Abstract
IbeA in extraintestinal pathogenic Escherichia coli (ExPEC) strains was previously described for its role in invasion. Here we investigated the role of IbeA and IbeT, encoded by a gene located downstream of ibeA, in the adhesion of the avian ExPEC strain BEN2908 to human brain microvascular endothelial cells (HBMEC). The ΔibeA mutant was less adhesive to HBMEC than the wild-type strain BEN2908 was. Because strain BEN2908 also expresses type 1 fimbriae, we measured the adhesion specifically due to IbeA by comparing the adhesive properties of a Δfim derivative of strain BEN2908 to those of a double Δfim ΔibeA mutant. No differences were observed, indicating that the reduction of adhesion in BEN2908 ΔibeA could be due to a decrease in type 1 fimbria expression. We indeed showed that the decreased adhesion of BEN2908 ΔibeA was correlated with a decrease in type 1 fimbria expression. Accordingly, more bacteria had a fim promoter orientated in the off position in a culture of BEN2908 ΔibeA than in a culture of BEN2908. Expression of fimB and fimE, two genes encoding recombinases participating in controlling the orientation of the fim promoter, was decreased in BEN2908 ΔibeA. A reduction of type 1 fimbria expression due to a preferential orientation of the fim promoter in the off position was also seen in an ibeT mutant of strain BEN2908. We finally suggest a role for IbeA and IbeT in modulating the expression of type 1 fimbriae through an as yet unknown mechanism.
Extraintestinal pathogenic Escherichia coli (ExPEC) strains are responsible for a wide range of diseases in humans and animals. In humans, almost 80% of urinary tract infections, as well as almost 20% of newborn meningitis cases, are caused by ExPEC strains (23, 45). In chickens, ExPEC strains are responsible for avian colibacillosis, which results in different syndromes, ranging from superficial dermatitis to respiratory infections that frequently develop into systemic infections (12, 32). Phylogenetic studies have shown that avian ExPEC strains are closely related to human ExPEC strains (40, 44). Additionally, avian ExPEC strains share common virulence genes with ExPEC strains isolated from patients with urinary tract infections or meningitis (16, 40, 44). Several potential virulence factors have been described for ExPEC strains. These virulence factors confer different properties, such as adhesion to host cells through type 1 and P fimbriae, iron acquisition, toxic activity toward eukaryotic cells (cytotoxins), and resistance to host defenses.
ibeA is an ExPEC virulence gene that was first described for strain RS218, isolated from a patient with human newborn meningitis (26, 46). ibeA encodes a 50-kDa protein and is located on the GimA genomic island. This island is inserted between yjiD and yjiE and is predicted to contain four operons, three of them potentially involved in carbohydrate metabolism (24). In addition to ibeA, the GimA4 operon is predicted to contain two other genes, ibeR and ibeT. ibeR encodes a potential transcriptional activator homologous to the DhaR protein of the Dha operon of E. coli, while ibeT is supposed to encode a protein belonging to the Na+/H+ antiporter family (24).
Huang et al. showed that the deletion of ibeA in strain RS218 decreases its virulence in an infant rat model of meningitis (26). Invasion assays with human brain microvascular endothelial cells (HBMEC) also indicated that the ibeA mutant strain is less invasive than the wild-type (WT) strain (25). Recent findings by Zou et al. indicate that ibeT is also involved in adhesion and invasion of HBMEC by strain RS218 and is required for the development of meningitis in infant rats (54).
We previously demonstrated that ibeA also plays a role in the pathogenicity of the avian ExPEC strain BEN2908 for chickens (18) and showed that its expression is increased in the early stage of chicken infection (8). ibeA was detected in approximately 20% of ExPEC strains of avian origin and is more prevalent in O2, O18, and O88 strains. In vivo studies showed that chickens inoculated with an ibeA mutant had significantly fewer bacteria in the blood and in the liver than did those inoculated with the WT strain. Furthermore, in HBMEC, an ibeA derivative of strain BEN2908 is less invasive than the WT strain (18).
Despite these numerous studies, the precise role of ibeA in the invasion process remains to be clarified. Based on the hypothesis that IbeA is an invasin, several putative cellular receptors for IbeA have been characterized, including Ibe10R on bovine BMEC (43) and vimentin (55) and the PSF protein (56) on HBMEC. Although these interactions with surface receptors were described for the invasion process, the possibility remains that they also participate in the adhesion of bacteria to eukaryotic cells. As an example, Khan et al. have recently shown that preventing adhesion of strain RS218 results in less invasion (28).
Paradoxically, IbeA is predicted to be a cytoplasmic protein by several prediction programs, such as Psort (see computed predictions at http://db.psort.org/), TMBETA-SVM (http://tmbeta-svm.cbrc.jp/), and BOMP (http://www.bioinfo.no/tools/bomp). No putative signal secretion sequences or lipoprotein signal sequence is found in the IbeA sequence (51). Additionally, IbeA contains a putative flavin adenine dinucleotide binding domain (E value = 6.8e−06 [http://pfam.sanger.ac.uk/]), which suggests an enzymatic activity. These predictions apparently contradict previous reports concerning interactions of IbeA with cell surface receptors, yet these interactions were described only in vitro, using purified glutathione S-transferase- or His6-labeled IbeA protein in pull-down or ligand overlay experiments. Despite a recent publication concerning protein folding intermediates of IbeA in solution (38), no report clearly demonstrating that IbeA is indeed located in the outer membrane of E. coli has been published so far. Concerning IbeT, bioinformatic analysis has shown that it belongs to the Na+/H+ antiporter family, a family of proteins located in the inner membranes of gram-negative bacteria (41).
Characterization of a putative adhesion mediated by IbeA will have to take into account the adhesion mediated by other known adhesins so that only the contribution of IbeA to adhesion is measured. Among adhesins characterized so far for ExPEC strains are type 1 fimbriae. Type 1 fimbriae are produced by many strains of E. coli, including nonpathogenic strains, but were proven to be an important determinant involved in binding and invasion of eukaryotic cells by various pathogenic E. coli strains (3, 6, 28, 35, 49). In vivo, type 1 fimbriae are involved in the colonization of the trachea in avian colibacillosis and might be required for interaction with lung epithelial cells (31, 36). Several reports have also shown their role in the development of urinary tract infections (19, 20, 37).
Nine genes, located on the fim operon, are necessary for the biosynthesis of type 1 fimbriae. Their synthesis requires, besides the assembly proteins FimC and FimD, the major subunit FimA and the minor subunits FimF, FimG, and FimH. FimH is the adhesin and is the component responsible for the mannose-sensitive binding ability of type 1 fimbriae (39). The expression of the fimAICDFGH operon is phase variable and is mediated by a 314-bp invertible element (IE) that contains the promoter of the fim operon (2). Depending on the orientation of this promoter (on or off), it drives (or not) the transcription of the fim genes and allows (or not) the expression of type 1 fimbriae. The percentage of bacteria with a promoter in the on orientation therefore determines the percentage of bacteria that express type 1 fimbriae within a bacterial culture. The switch of the IE is regulated by several environmental signals, such as temperature, osmolarity, pH, and amino acids, and is mediated by site-specific recombinases, with the contributions of other DNA-binding proteins, such as H-NS, IHF, and Lrp (5). FimB and FimE are the most-studied site-specific recombinases involved in IE switching (30). Much less is known concerning more recently described recombinases, such as IpuA, IpuB, and IpbA (also known as HbiF or FimX) (7, 21, 53).
In this work, the real contribution of IbeA to the adhesion of strain BEN2908 to HBMEC was investigated. Our results led us to consider the possibility that expression of type 1 fimbriae is modified when ibeA or ibeT is deleted.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are described in Table 1. Bacteria were routinely grown in Luria-Bertani (LB) broth at 37°C. Ampicillin (100 μg ml−1), kanamycin (50 μg ml−1), and nalidixic acid (30 μg ml−1) were used when necessary.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristics or sequence | Reference or source |
|---|---|---|
| E. coli strains | ||
| BEN2908 | Avian ExPEC O2:K1:H5 Nalr | 13 |
| BEN2908 ΔibeA | This study | |
| BEN2908 ΔibeT | This study | |
| DM34 | BEN2908 Δfim | 35 |
| DM34 ΔibeA | BEN2908 Δfim ΔibeA | This study |
| Plasmids | ||
| pUC13 | 52 | |
| pUC23A | ibeA gene cloned into pUC13 | 25 |
| pKD4 | 11 | |
| pKD46 | 11 | |
| pCP20 | 11 | |
| Primers | ||
| ibeA primers | ||
| ibeA-5F | TATTAGCATGATGTTGCTTG | |
| ibeA-5R | TGCCAACAACCAACACGATC | |
| ibeT primers | ||
| MC 6 | AAAATAACACTACCGTTTCACAGG | |
| MC 49 | GACATTAACGCGGGATGG | |
| IE primers | ||
| Ch1-F | AGTAATGCTGCTCGTTTTGC | 49 |
| Ch1-R | GACAGAGCCGACAGAACAAC | 49 |
| frr primers | ||
| PG 198 | GCGTAGAAGCGTTCAAAACC | |
| PG 199 | CAAGATCGGACGCCATAATC | |
| fimB primers | ||
| PG 340 | CCTGACCCATAGTGAAATCG | |
| PG 341 | CTTTGCCTTAAGATCAATATC | |
| fimE primers | ||
| PG 342 | GGCATATCGGCATGGGATGC | |
| PG 343 | CGTTCCTGGGTCCAGCGTTC | |
| ipbA primers | ||
| MC 38 | CACACAGGATGAAGTCTACAG | |
| MC 39 | CCGTGAATGAAAGCC | |
| ipuA primers | ||
| MC 40 | AGTTTCTGCTGCGGG | |
| MC 41 | TAAGACGGGTATCCACAC | |
| ipuB primers | ||
| MC 42 | CTCTTTGTTTCTTTCTTGTC | |
| MC 43 | TATTAGTCTGGTGTC | |
| ibeA mutant primers | ||
| PG 1 | GTATGGAATTTTATCTGGAACCCGCTCGTAATATACCTGTACTGGCAACC | |
| PG 2 | AAGACTTTTACGCCATTTTGCTGTAAGCGTTCCTGTACCGTCTGAAT | |
| ibeT mutant primers | ||
| MC 36 | CAGTTTTTATGTCGCGCTTACACCGATCATTTTTATGATGATCGTGTTGGGTGTAGGCTGGAGCTGCTTC | |
| MC 37 | CGGGATGGAGATTAATCCATCCGGCAAACGCATTTATTATCTTGAATAACCATATGAATATCCTCCTTAG |
Cell lines and culture.
HBMEC were kindly provided by C. Kieda (CNRS, Orléans, France) (29). HBMEC were grown in Opti-MEM (Invitrogen) containing 2% heat-inactivated fetal bovine serum at 37°C and 5% CO2.
DNA techniques.
Restriction endonucleases and modification enzymes (New England Biolabs) were used according to the manufacturer's instructions. DNA fragments were purified from agarose gels by use of a Nucleo-spin Extract II purification kit (Macherey-Nagel).
Primers used in this study are described in Table 1. PCRs were performed with an Applied Biosystems model 9700 apparatus, using 1 U Taq DNA polymerase from New England Biolabs in 1× buffer, a 200 μM concentration of each deoxynucleoside triphosphate, a 0.8 μM concentration of each primer, and 10 ng chromosomal DNA in a 50-μl reaction volume. Cycling conditions were as follows: 1 cycle of 5 min at 95°C; 30 cycles of 10 s at 95°C, 10 s at 52°C, and 1 min/kb at 72°C; and a final extension of 5 min at 72°C. PCR products were separated in 1% agarose gels for 1 h at 10 V/cm of gel.
Deletions of ibeA and ibeT were obtained as described by Datsenko and Wanner, using primers PG1 and PG2 for deletion of ibeA and MC36 and MC37 for deletion of ibeT (11). The replacement of ibeA was confirmed by PCR, using primers IbeA-5F and IbeA-5R; that of ibeT was confirmed with primers MC6 and MC49. The Kanr cassette was then removed using plasmid pCP20, leaving a scar of 80 bp. The deletions of ibeA and ibeT were confirmed by PCR and sequencing, using primers ibeA-5F/ibeA-5R and MC6/MC49, respectively.
Adhesion assay.
E. coli adhesion assays were performed on HBMEC. Cells were distributed in 24-well plates. When the cell density reached 5 × 105 cells per well, the cells were infected by the studied strain at a multiplicity of infection (MOI) of 10. Bacterial inocula were prepared from an exponentially growing culture grown at 37°C with agitation. Upon reaching exponential phase (optical density at 600 nm of 0.5), bacteria were diluted to the desired concentration in Opti-MEM medium and immediately used to infect cell cultures. The inoculum size was determined by colony plate counting for each experiment. A final concentration of 2% d-mannose was added to the inoculum, when required, before bacteria were added to the HBMEC. After 90 min of infection, monolayers were washed with Opti-MEM and then lysed with cold sterile demineralized water for 30 min at 4°C. The growth in Opti-MEM of strain BEN2908 and its ibeA and ibeT derivatives was equivalent. Adherent and intracellular bacteria were enumerated by plating 10-fold serial dilutions on LB agar. From previous experiments, intracellular bacteria account for only approximately 2% of bacteria recovered (18). We thus considered that counted bacteria represented mostly adherent bacteria. The percentage of adhesion was thus calculated as follows: 100 × (number of recovered bacteria after 90 min of infection/number of bacteria inoculated). The results were expressed as relative adhesion (percentage of adhesion for experimental strain versus percentage of adhesion of BEN2908 strain). Each assay was performed in triplicate and repeated at least three times.
Immunodot blot assay.
Approximately 3 × 108 bacteria in exponential phase were pelleted and resuspended in 1 ml of phosphate-buffered saline (PBS). Five microliters of the bacterial suspension was then spotted onto a nitrocellulose membrane. After being blocked with PBS-1% bovine serum albumin (BSA), the membrane was incubated in PBS-1% BSA in the presence of the anti-type 1 fimbria antiserum 4168 at a 1/2,500 dilution (14) for 1 h, washed, and then incubated in PBS-1% BSA with a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (Sigma) used at a dilution of 1/10,000. The signals were detected using a Chemi-Smart 5000 cooled charge-coupled device camera (Vilber-Lourmat, France) and Supersignal West Pico chemiluminescent substrate (Pierce). This experiment was performed in triplicate. Quantification of the spots was done with Bio-Profil Bio-1D++ software (Vilber-Lourmat, France), and the background signal obtained with the afimbriate strain DM34 was subtracted from each intensity. The results were expressed as relative intensities (percentage of intensity for the tested strain versus percentage of intensity for strain BEN2908).
IE orientation assay.
The orientation of the IE was tested as previously described (49). A PCR was performed with primers Ch1-F and Ch1-R, yielding a 602-bp fragment containing the IE. This product was subjected to restriction digestion by SnaBI (New England Biolabs). The digested PCR products were separated in a 2% agarose gel. Fragments of 404 bp and 198 bp (on orientation) or 442 bp and 160 bp (off orientation) were observed.
Real-time quantitative PCR (qPCR) analysis.
Total RNA was extracted from 200 microliters of bacteria in liquid culture (optical density at 600 nm of 0.5). Bacteria were mixed with 1 ml of Trizol (Invitrogen) and placed in lysing matrix B tubes (MP Biomedicals). Bacteria were lysed by shaking tubes for 45 s at speed 6 in a Fastprep FP-120 apparatus (MP Biomedicals). One hundred microliters of 1-bromo-3-chloro-propane was then added, and tubes were centrifuged for 10 min at 10,000 × g. The aqueous phase was recovered, and RNAs were precipitated with 500 μl of isopropyl alcohol and washed with 500 μl of 75% ethanol. RNAs were then resuspended in 50 μl of diethyl pyrocarbonate-treated water, treated with DNase I, and purified on RNeasy minicolumns (Qiagen). The quality of the RNAs was verified by agarose gel electrophoresis.
Quantitative reverse transcriptase PCR (RT-PCR) was performed as described by Chouikha et al. (8). Briefly, gene-specific reverse transcription of RNAs was performed with Superscript RT III (Invitrogen), using primer PG 199 for frr (housekeeping gene), PG 341 for fimB, and PG 343 for fimE. Samples without RT were concurrently prepared and analyzed for the absence of contaminating genomic DNA. Real-time qPCR analysis was performed in an iCycler system (Bio-Rad), using Absolute qPCR SYBR green mix (ABgene). cDNAs obtained as described above were diluted twofold in nuclease-free distilled water. Four microliters was used for qPCR. The PCR program consisted of 35 amplification/quantification cycles of 95°C for 15 s and 60°C for 1 min, with signal acquisition at the end of each cycle. Primers used for qPCR were as follows: PG198/PG199 for frr, PG340/PG341 for fimB, and PG342/PG343 for fimE. Equation 1 from Pfaffl was used to analyze the statistical significance of the data, using frr as a housekeeping gene standard (34, 42).
Statistical analysis.
Statistical analysis of adhesion and type 1 fimbria quantification were done by applying the Mann-Whitney test. Exact P values were calculated with StatXact software (version 5.0; Cytel Software, Cambridge, MA).
RESULTS
ibeA is required for adhesion of strain BEN2908 to HBMEC.
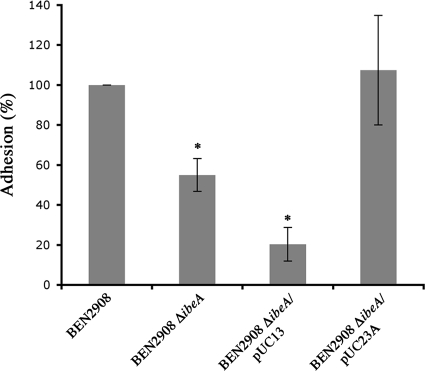
A previous report has shown that inactivation of ibeA reduces the invasion of HBMEC by strain BEN2908 (18). Given that IbeA might directly interact with cell surface receptors, we tested the possibility that the reduced invasion of an ibeA mutant could be due to differences in adhesion. The adhesion to HBMEC of strain BEN2908 was thus compared to that of strain BEN2908 ΔibeA. Figure 1 shows that adhesion of strain BEN2908 ΔibeA was reduced approximately twofold compared to that of strain BEN2908 (55% ± 8.17% versus 100%; P < 0.01). To confirm that the decreased adhesion was linked to the deletion of ibeA, we transformed strain BEN2908 ΔibeA with plasmid pUC23A (25), carrying ibeA, and with plasmid pUC13 (empty vector). Adhesion was restored by expression of ibeA from plasmid pUC23A (107.46% ± 27.41%), while the empty plasmid pUC13 had a negative effect on adhesion, reducing it about 2.7 times (20.35% ± 8.35%). Altogether, these results indicate that IbeA is involved in the adhesion of strain BEN2908 to HBMEC.
FIG. 1.
Adhesion of BEN2908 ΔibeA to HBMEC is reduced. Confluent HBMEC were incubated with different strains at an MOI of 10. After 90 min, cells were washed and bacteria were counted after lysis of HBMEC with cold water. The percentage of adhesion of each strain was calculated from three independent experiments, as follows: 100 × (number of adhesive bacteria/number of bacteria inoculated). The results are expressed as relative adhesion (percentage of adhesion of test strain versus percentage of adhesion of BEN2908 strain). Strains tested are indicated on the graph. The assay was performed three times in triplicate. *, adhesion was significantly different (Mann-Whitney test; P < 0.01) from that of strain BEN2908.
Inactivation of ibeA in a fim mutant does not decrease adhesion.
From previous studies concerning the adhesive properties of strain BEN2908, we knew that type 1 fimbriae are important for the adhesion of this strain to eukaryotic cells (35, 36). Therefore, we hypothesized that the adhesion measured above was not only mediated by IbeA but could also involve type 1 fimbriae.
In order to analyze only the contribution of IbeA to the adhesion of BEN2908, we measured the percentage of adhesion specifically due to IbeA by using strain DM34, a derivative of strain BEN2908 deleted of the fim operon (35, 36). As for strain BEN2908, we deleted ibeA from DM34, yielding strain DM34 ΔibeA. The adhesion activities on HBMEC of strains BEN2908, DM34, and DM34 ΔibeA were then compared.
As expected, the adhesion of strain DM34 was reduced almost fivefold compared to that of strain BEN2908 (100% versus 23.98% ± 9.66%; P < 0.01) (Fig. 2A). These results confirmed that the loss of type 1 fimbriae was associated with a loss of the adhesive properties of strain BEN2908. Surprisingly, adhesion of strain DM34 ΔibeA was not significantly different from that of strain DM34 (25.28% ± 10.41% versus 23.98% ± 9.66%) (Fig. 2A). This suggests that in the absence of type 1 fimbriae, deletion of ibeA did not affect adhesion on HBMEC. We confirmed these results by performing adhesion assays with strains BEN2908 and BEN2908 ΔibeA in the presence of 2% d-mannose to prevent type 1 fimbria-mediated adhesion, and we found no difference in adhesion between strains BEN2908 and BEN2908 ΔibeA (Fig. 2B), showing that when we prevent adhesion through type 1 fimbriae, IbeA plays very little, if any, role in the adhesive process.
FIG. 2.
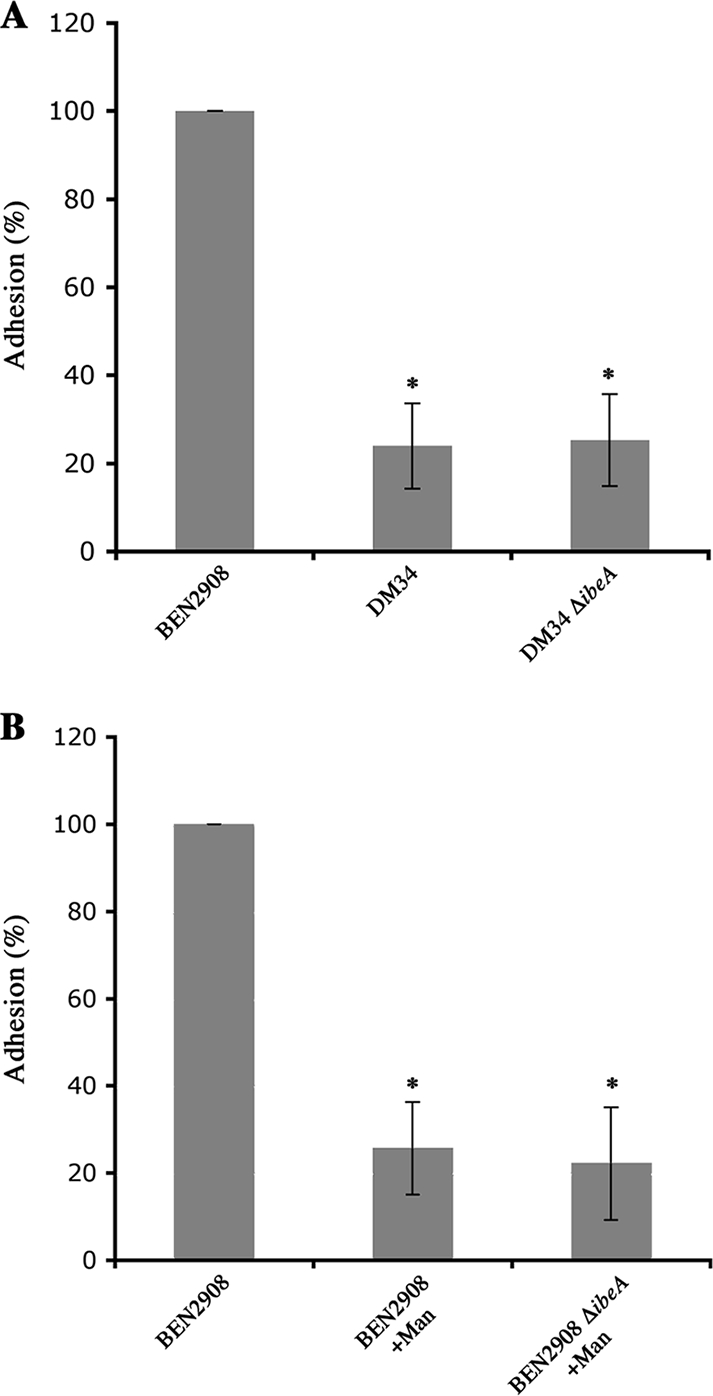
Adhesion of bacteria to HBMEC in the presence or absence of d-mannose. Confluent HBMEC were incubated with different strains at an MOI of 10 in the absence (A) or presence (B) of 2% d-mannose. After 90 min, cells were washed and bacteria were counted after lysis of HBMEC with cold water. The percentage of adhesion of each strain was calculated from three independent experiments, as follows: 100 × (number of adhesive bacteria/number of bacteria inoculated). The results are expressed as relative adhesion (percentage of adhesion of test strain versus percentage of adhesion of BEN2908 strain). Strains tested are indicated on the graphs. The assay was performed three times in triplicate. *, adhesion was significantly different (Mann-Whitney test; P < 0.01) from that of strain BEN2908.
This result led us to suspect that the loss of the adhesive properties of BEN2908 ΔibeA was not the consequence of the loss of an interaction between IbeA and HBMEC surface receptors but was rather due to a decrease of type 1 fimbria expression in this mutant.
Decreased expression of type 1 fimbriae on the surface of BEN2908 ΔibeA.
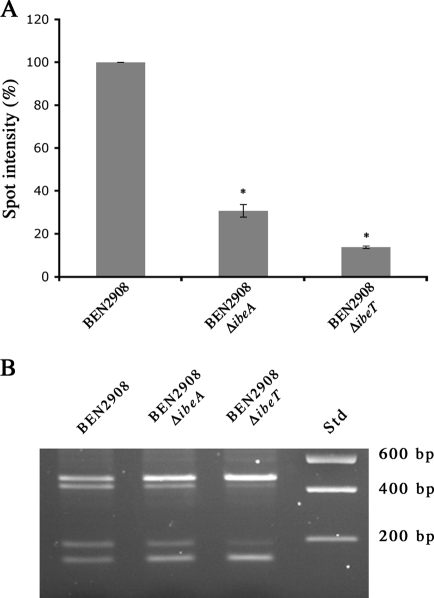
We thus compared the expression of type 1 fimbriae in strains BEN2908 and BEN2908 ΔibeA by using a type 1 fimbria antiserum previously obtained in our laboratory (14). Figure 3 shows that more type 1 fimbriae were detected in a culture of strain BEN2908 than in a culture of strain BEN2908 ΔibeA (100% versus 22.6% ± 8.5%; P < 0.01). This phenotype was complemented, albeit partially, by plasmid pUC23A (41.5% ± 15.7%): although such a level is significantly lower than that observed with strain BEN2908 (P < 0.01), it is significantly higher than that of the ibeA mutant (P < 0.02). In agreement with the adhesion results, the number of type 1 fimbriae in strain BEN2908 ΔibeA containing plasmid pUC13 (1% ± 5.60%) was almost null, as the signal detected was equivalent to that of the negative control strain DM34. Introduction of plasmid pUC13 into strain BEN2908 also reduced type 1 fimbria expression, but only to a level similar to that of strain BEN2908 ΔibeA containing plasmid pUC23A (data not shown). Since expression of type 1 fimbriae is phase variable, these data suggest that a larger number of bacteria express type 1 fimbriae on their surfaces in a culture of strain BEN2908 than in a culture of BEN2908 ΔibeA.
FIG. 3.
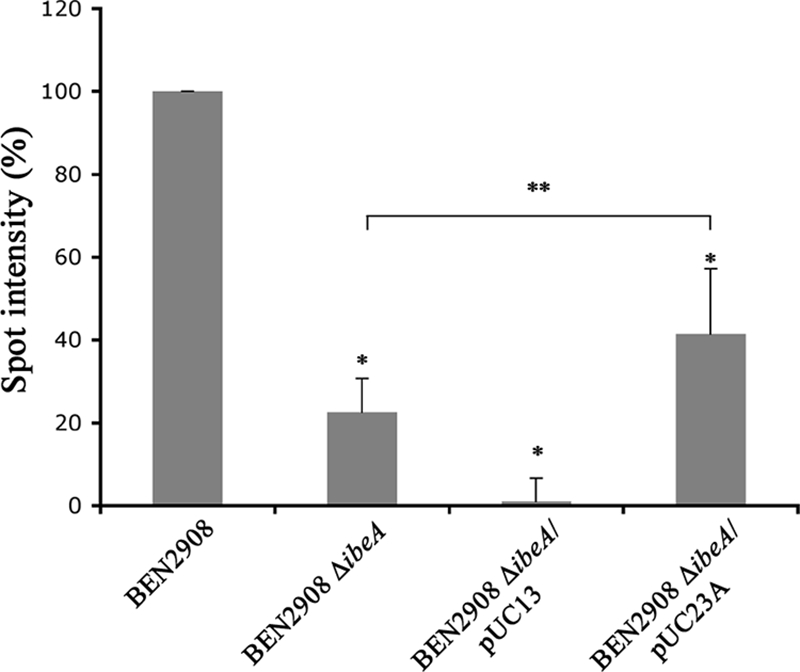
Surface expression of type 1 fimbriae is decreased in BEN2908 ΔibeA. Five-microliter samples of exponential-phase cultures of strains BEN2908, BEN2908 ΔibeA, BEN2908 ΔibeA/pUC13, BEN2908 ΔibeA/pUC23A, and DM34 were spotted on a nitrocellulose membrane. Type 1 fimbriae were detected using an anti-type 1 fimbriae polyclonal antibody. Quantification of the spots was performed using Bio1D++, and the background value obtained with the fim mutant DM34 was subtracted. Quantification results are expressed as percentages of the quantification obtained with strain BEN2908 and are averages for three experiments. *, spot intensity was significantly different (Mann-Whitney test; P < 0.01) from that of strain BEN2908; **, spot intensity was significantly different (Mann-Whitney test; P < 0.02) from that of strain BEN2908 ΔibeA.
The IE is preferentially in the off orientation in BEN2908 ΔibeA.
Phase variation of type 1 fimbria expression is controlled by switching the orientation of an IE containing the fim promoter. To confirm that the decreased quantity of type 1 fimbriae in BEN2908 ΔibeA was due to a larger proportion of bacteria not expressing these fimbriae, we used a PCR-based assay to characterize the orientation of the IE (33, 49).
In a population of WT strain BEN2908 cells, we observed digestion patterns indicating that the IE was present in both orientations (Fig. 4). In a population of strain BEN2908 ΔibeA cells, we observed both restriction profiles, but digestion fragments corresponding to the off orientation were more intense than the one corresponding to the on orientation. This indicates that there was more IE in the off orientation in the ibeA mutant than in the WT strain. This phenotype was complemented by introduction of plasmid pUC23A into strain BEN2908 ΔibeA. In agreement with the results presented above, introduction of pUC13 into the mutant strain BEN2908 ΔibeA increased the amount of IE in the off orientation.
FIG. 4.
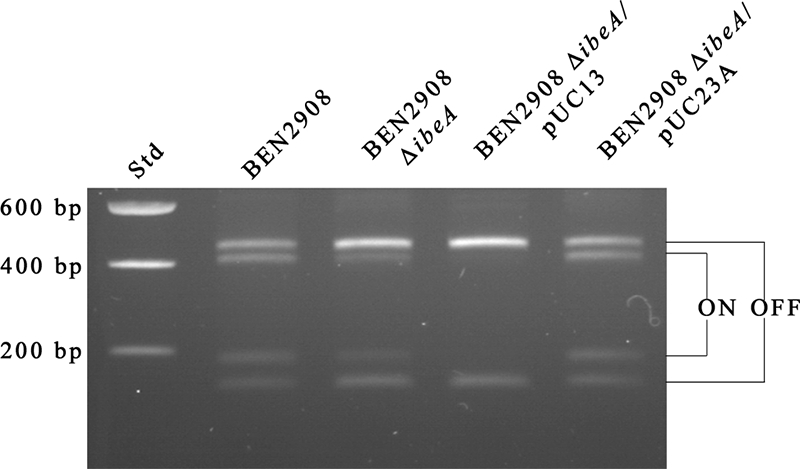
The fim promoter is preferentially in the off position in an ΔibeA mutant. The IE was PCR amplified with primers Ch1-F and Ch1-R from chromosomal DNAs of strains BEN2908, BEN2908 ΔibeA, BEN2908 ΔibeA/pUC13, and BEN2908 ΔibeA/pUC23A. The PCR products were digested by SnaBI, and restriction fragments were separated in 2% agarose gels. Std, DNA size standard.
These results clearly demonstrated that a deletion of ibeA increased the proportion of IE in the off orientation, resulting in an overall decreased expression of type 1 fimbriae at the surfaces of the bacteria.
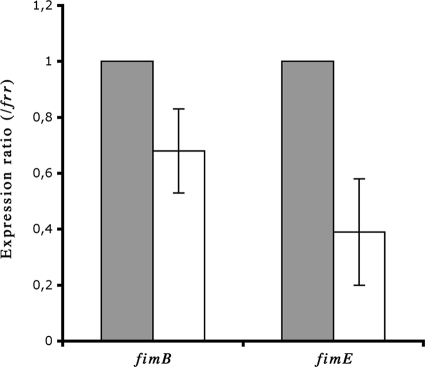
Expression of recombinases FimB and FimE is reduced in strain BEN2908 ΔibeA.
Since the orientation of the IE is in part determined by the recombinases FimB and FimE, we tested whether the expression of these recombinases might be disturbed in an ibeA mutant. We quantified the expression of fimB and fimE by quantitative RT-PCR, using frr as a housekeeping gene (8, 34). The expression of fimB and fimE in strain BEN2908 ΔibeA was reduced compared to their expression in strain BEN2908 (Fig. 5). Using specific primers, we showed that strain BEN2908 also possessed the ipbA gene but not the ipuA or ipuB gene, all of which encode recently described recombinases. However, expression of ipbA was too low to detect any significant change (data not shown).
FIG. 5.
Quantification of fimB and fimE expression. Expression of fimB and fimE in strains BEN2908 and BEN2908 ΔibeA was measured by quantitative real-time PCR as described in Materials and Methods, using the frr gene as a housekeeping gene standard. Results are relative expression ratios compared to expression in the WT strain. Gray bars, BEN2908; white bars, BEN2908 ΔibeA.
Inactivation of ibeT also leads to decreased expression of type 1 fimbriae.
Recently, Zou et al. showed that inactivation of ibeT also causes less adhesion of strain RS218 to HBMEC (54). We therefore assessed whether deletion of ibeT in strain BEN2908 also results in a decrease in type 1 fimbria expression. Inactivation of ibeT reduced the number of type 1 fimbriae on the surfaces of the bacteria, to only 13.5% of the level for strain BEN2908 (Fig. 6A). Furthermore, the IE was almost exclusively in the off orientation (Fig. 6B). Inactivation of ibeT therefore had a more pronounced effect on type 1 fimbria expression than did inactivation of ibeA.
FIG. 6.
Deletion of ibeT decreases type 1 fimbria expression by increasing the proportion of bacteria with the IE in the off orientation. (A) Five-microliter samples of exponential-phase cultures of strains BEN2908, BEN2908 ΔibeT, BEN2908 ΔibeA, and DM34 were spotted on a nitrocellulose membrane. Type 1 fimbriae were detected using an anti-type 1 fimbriae polyclonal antibody. *, spot intensity was significantly different (Mann-Whitney test; P < 0.01) from that of strain BEN2908. (B) The IE was PCR amplified with primers Ch1-F and Ch1-R from chromosomal DNAs of strains BEN2908, BEN2908 ΔibeT, and BEN2908 ΔibeA. The PCR products were digested by SnaBI, and restriction fragments were separated in 2% agarose gels. Std, DNA size standard.
DISCUSSION
Previous studies have demonstrated the importance of IbeA in invasion of HBMEC and in the crossing of the blood-brain barrier (24-26). We also showed that, as in meningitic E. coli strains, IbeA is involved in the virulence of strain BEN2908 for chickens (18). However, the role of IbeA in the virulence of ExPEC remains unclear, despite the characterization of proteins interacting with IbeA in vitro (43, 55, 56).
In this report, we analyzed in more detail the invasion-deficient phenotype of an ibeA derivative of the avian ExPEC strain BEN2908. We found that this decreased invasion was linked to decreased adhesion to HBMEC. Because strain BEN2908 also expresses type 1 fimbriae that could mediate adhesion to HBMEC, we measured the contribution solely of IbeA to the adhesion of BEN2908 to HBMEC by preventing type 1 fimbria-mediated adhesion through a Δfim mutation or by the use of mannose. Surprisingly, our results showed that deletion of ibeA in the absence of type 1 fimbriae did not have any consequences on adhesion. Based on these data, we hypothesized that a decreased level of type 1 fimbria expression might be responsible for the lower adhesion level of BEN2908 ΔibeA. Notably, adhesion of strain DM34 remained relatively high in these assays, suggesting that type 1 fimbriae are not the only adhesins expressed by BEN2908; the high motility of strain BEN2908 (data not shown) could indeed be involved in its adhesion to HBMEC (4). We then showed that fewer type 1 fimbriae were found in a culture of BEN2908 ΔibeA, due to a larger proportion of bacteria with an IE in the off orientation. This might be explained by the lower expression of fimB and fimE measured in BEN2908 ΔibeA. Type 1 fimbria expression was only partially complemented by plasmid pUC23A, while adhesion and the proportion of bacteria expressing type 1 fimbriae were restored to levels similar to those of the WT strain. A possible explanation for these apparently inconsistent results could be that bacteria that express fimbriae in the complemented strain do so at a level lower than that of bacteria that express type 1 fimbriae in the WT strain, with the proportion of bacteria expressing type 1 fimbriae being the same in both strains.
The major finding of this report is that no difference in adhesion was observed between ibeA+ and ibeA mutant strains when type 1 fimbria-mediated adhesion was prevented. This suggests that very few, if any, adhesive properties of strain BEN2908 are due to direct interaction between IbeA and HBMEC surface proteins. In fact, the lower adhesion level of BEN2908 ΔibeA was mainly due to a decrease of the proportion of fimbriated bacteria with this strain. In the original description of the ibeA gene, Huang et al. indicated that expression of S fimbriae and type 1 fimbriae was not reduced in their original RS218 ibeA mutant (10A23 mutant) (26). Our data seem to contradict this first report. One reason why the decrease in type 1 fimbriae in the ibeA derivative may not have been detected by Huang et al. might be the lower expression of type 1 fimbriae in strain RS218 than in BEN2908 (our unpublished observation), hence the lesser adhesive properties of strain RS218 than of the strain used in this study, BEN2908 (18).
Furthermore, we found that the differences in adhesion between strains BEN2908 and BEN2908 ΔibeA were not observed under certain growth conditions, i.e., when bacteria were grown to stationary phase without agitation (data not shown). These conditions are often used to characterize type 1 fimbria expression and may have been used by Huang et al. to conclude that there was normal expression of type 1 fimbriae in their ibeA mutant (26).
Our study also showed that an ibeT derivative of strain BEN2908 expressed fewer type 1 fimbriae at its surface and presented a larger proportion of IE in the off orientation than the WT strain BEN2908 did. These results are in agreement with the lower adhesion level of an ibeT derivative of strain RS218 reported by Zou et al. (54). The number of type 1 fimbriae was lower in BEN2908 ΔibeT than in BEN2908 ΔibeA, and the proportion of IE in the off orientation seemed to be increased in BEN2908 ΔibeT compared to that in BEN2908 ΔibeA. Nevertheless, given their genetic location and the somehow similar consequences of their deletion, we suggest that ibeA and ibeT alter the expression of type 1 fimbriae through a common but so far unknown mechanism. Our results suggest that the larger proportion of bacteria with an IE in the off orientation in BEN2908 ΔibeA might be related to decreased expression of fimB and fimE. Expression of type 1 fimbriae is regulated through sophisticated mechanisms, as reviewed by Blomfield and Van Der Woude (5). Whether the decreased expression of fimB and fimE is sufficient to explain the observed phenotype of BEN2908 ΔibeA will have to be investigated. It has been shown that when the IE is in the off orientation, the RNA encoding fimE is unstable due to early rho-dependent termination (22, 27). It is therefore possible that the reduced level of fimE RNA that we observed was only a consequence, and not the cause, of the larger proportion of IE in the off orientation. Strain BEN2908 also carries the ipbA gene but not the ipuA and ipuB genes (data not shown). However, from our results, we could not conclude whether there was a modification of ipbA expression in BEN2908 ΔibeA. Interestingly, the phenotype of our BEN2908 ΔibeA mutant is very similar to that of an ompA mutant of strain RS218 (50). In particular, the reduction of fimB and fimE expression observed in our report is strikingly similar to that observed by Teng et al. for their ompA mutant. It is thus possible that these modifications of type 1 fimbriae involve the same mechanisms.
In strain RS218, ibeA is located inside a genomic island that contains several putative operons. Our data (Southern blots and PCR [data not shown]) indicate that ibeA is also located in such a genomic island in strain BEN2908 and that operons GimA1, GimA2, GimA3, and GimA4 are present in strain BEN2908. Huang et al. suggested that this island contributes to invasion through a carbon source-regulated mechanism (24). For example, genes from GimA1 encode proteins that have homologies with the different subunits of a putative phosphotransferase-dependent dihydroxyacetone kinase. cglD from operon GimA2 encodes a putative glycerol dehydrogenase that could yield dihydroxyacetone from glycerol. Operon GimA3 encodes four proteins potentially involved in the formation of 3-phospho-d-glycerate and/or hydroxypyruvate from glyoxylate. Indeed, genes of GimA3 encode proteins that are very similar to those involved in the d-glycerate pathway participating in the degradation of allantoin (10).
Based on our results regarding the above predictions and the predictions of IbeA as a cytoplasmic protein and IbeT as an inner membrane transporter, an alternative hypothesis to current models is that IbeA does not directly mediate interaction with HBMEC but rather acts upon expression of type 1 fimbriae through modulation of a metabolic pathway. Unbalance of this metabolic pathway by inactivation of ibeA or ibeT would, in turn, have consequences on the expression of various bacterial components, such as type 1 fimbriae. Interestingly, regulation of type 1 fimbriae by metabolic pathways is suggested by several reports. Eisenstein and colleagues demonstrated that growth in glucose inhibited the expression of type 1 fimbriae (15). Gally and coworkers also showed that the fim switch was regulated by environmental conditions, such as temperature or growth medium. FimB-mediated switching is thus increased when bacteria are grown in minimal medium at a high temperature (42°C), while FimE-dependent switching is higher in rich medium and at a low temperature (28°C) (17).
Several reports have unraveled a series of regulators of fimB and fimE expression. Inactivation of these regulatory pathways leads to altered type 1 fimbria expression. For example, expression of fimB is regulated by N-acetylglucosamine and N-acetyl neuraminic acid, through the NagC and NanR regulators, respectively (47, 48). Aberg et al. also recently described the control of fimB expression by the modification of ppGpp levels (1), and cyclic-di-GMP was recently shown to be involved in type 1 fimbria expression (9).
One possibility is therefore that expression of Fim recombinases is controlled by an as yet unknown regulator, with this regulator acting as a sensor of a component that is either a substrate or a product of IbeA; inactivation of ibeA would in turn modify the concentration of this compound and consequently alter the activity of this putative regulator. As a consequence, type 1 fimbriae, and hence adhesive properties of the ibeA mutant, would be decreased. Another possibility is that the inactivation of the putative pathway involving IbeA and/or IbeT leads to changes in the concentration of secondary messengers, such as ppGpp or cyclic-di-GMP.
The exact role of IbeA in the infectious process remains to be clarified. We have shown that it is required for full virulence of strain BEN2908 for chickens (18). Reduction of the adhesive properties by an ibeA mutation is likely to participate in this reduced virulence, in particular in the first stages of colibacillosis, such as interaction with lung epithelial cells. Yet one cannot exclude that IbeA is also involved in another process that is required for full virulence, and future research will have to address this possibility.
In conclusion, this report opens up new possibilities concerning the exact role of IbeA in the virulence of ExPEC. Future studies will have to address the exact mechanism by which GimA4 regulates type 1 fimbria expression. In particular, experiments with bacteria having a locked-on or locked-off fim switch will answer the question of whether the effect seen in this study is mediated only through regulation of fimB and fimE expression. Altogether, new experiments should help to validate or not validate the hypothesis that the lower proportions of bacteria expressing type 1 fimbriae in BEN2908 ΔibeA and BEN2908 ΔibeT might be the result of an unbalanced metabolic pathway.
Acknowledgments
We thank Sheng-He Huang for providing us with plasmid pUC23A. HBMEC were kindly provided by Claudine Kieda (Centre de Biophysique Moléculaire, CNRS, Orléans, France). We thank C. Schouler for fruitful discussions and a critical reading of the manuscript. Helpful discussions with C. Le Bouguenec, A. Wiedemann, and J. L. Dacheux are acknowledged.
This project was supported by the EC under FP5 (COLIRISK grant no. QLK2-CT-2002-00944) and by ERA-NET Pathogenomics (ANR-06-PATHO-002-01). M.C. was funded by a grant from the Ministère de la Recherche.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 30 June 2008.
REFERENCES
- 1.Aberg, A., V. Shingler, and C. Balsalobre. 2006. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol. Microbiol. 601520-1533. [DOI] [PubMed] [Google Scholar]
- 2.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 825724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 451079-1093. [DOI] [PubMed] [Google Scholar]
- 4.Barnich, N., J. Boudeau, L. Claret, and A. Darfeuille-Michaud. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol. Microbiol. 48781-794. [DOI] [PubMed] [Google Scholar]
- 5.Blomfield, I., and M. Van Der Woude 13 September 2007, posting date. Chapter 2.4.2.2, Regulation of fimbrial expression. In R. Curtis III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [DOI] [PubMed]
- 6.Boudeau, J., N. Barnich, and A. Darfeuille-Michaud. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 391272-1284. [DOI] [PubMed] [Google Scholar]
- 7.Bryan, A., P. Roesch, L. Davis, R. Moritz, S. Pellett, and R. A. Welch. 2006. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 741072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chouikha, I., A. Brée, M. Moulin-Schouleur, P. Gilot, and P. Germon. 2008. Differential expression of iutA and ibeA in the early stages of infection by extra-intestinal pathogenic E. coli. Microbes Infect. 10432-438. http://dx.doi.org/10.1016/j.micinf.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Claret, L., S. Miquel, N. Vieille, D. A. Ryjenkov, M. Gomelsky, and A. Darfeuille-Michaud. 2007. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J. Biol. Chem. 28233275-33283. [DOI] [PubMed] [Google Scholar]
- 10.Cusa, E., N. Obradors, L. Baldoma, J. Badia, and J. Aguilar. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 1817479-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30299-316. [PubMed] [Google Scholar]
- 13.Dho-Moulin, M., and J.-P. Lafont. 1982. Escherichia coli colonization of the trachea in poultry: comparison of virulent and avirulent strains in gnotoxenic chickens. Avian Dis. 26787-797. [PubMed] [Google Scholar]
- 14.Dho-Moulin, M., J. F. van den Bosch, J. P. Girardeau, A. Brée, T. Barat, and J. P. Lafont. 1990. Surface antigens from Escherichia coli O2 and O78 strains of avian origin. Infect. Immun. 58740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenstein, B. I., and D. C. Dodd. 1982. Pseudocatabolite repression of type 1 fimbriae of Escherichia coli. J. Bacteriol. 1511560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewers, C., G. Li, H. Wilking, S. Kiessling, K. Alt, E. M. Antao, C. Laturnus, I. Diehl, S. Glodde, T. Homeier, U. Bohnke, H. Steinruck, H. C. Philipp, and L. H. Wieler. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297163-176. [DOI] [PubMed] [Google Scholar]
- 17.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 1756186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Germon, P., Y. H. Chen, L. He, J. E. Blanco, A. Brée, C. Schouler, S. H. Huang, and M. Moulin-Schouleur. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 1511179-1186. [DOI] [PubMed] [Google Scholar]
- 19.Gunther, I. N., J. A. Snyder, V. Lockatell, I. Blomfield, D. E. Johnson, and H. L. Mobley. 2002. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect. Immun. 703344-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunther, N. I., V. Lockatell, D. E. Johnson, and H. L. Mobley. 2001. In vivo dynamics of type 1 fimbria regulation in uropathogenic Escherichia coli during experimental urinary tract infection. Infect. Immun. 692838-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannan, T. J., I. U. Mysorekar, S. L. Chen, J. N. Walker, J. M. Jones, J. S. Pinkner, S. J. Hultgren, and P. C. Seed. 2007. LeuX tRNA-dependent and -independent mechanisms of Escherichia coli pathogenesis in acute cystitis. Mol. Microbiol. 76116-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinde, P., P. Deighan, and C. J. Dorman. 2005. Characterization of the detachable Rho-dependent transcription terminator of the fimE gene in Escherichia coli K-12. J. Bacteriol. 1878256-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooton, T. M., and W. E. Stamm. 1997. Diagnosis and treatment of uncomplicated urinary tract infection. Infect. Dis. Clin. N. Am. 11551-581. [DOI] [PubMed] [Google Scholar]
- 24.Huang, S. H., Y. H. Chen, G. Kong, S. H. Chen, J. Besemer, M. Borodovsky, and A. Jong. 2001. A novel genetic island of meningitic Escherichia coli K1 containing the ibeA invasion gene (GimA): functional annotation and carbon-source-regulated invasion of human brain microvascular endothelial cells. Funct. Integr. Genomics 1312-322. [DOI] [PubMed] [Google Scholar]
- 25.Huang, S. H., Z. S. Wan, Y. H. Chen, A. Y. Jong, and K. S. Kim. 2001. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J. Infect. Dis. 1831071-1078. [DOI] [PubMed] [Google Scholar]
- 26.Huang, S. H., C. Wass, Q. Fu, N. V. Prasadarao, M. Stins, and K. S. Kim. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 634470-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joyce, S. A., and C. J. Dorman. 2002. A Rho-dependent phase-variable transcription terminator controls expression of the FimE recombinase in Escherichia coli. Mol. Microbiol. 451107-1117. [DOI] [PubMed] [Google Scholar]
- 28.Khan, N. A., Y. Kim, S. Shin, and K. S. Kim. 2007. FimH-mediated Escherichia coli K1 invasion of human brain microvascular endothelial cells. Cell. Microbiol. 9169-178. [DOI] [PubMed] [Google Scholar]
- 29.Kieda, C., M. Paprocka, A. Krawczenko, P. Zalecki, P. Dupuis, M. Monsigny, C. Radzikowski, and D. Dus. 2002. New human microvascular endothelial cell lines with specific adhesion molecule phenotypes. Endothelium 9247-261. [DOI] [PubMed] [Google Scholar]
- 30.Klemm, P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 51389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.La Ragione, R. M., A. R. Sayers, and M. J. Woodward. 2000. The role of fimbriae and flagella in the colonization, invasion and persistence of Escherichia coli O78:K80 in the day-old-chick model. Epidemiol. Infect. 124351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Ragione, R. M., and M. J. Woodward. 2002. Virulence factors of Escherichia coli serotypes associated with avian colisepticaemia. Res. Vet. Sci. 7327-35. [DOI] [PubMed] [Google Scholar]
- 33.Lim, J. K., N. W. T. Gunther, H. Zhao, D. E. Johnson, S. K. Keay, and H. L. Mobley. 1998. In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect. Immun. 663303-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, M., T. Durfee, J. E. Cabrera, K. Zhao, D. J. Jin, and F. R. Blattner. 2005. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J. Biol. Chem. 28015921-15927. [DOI] [PubMed] [Google Scholar]
- 35.Marc, D., P. Arné, A. Brée, and M. Dho-Moulin. 1998. Colonization ability and pathogenic properties of a fim− mutant of an avian strain of Escherichia coli. Res. Microbiol. 149473-485. [DOI] [PubMed] [Google Scholar]
- 36.Marc, D., and M. Dho-Moulin. 1996. Analysis of the fim cluster of an avian O2 strain of Escherichia coli: serogroup-specific sites within fimA and nucleotide sequence of fimI. J. Med. Microbiol. 44444-452. [DOI] [PubMed] [Google Scholar]
- 37.Martinez, J. J., M. A. Mulvey, J. D. Schilling, J. S. Pinkner, and S. J. Hultgren. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 192803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendu, D. R., V. R. Dasari, M. Cai, and K. S. Kim. 2008. Protein folding intermediates of invasin protein IbeA from Escherichia coli. FEBS J. 275458-469. [DOI] [PubMed] [Google Scholar]
- 39.Mol., O., and B. Oudega. 1996. Molecular and structural aspects of fimbriae biosynthesis and assembly in Escherichia coli. FEMS Microbiol. Rev. 1925-52. [DOI] [PubMed] [Google Scholar]
- 40.Moulin-Schouleur, M., M. Reperant, S. Laurent, A. Brée, S. Mignon-Grasteau, P. Germon, D. Rasschaert, and C. Schouler. 2007. Extra-intestinal pathogenic Escherichia coli of avian and human origin: link between phylogenetic relationships and common virulence patterns. J. Clin. Microbiol. 453366-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padan, E., T. Tzubery, K. Herz, L. Kozachkov, A. Rimon, and L. Galili. 2004. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+ antiporter. Biochim. Biophys. Acta 16582-13. [DOI] [PubMed] [Google Scholar]
- 42.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prasadarao, N. V., C. A. Wass, S. H. Huang, and K. S. Kim. 1999. Identification and characterization of a novel Ibe10 binding protein that contributes to Escherichia coli invasion of brain microvascular endothelial cells. Infect. Immun. 671131-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 1512097-2110. [DOI] [PubMed] [Google Scholar]
- 45.Russo, T. A., and J. R. Johnson. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5449-456. [DOI] [PubMed] [Google Scholar]
- 46.Silver, R. P., W. Aaronson, A. Sutton, and R. Schneerson. 1980. Comparative analysis of plasmids and some metabolic characteristics of Escherichia coli K1 from diseased and healthy individuals. Infect. Immun. 29200-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sohanpal, B. K., S. El-Labany, M. Lahooti, J. A. Plumbridge, and I. C. Blomfield. 2004. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 10116322-16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sohanpal, B. K., S. Friar, J. Roobol, J. A. Plumbridge, and I. C. Blomfield. 2007. Multiple co-regulatory elements and IHF are necessary for the control of fimB expression in response to sialic acid and N-acetylglucosamine in Escherichia coli K-12. Mol. Microbiol. 631223-1236. [DOI] [PubMed] [Google Scholar]
- 49.Teng, C. H., M. Cai, S. Shin, Y. Xie, K. J. Kim, N. A. Khan, F. Di Cello, and K. S. Kim. 2005. Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect. Immun. 732923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teng, C. H., Y. Xie, S. Shin, F. Di Cello, M. Paul-Satyaseela, M. Cai, and K. S. Kim. 2006. Effects of ompA deletion on expression of type 1 fimbriae in Escherichia coli K1 strain RS218 and on the association of E. coli with human brain microvascular endothelial cells. Infect. Immun. 745609-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 16935-13. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Y., S. H. Huang, C. A. Wass, M. F. Stins, and K. S. Kim. 1999. The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infect. Immun. 674751-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie, Y., Y. Yao, V. Kolisnychenko, C. H. Teng, and K. S. Kim. 2006. HbiF regulates type 1 fimbriation independently of FimB and FimE. Infect. Immun. 744039-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou, Y., L. He, F. Chi, A. Jong, and S. H. Huang. 27 November 2007. Involvement of Escherichia coli K1 ibeT in bacterial adhesion that is associated with the entry into human brain microvascular endothelial cells. Med. Microbiol. Immunol. doi: 10.1007/s00430-007-0065-y. [DOI] [PubMed]
- 55.Zou, Y., L. He, and S. H. Huang. 2006. Identification of a surface protein on human brain microvascular endothelial cells as vimentin interacting with Escherichia coli invasion protein IbeA. Biochem. Biophys. Res. Commun. 351625-630. [DOI] [PubMed] [Google Scholar]
- 56.Zou, Y., L. He, C. H. Wu, H. Cao, Z. H. Xie, Y. Ouyang, Y. Wang, A. Jong, and S. H. Huang. 2007. PSF is an IbeA-binding protein contributing to meningitic Escherichia coli K1 invasion of human brain microvascular endothelial cells. Med. Microbiol. Immunol. 196135-143. [DOI] [PubMed] [Google Scholar]