler, A. J. Roe, K. Catherwood, K. Lindner, U. Dobrindt, and D. L. Gally. 2006. Demonstration of regulatory cross-talk between P fimbriae and type 1 fimbriae in uropathogenic Escherichia coli. Microbiology 1521143-1153. [DOI] [PubMed] [Google Scholar]
- 20.Hornick, D. B., B. L. Allen, M. A. Horn, and S. Clegg. 1991. Fimbrial types among respiratory isolates belonging to the family Enterobacteriaceae. J. Clin. Microbiol. 291795-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornick, D. B., B. L. Allen, M. A. Horn, and S. Clegg. 1992. Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect. Immun. 601577-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hultgren, S. J., W. R. Schwan, A. J. Schaeffer, and J. L. Duncan. 1986. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect. Immun. 54613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hvidberg, H., C. Struve, K. A. Krogfelt, N. Christensen, S. N. Rasmussen, and N. Frimodt-Moller. 2000. Development of a long-term ascending urinary tract infection mouse model for antibiotic treatment studies. Antimicrob. Agents Chemother. 44156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagnow, J., and S. Clegg. 2003. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 1492397-2405. [DOI] [PubMed] [Google Scholar]
- 25.Keane, W. F., and L. R. Freedman. 1967. Experimental pyelonephritis. XIV. Pyelonephritis in normal mice produced by inoculation of E. coli into the bladder lumen during water diuresis. Yale J. Biol. Med. 40231-237. [PMC free article] [PubMed] [Google Scholar]
- 26.Keynan, Y., and E. Rubinstein. 2007. The changing face of Klebsiella pneumoniae infections in the community. Int. J. Antimicrob. Agents 30385-389. [DOI] [PubMed] [Google Scholar]
- 27.Klemm, P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 51389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ko, W. C., D. L. Paterson, A. J. Sagnimeni, D. S. Hansen, A. Von Gottberg, S. Mohapatra, J. M. Casellas, H. Goossens, L. Mulazimoglu, G. Trenholme, K. P. Klugman, J. G. McCormack, and V. L. Yu. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 8160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane, M. C., A. N. Simms, and H. L. Mobley. 2007. Complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J. Bacteriol. 1895523-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langermann, S., S. Palaszynski, M. Barnhart, G. Auguste, J. S. Pinkner, J. Burlein, P. Barren, S. Koenig, S. Leath, C. H. Jones, and S. J. Hultgren. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276607-611. [DOI] [PubMed] [Google Scholar]
- 31.Langstraat, J., M. Bohse, and S. Clegg. 2001. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect. Immun. 695805-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawlor, M. S., J. Hsu, P. D. Rick, and V. L. Miller. 2005. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol. Microbiol. 581054-1073. [DOI] [PubMed] [Google Scholar]
- 33.Lawlor, M. S., C. O'Connor, and V. L. Miller. 2007. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 751463-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lederman, E. R., and N. F. Crum. 2005. Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am. J. Gastroenterol. 100322-331. [DOI] [PubMed] [Google Scholar]
- 35.Licht, T. R., K. A. Krogfelt, P. S. Cohen, L. K. Poulsen, J. Urbance, and S. Molin. 1996. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect. Immun. 643811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim, J. K., N. W. Gunther, H. Zhao, D. E. Johnson, S. K. Keay, and H. L. Mobley. 1998. In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect. Immun. 663303-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livrelli, V., C. De Champs, P. Di Martino, A. Darfeuille-Michaud, C. Forestier, and B. Joly. 1996. Adhesive properties and antibiotic resistance of Klebsiella, Enterobacter, and Serratia clinical isolates involved in nosocomial infections. J. Clin. Microbiol. 341963-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maayan, M. C., I. Ofek, O. Medalia, and M. Aronson. 1985. Population shift in mannose-specific fimbriated phase of Klebsiella pneumoniae during experimental urinary tract infection in mice. Infect. Immun. 49785-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madison, B., I. Ofek, S. Clegg, and S. N. Abraham. 1994. Type 1 fimbrial shafts of Escherichia coli and Klebsiella pneumoniae influence sugar-binding specificities of their FimH adhesins. Infect. Immun. 62843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClain, M. S., I. C. Blomfield, and B. I. Eisenstein. 1991. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 1735308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCormick, B. A., D. P. Franklin, D. C. Laux, and P. S. Cohen. 1989. Type 1 pili are not necessary for colonization of the streptomycin-treated mouse large intestine by type 1-piliated Escherichia coli F-18 and E. coli K-12. Infect. Immun. 573022-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCormick, B. A., P. Klemm, K. A. Krogfelt, R. L. Burghoff, L. Pallesen, D. C. Laux, and P. S. Cohen. 1993. Escherichia coli F-18 phase locked ‘on’ for expression of type 1 fimbriae is a poor colonizer of the streptomycin-treated mouse large intestine. Microb. Pathog. 1433-43. [DOI] [PubMed] [Google Scholar]
- 43.Merino, S., M. Altarriba, L. Izquierdo, M. M. Nogueras, M. Regue, and J. M. Tomas. 2000. Cloning and sequencing of the Klebsiella pneumoniae O5 wb gene cluster and its role in pathogenesis. Infect. Immun. 682435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montgomerie, J. Z. 1979. Epidemiology of Klebsiella and hospital-associated infections. Rev. Infect. Dis. 1736-753. [DOI] [PubMed] [Google Scholar]
- 45.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 2821494-1497. [DOI] [PubMed] [Google Scholar]
- 46.Nassif, X., and P. J. Sansonetti. 1986. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oelschlaeger, T. A., and B. D. Tall. 1997. Invasion of cultured human epithelial cells by Klebsiella pneumoniae isolated from the urinary tract. Infect. Immun. 652950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Podschun, R., P. Heineken, and H. G. Sonntag. 1987. Haemagglutinins and adherence properties to HeLa and intestine 407 cells of Klebsiella pneumoniae and Klebsiella oxytoca isolates. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 263585-593. [DOI] [PubMed] [Google Scholar]
- 49.Podschun, R., and H. Sahly. 1991. Hemagglutinins of Klebsiella pneumoniae and K. oxytoca isolated from different sources. Zentralbl. Hyg. Umweltmed. 19146-52. [PubMed] [Google Scholar]
- 50.Podschun, R., D. Sievers, A. Fischer, and U. Ullmann. 1993. Serotypes, hemagglutinins, siderophore synthesis, and serum resistance of Klebsiella isolates causing human urinary tract infections. J. Infect. Dis. 1681415-1421. [DOI] [PubMed] [Google Scholar]
- 51.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell, B. K., and S. Clegg. 1983. Construction and expression of recombinant plasmids encoding type 1 fimbriae of a urinary Klebsiella pneumoniae isolate. Infect. Immun. 391122-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9218-228. [DOI] [PubMed] [Google Scholar]
- 54.Rosen, D. A., J. S. Pinkner, J. M. Jones, J. N. Walker, S. Clegg, and S. J. Hultgren. 14 April 2008. Utilization of an IBC pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect. Immun. doi: 10.1128/IAI.00090-08. [DOI] [PMC free article] [PubMed]
- 55.Saeland, E., G. Vidarsson, and I. Jonsdottir. 2000. Pneumococcal pneumonia and bacteremia model in mice for the analysis of protective antibodies. Microb. Pathog. 2981-91. [DOI] [PubMed] [Google Scholar]
- 56.Sahly, H., R. Podschun, T. A. Oelschlaeger, M. Greiwe, H. Parolis, D. Hasty, J. Kekow, U. Ullmann, I. Ofek, and S. Sela. 2000. Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect. Immun. 686744-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwan, W. R., M. T. Beck, S. J. Hultgren, J. Pinkner, N. L. Woolever, and T. Larson. 2005. Down-regulation of the kps region 1 capsular assembly operon following attachment of Escherichia coli type 1 fimbriae to d-mannose receptors. Infect. Immun. 731226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shankar-Sinha, S., G. A. Valencia, B. K. Janes, J. K. Rosenberg, C. Whitfield, R. A. Bender, T. J. Standiford, and J. G. Younger. 2004. The Klebsiella pneumoniae O antigen contributes to bacteremia and lethality during murine pneumonia. Infect. Immun. 721423-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharon, N. 1987. Bacterial lectins, cell-cell recognition and infectious disease. FEBS Lett. 217145-157. [DOI] [PubMed] [Google Scholar]
- 60.Simoons-Smit, A. M., A. M. Verweij-van Vught, and D. M. MacLaren. 1986. The role of K antigens as virulence factors in Klebsiella. J. Med. Microbiol. 21133-137. [DOI] [PubMed] [Google Scholar]
- 61.Sohanpal, B. K., S. El Labany, M. Lahooti, J. A. Plumbridge, and I. C. Blomfield. 2004. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 10116322-16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sokurenko, E. V., V. Chesnokova, D. E. Dykhuizen, I. Ofek, X. R. Wu, K. A. Krogfelt, C. Struve, M. A. Schembri, and D. L. Hasty. 1998. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. USA 958922-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Struve, C., and K. A. Krogfelt. 1999. In vivo detection of Escherichia coli type 1 fimbrial expression and phase variation during experimental urinary tract infection. Microbiology 1452683-2690. [DOI] [PubMed] [Google Scholar]
- 64.Struve, C., C. Forestier, and K. A. Krogfelt. 2003. Application of a novel multi-screening signature-tagged mutagenesis assay for identification of Klebsiella pneumoniae genes essential in colonization and infection. Microbiology 149167-176. [DOI] [PubMed] [Google Scholar]
- 65.Struve, C., and K. A. Krogfelt. 2003. Role of capsule in Klebsiella pneumoniae virulence: lack of correlation between in vitro and in vivo studies. FEMS Microbiol. Lett. 218149-154. [DOI] [PubMed] [Google Scholar]
- 66.Struve, C., and K. A. Krogfelt. 2004. Pathogenic potential of environmental Klebsiella pneumoniae isolates. Environ. Microbiol. 6584-590. [DOI] [PubMed] [Google Scholar]
- 67.Tarkkanen, A. M., B. L. Allen, B. Westerlund, H. Holthofer, P. Kuusela, L. Risteli, S. Clegg, and T. K. Korhonen. 1990. Type V collagen as the target for type-3 fimbriae, enterobacterial adherence organelles. Mol. Microbiol. 41353-1361. [DOI] [PubMed] [Google Scholar]
- 68.Tarkkanen, A. M., B. L. Allen, P. H. Williams, M. Kauppi, K. Haahtela, A. Siitonen, I. Orskov, F. Orskov, S. Clegg, and T. K. Korhonen. 1992. Fimbriation, capsulation, and iron-scavenging systems of Klebsiella strains associated with human urinary tract infection. Infect. Immun. 601187-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Valenski, M. L., S. L. Harris, P. A. Spears, J. R. Horton, and P. E. Orndorff. 2003. The product of the fimI gene is necessary for Escherichia coli type 1 pilus biosynthesis. J. Bacteriol. 1855007-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, J. H., Y. C. Liu, S. S. Lee, M. Y. Yen, Y. S. Chen, J. H. Wang, S. R. Wann, and H. H. Lin. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 261434-1438. [DOI] [PubMed] [Google Scholar]
- 71.Williams, P., and J. M. Tomas. 1990. The pathogenicity of Klebsiella pneumoniae. Rev. Med. Microbiol. 1196-204. [Google Scholar]