Abstract
Proteins with tandem repeat (TR) domains have been found in various protozoan parasites, often acting as targets of B-cell responses. However, the extent of the repeats within Trypanosoma cruzi, the causative agent of Chagas’ disease, has not been examined well. Here, we present a systematic survey of the TR genes found in T. cruzi, in comparison with other organisms. Although the characteristics of TR genes varied from organism to organism, the presence of genes having large TR domains was unique to the trypanosomatids examined, including T. cruzi. Sequence analyses of T. cruzi TR genes revealed their divergency; they do not share such characteristics as sequence similarity or biased cellular location predicted by the presence of a signal sequence or transmembrane domain(s). In contrast, T. cruzi TR proteins seemed to possess significant antigenicity. A number of previously characterized T. cruzi antigens were detected by this computational screening, and several of those antigens contained a large TR domain. Further analyses of the T. cruzi genome demonstrated that previously uncharacterized TR proteins in this organism may also be immunodominant. Taken together, T. cruzi is rich in large TR domain-containing proteins with immunological significance; it is worthwhile further analyzing such characteristics of TR proteins as the copy number and consensus sequence of the repeats to determine whether they might contribute to the biological variability of T. cruzi strains with regard to induced immunological responses, host susceptibility, disease outcomes, and pathogenicity.
Chagas’ disease results from infection, generally via contaminated blood or the bite of an infected insect, by the protozoan parasite Trypanosoma cruzi, which can live in a variety of tissues in the mammalian host. According to the World Health Organization Special Programme for Research and Training in Tropical Disease Report 2002, the disease is endemic in 18 countries in Central and South America. It is estimated that 16 to 18 million individuals are infected with T. cruzi, with 300,000 new cases per year, and the infection causes 21,000 deaths annually. Chagas’ disease has an acute phase and a chronic phase. Manifestations in the acute phase include swelling at the infection site, fever, and hepatosplenomegaly. Following an asymptomatic period after the acute phase, an estimated 32% of infected individuals develop chronic Chagas’ disease, often leading to fatal damage to the heart and digestive tract.
Genes encoding proteins with tandem repeat (TR) domains, defined here as two or more copies of an amino acid pattern, have been found in a variety of organisms, from prokaryotes to higher animals. TRs appear to be diverse and provide regular arrays of spatial and functional groups (38). They are conspicuous in structural and cell surface proteins in some organisms (38, 60). In contrast, previously characterized T. cruzi TR proteins include trans-sialidase, ribosomal protein, flagellar protein FRA, and cytoplasmic protein CRA (14, 42, 49), which do not seem to share functional characteristics. Also, the functions of many T. cruzi TR proteins remain unknown due to a lack of systematic characterization. Although the functions of TR proteins are disparate and not confined to a single type of protein, and a common time of expression or cellular localization is not consistently observed, one feature appears to be shared: they are often potent B-cell antigens. The immunological significance of TR proteins during bacterial infections has been reported (3, 31), and even some cancer antigens to which patients show antibody responses contain TR domains (41, 47). In some organisms, having a variety of TRs within a given protein may play an important role in generating variability in cell surface immunogens and adhesion molecules, thereby evading the immune system or enhancing pathogenicity (27, 37, 43, 44, 60). In protozoan parasites, TR proteins often serve as targets of B-cell responses (39, 54). Antibody responses to TR proteins have been found in Chagas’ disease (14, 28, 34) and other parasitic diseases such as leishmaniasis (10, 13) and malaria (16, 17, 40). However, because the immunological dominance of TR proteins is not restricted to protozoan parasites, systematic analyses of TR genes and proteins are required to see (i) if T. cruzi has more or fewer TR proteins than other pathogens or organisms do and (ii) whether these TR proteins have sequence similarity, a biased cellular location, or shared immunological recognition motifs. For example, a genome scale analysis of Saccharomyces cerevisiae has revealed that most genes containing TRs encode cell wall proteins (60). In a previous study, we have demonstrated that TR proteins of Leishmania infantum share immunological dominance (26). This is the only systematic study of the immunological properties of protozoan parasite TR proteins, and it still remains unclear whether other protozoan parasites, including T. cruzi, possess TR proteins with such characteristics.
Here we performed a computational search for TR genes in T. cruzi, in comparison with various parasitic protozoan (Leishmania major, L. infantum, Trypanosoma brucei, Plasmodium falciparum, Toxoplasma gondii, and Entamoeba histolytica), fungal (Candida albicans), bacterial (Salmonella enterica and Mycobacterium tuberculosis), and human genomes. The analysis revealed no biochemical but immunological characteristics common in the T. cruzi TR proteins. As an indication of its detection sensitivity, this computational method captured a number of previously characterized antigens from T. cruzi suggesting the immunological dominance of TR proteins. To further validate the immunological significance of TR proteins from protozoan parasites, we evaluated the antigenicity of previously uncharacterized T. cruzi TR proteins. The results demonstrated that immunological recognition was a feature common to the T. cruzi TR proteins, whereas there were no apparent similarities or biases in their sequences or predicted cellular locations.
MATERIALS AND METHODS
Bioinformatic screening of TR genes.
We obtained DNA sequence data for P. falciparum 3D7 CDS (coding sequence) version 2.1.4. (without pseudogenes) (22), L. major CDS version 5.2 (35), L. infantum CDS version 3.0 (51), and T. brucei Tb927_CDSs_v4_nopseudo (9) from GeneDB (www.genedb.org) (30); Trypanosoma cruzi Annotated CDS Release 5.1 (20) from TcruziDB (www.tcruzidb.org/tcruzidb) (2); T. gondii Annotated CDS Release 4.2 from ToxoDB (www.toxodb.org/toxo) (21); C. albicans open reading frame coding assembly 21 (36, 58) from The Candida Genome Database (www.candidagenome.org) (5); M. tuberculosis Release R7 (15) from TubercuList (http://genolist.pasteur.fr/TubercuList/); S. enterica serovar Typhi CT18 (50); and Homo sapiens (59) Hs36.2 CCDS nucleotide 20070227 from the NCBI database (www.ncbi.nlm.nih.gov/projects/CCDS/). Tandem Repeats Finder, a program to locate and display TRs in DNA sequences, was used for these analyses (http://tandem.bu.edu/trf/trf.html) (8). The program calculates the score according to selected characteristics of the TR genes such as the period size of the repeat (i.e., the length of the repeat unit), the number of copies aligned with the consensus pattern, and the overall percentage of matches between adjacent copies of a pattern. Most likely, a high score indicates that the gene possesses a large TR domain. In this study, the genes were regarded as TR genes if the scores from the Tandem Repeats Finder analysis were 150 or higher. The cutoff value of 150 is likely to eliminate genes with repeat domains shorter than 75 bp. When more than one TR domain was found within a gene, only the domain with the highest score was listed or used for further analyses and protein production.
Analyses of the TR genes of T. cruzi.
The biochemical properties of each of the bioinformatically selected T. cruzi TR proteins were further analyzed virtually for (i) a protein's molecular mass, isoelectric point, and hydrophobicity and the presence of a signal sequence and a transmembrane domain; (ii) its known antigenicity and/or functions by BLAST searches with both DNA and deduced amino acid sequences against the NCBI database; and (iii) a mass spectrometry-evidenced protein expression profile (6), available through the database TcruziDB. Biochemical characteristics such as average hydrophobicity, isoelectric point, and molecular weight were calculated with the ProteinMachine software package from Protein Advances Inc., Seattle, WA. To analyze the entire database, a software interface programmed in C# created protein data files as comma-separated values for export to Excel. Average hydrophobicity or hydrophilicity plots of each sequence were determined with a modified Kyte-Doolittle algorithm with scores ranging from 0.6 (most hydrophilic score possible) to 9.0 (most hydrophobic score possible). T. cruzi TR genes were analyzed for their specificity for T. cruzi, i.e., whether a homologous gene or protein is found in Leishmania or other organisms, by blasting the DNA and deduced amino acid sequences against the NCBI database and GeneDB.
Expression of T. cruzi TR proteins.
Partial TR domains containing multiple repeat units were either PCR amplified or synthesized. Partial TR domains of Tc00.1047053510827.40 (designated Tc2 in this study), Tc00.1047053511821.179 (Tc3), Tc00.1047053509157.120 (Tc4), and Tc00.1047053508119.200 (Tc6) were amplified by PCR with T. cruzi total DNA and the following primer sets: Tc2, 5′ CAA TTA CAT ATG AGC GCG AGC ACC GCC TGG and 3′ CAA TTA AAG CTT CTA GTC GCT CAA CAA CCG CAT G; Tc3, 5′ CAA TTA CAT ATG GAG AAC GAG GAG CTG CGT G and 3′ CAA TTA AAG CTT CTA CGC ACG AAG CTC CTC CAG; Tc4, 5′ CAA TTA CAT ATG CCG GAG ACA GCC TCA GTC and 3′ CAA TTA AAG CTT CTA CGC GTG ACC GTC CTC GTC; Tc6, 5′ CAA TTA CAT ATG GCA ACG GAC GAG TTG and 3′ CAA TTA AAG CTT CTA GAG CGC AGT CGC ATC CCT G. Partial TR domains of Tc00.1047053511557.50 (Tc1), Tc00.1047053510217.10 (Tc8), Tc00.1047053504019.3 (Tc9), Tc00.1047053506495.40 (Tc10), Tc00.1047053506491.20 (Tc12), Tc00.1047053506559.559 (Tc13), and Tc00.1047053507049.119 (Tc15) were synthesized by Blue Heron Biotechnology, Inc. (Bothell, WA). The amplified PCR products or synthesized oligonucleotides were inserted in frame with the six-His tag of vector pET-28a. The vectors were then transformed into the Escherichia coli Rosetta strain. The transformed E. coli cells were grown in 2× yeast extract-Tryptone medium, and expression of the recombinant proteins was induced by cultivation with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h. After lysing cells by sonication and centrifuging them at 10,000 × g, the supernatants were used for purifying the proteins as six-His-tagged proteins with Ni-nitrilotriacetic acid agarose (Qiagen Inc., Valencia, CA). Proteins were bound to the resin, washed with sodium deoxycholate-containing buffer, and eluted with buffer containing 250 μM imidazole. The eluted protein was dialyzed against phosphate-buffered saline (pH 7.4), and the concentration of the purified protein was measured by the bicinchoninic acid protein assay (Pierce Biotechnology Inc., Rockford, IL). The purity of the proteins was assessed by Coomassie blue staining following sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Antibody ELISA.
The expressed T. cruzi TR proteins were analyzed for seroreactivity with sera from Brazilian or Ecuadorian Chagas’ disease patients (n = 24). Sera from Brazilian visceral leishmaniasis (VL) patients (n = 16) and healthy Brazilian people were used as controls. Proteins were diluted in enzyme-linked immunosorbent assay (ELISA) coating buffer, and 96-well plates were coated with 200 ng of individual recombinant antigens, followed by blocking with phosphate-buffered saline containing 0.05% Tween 20 and 1% bovine serum albumin. Plates were incubated sequentially with human serum samples (1:200 dilution) and with horseradish peroxidase-conjugated anti-human immunoglobulin G (Rockland Immunochemicals, Inc., Gilbertsville, PA). The plates were developed with tetramethylbenzidine peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, MD) and scanned with a microplate reader at 450 nm (570-nm reference). Three additional recombinant proteins were tested as controls: T. cruzi sterol 24-c methyltransferase (TcSMT) as a conserved antigen between Trypanosoma and Leishmania species (24), rK39 as a Leishmania-specific TR antigen (13), and CRA as a T. cruzi-specific TR antigen (42). Statistical analyses were performed to compare the reactivity of Chagas’ disease patient sera to individual antigens with that of VL patients or healthy donors by either unpaired t test or Mann-Whitney test based on whether the data sets have a Gaussian distribution.
RESULTS
Search of TR genes from the T. cruzi genome database.
With the exception of S. enterica, all of the organisms examined in this study had TR genes comprising >1% of their genomes (Table 1). A markedly higher prevalence of TR genes was found in P. falciparum and T. gondii (24.61 and 5.70%, respectively). In contrast, the prevalence of TR genes in the trypanosomatid parasites was no greater than in E. histolytica, C. albicans, M. tuberculosis, and H. sapiens. TR genes with a score of ≥2,000 are likely to have a TR domain ≥1,000 bp long. The prevalence of these genes in the whole genomes was higher in the trypanosomatid and the apicomplexan parasites than in the other examined pathogens or H. sapiens. The trypanosomatid parasites, including T. cruzi, had a higher prevalence of TR genes scoring ≥2,000 in the whole genomes than the apicomplexans, although the apicomplexan parasites, especially P. falciparum, were richer in total TR genes. The trypanosomatid parasites seemed to have a preference for such large TR genes according to a higher prevalence of TR genes, scoring ≥2,000 in all TR genes, and higher mean and median TR scores in these species compared with others examined in this study.
TABLE 1.
Numbers of TR genes in selected pathogens and H. sapiens
| Species | No. of genes | No. with TR score of:
|
Total TR genes
|
Genes with TR score of ≥2,000
|
Mean TR score | Median TR Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 150-499 | 500-999 | 1,000-1,999 | 2,000-4,999 | 5,000-9,999 | ≥10,000 | No. | % of total genes | No. | % of total genes | % of total TR genes | ||||
| L. major | 9,218 | 44 | 15 | 16 | 15 | 10 | 3 | 103 | 1.12 | 28 | 0.30 | 27.20 | 2,846 | 715 |
| L. infantum | 8,184 | 35 | 9 | 12 | 21 | 7 | 7 | 91 | 1.11 | 35 | 0.43 | 38.50 | 2,651 | 1,055 |
| T. brucei | 8,161 | 39 | 18 | 21 | 30 | 9 | 5 | 122 | 1.49 | 44 | 0.54 | 36.10 | 2,482 | 1,122 |
| T. cruzi | 19,605 | 154 | 48 | 83 | 69 | 3 | 0 | 357 | 1.82 | 72 | 0.37 | 20.20 | 1,183 | 717 |
| P. falciparum | 5,387 | 1,153 | 135 | 29 | 7 | 2 | 0 | 1,326 | 24.61 | 9 | 0.17 | 0.70 | 356 | 277 |
| T. gondii | 7,793 | 340 | 53 | 34 | 17 | 0 | 0 | 444 | 5.70 | 17 | 0.22 | 3.80 | 481 | 253 |
| E. histolytica | 9,905 | 180 | 37 | 2 | 0 | 0 | 0 | 219 | 2.21 | 0 | 0.00 | 0.00 | 342 | 273 |
| C. albicans | 6,107 | 72 | 7 | 5 | 3 | 0 | 0 | 87 | 1.42 | 3 | 0.05 | 3.40 | 448 | 226 |
| S. enterica | 4,395 | 5 | 1 | 1 | 0 | 0 | 0 | 7 | 0.16 | 0 | 0.00 | 0.00 | 594 | 479 |
| M. tuberculosis | 4,005 | 43 | 4 | 2 | 0 | 0 | 0 | 49 | 1.22 | 0 | 0.00 | 0.00 | 334 | 230 |
| H. sapiens | 17,751 | 235 | 141 | 46 | 6 | 1 | 1 | 430 | 2.42 | 8 | 0.05 | 1.90 | 608 | 433 |
aNumber of genes with TR scores of ≥2,000.
The high prevalence of large genes in the trypanosomatid and the apicomplexan parasites may reflect the high prevalence of large TR regions in the genes of those parasites. S. enterica and M. tuberculosis had average gene sizes smaller than those of the others examined in this study (Fig. 1). C. albicans and H. sapiens showed gene size distributions overlapping those of the trypanosomatid and the apicomplexans, up to 10,000 bp. The ratios of genes over 10,000 bp, however, were much higher in the trypanosomatid and the apicomplexan parasites (0.75 to 1.95%) than any other species, including C. albicans and H. sapiens (0.01 to 0.05%).
FIG. 1.
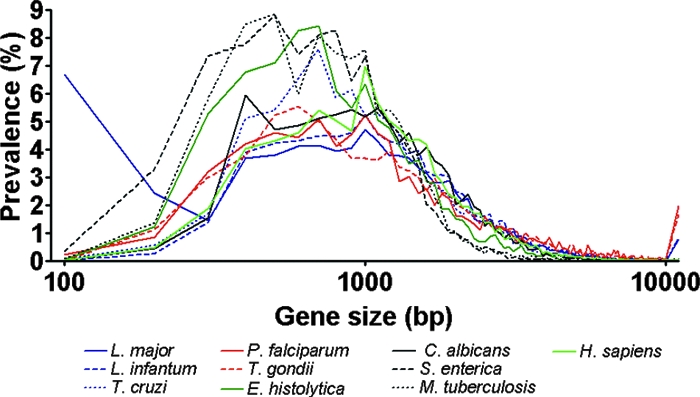
Gene size distribution of protozoan parasites and other organisms. All genes in the genome of were sorted by their sizes with 100-bp intervals.
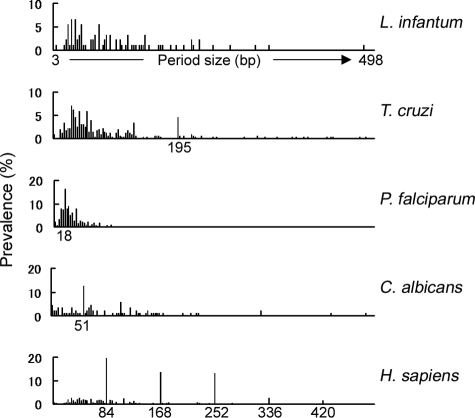
The period size of the TR refers to the length of the repeat unit. The distribution of the TR period sizes varied among these organisms (Fig. 2). TR genes in L. infantum were divergent in their repeat motifs, as their period sizes ranged widely, from 3 to 498, and no particular period size dominated (i.e., constituted more than 10%) in any of the TR genes. A similar pattern was found in L. major and T. brucei (data not shown). T. cruzi also had a wide distribution of period sizes, although the overall distribution was shifted to the left compared to L. infantum, indicating that T. cruzi TR genes are also divergent. In contrast, 76.2% of the P. falciparum TR genes had period sizes of ≤36 bp and 93.4% had period sizes of ≤72 bp. Although >10% of the TR genes in C. albicans had a period size of 51 (single peak, Fig. 2), the period sizes in other TR genes were widely divergent, as seen in Leishmania and Trypanosoma. There was a more restricted pattern in the period sizes of human TR genes, which will be discussed below.
FIG. 2.
Unique patterns in period sizes of TR genes. The y axis shows the prevalence of TR genes having a particular period size among all of the TR genes in the same species.
Search for functional similarity in T. cruzi TR proteins.
We examined TR genes in T. cruzi, C. albicans, M. tuberculosis, and H. sapiens for functional similarities. There were 87 TR genes identified in C. albicans, 25 (28.7%) of which encoded cell surface proteins such as the agglutinin-like sequence family (33). C. albicans TR genes with higher scores were more likely to encode cell surface proteins: 10 of 15 TR genes with a score of ≥500 and 7 of 8 TR genes with a score of ≥1,000 encoded cell surface proteins (the one exception was polyubiquitin). Such a high prevalence of TR genes encoding cell surface proteins has been shown in another fungal species, S. cerevisiae (60). In M. tuberculosis, 49 genes were identified as containing TRs. Thirty-five (71.4%) of those were categorized in either the PE family polymorphic GC-rich repetitive sequence (23 genes, 46.9%) or the PPE family (12 genes, 24.5%), values that were considerably higher than the prevalence in all M. tuberculosis genes (1.6 and 1.7% for the PE family polymorphic GC-rich repetitive sequence and the PPE family, respectively). The PE and PPE families, the major source of divergence between the genomes of M. tuberculosis and M. bovis, which are otherwise >99% similar, often have multiple copies of repeat motifs and participate in antigenic variation and intramacrophage survival of M. tuberculosis (15, 29, 45, 52). As described above, 215 (50%) of 430 TR genes of H. sapiens had period sizes of 84 bp or multiples of 84 bp (164, 252, 368, and 420 bp). It was revealed that most of these (209 genes, 48.6% of all H. sapiens TR proteins) were zinc finger proteins, one of the largest families of human proteins, composing ∼2% of the human proteome. The zinc finger proteins have the characteristic, approximately 30-amino-acid-long, zinc finger motifs as sites of binding to typically DNA, often repeated motifs within a molecule (7, 59).
In contrast, such functional similarities as seen in C. albicans, M. tuberculosis, or H. sapiens were not evident in TR proteins of T. cruzi, especially in those with high TR scores. Of 357 TR genes identified from T. cruzi (Table 1), 14 were apparently not full length, as they lacked either a start or a stop codon, and were not analyzed further. The mean and median CDS lengths of the remaining 343 T. cruzi TR genes were 2,220 and 1,920 bp, respectively, and were larger than those of the average for all T. cruzi genes (1,513 and 1,152 bp, respectively (20). The prevalences of proteins having predicted signal sequences or transmembrane domains among these T. cruzi TR proteins were 27.4 and 31.1%, respectively, and were slightly higher than those in all T. cruzi proteins (16.0 and 26.4%, respectively [data obtained from TcruziDB]). Higher prevalences of TR proteins with predicted signal sequences (41.3%) or transmembrane domains (40.1%) were found in those having scores of <500, but they were not apparent in those with scores of ≥500 (16.6 and 23.8%, respectively). As shown in Table 2, predicted trans-sialidase, mucins, and mucin-associated surface proteins, the three largest gene families in T. cruzi (20), constituted >40% of the T. cruzi TR proteins with a score of <500. In contrast, the category of TR proteins scoring ≥500 did not have a high prevalence of any particular family and the majority was categorized as hypothetical proteins lacking known functional domains. Unlike TR proteins with lower TR values (<500), those with a ≥500 score had a higher mean hydrophilicity compared with that of all T. cruzi proteins (Table 2, P < 0.0001 by Mann-Whitney test).
TABLE 2.
Characteristics of T. cruzi TR proteins
| Parameter | No. of proteins (% of total) with:
|
Total no. of T. cruzi proteins (% of total)c | |
|---|---|---|---|
| 150-499 TRsa | ≥500 TRsb | ||
| Presence of: | |||
| Signal peptide | 62 (41.3) | 32 (16.6) | 3,141 (16.0) |
| Transmembrane domain(s) | 61 (40.1) | 46 (23.8) | 5,169 (26.4) |
| Gene products: | |||
| trans-Sialidase | 20 (13.3) | 14 (7.3) | 735 (3.7) |
| MASPd | 24 (16.0) | 3 (1.6) | 938 (4.8) |
| Mucin | 22 (14.7) | 2 (1.0) | 662 (3.4) |
| Hypothetical protein | 38 (25.3) | 131 (67.9) | 11,171 (57.0) |
No. of genes, 150. Mean hydrophobicity score, 4.124.
No. of genes, 193. Mean hydrophobicity score, 3.868 (P < 0.0001 by Mann-Whitney test compared with hydrophobicity of all T. cruzi proteins).
No. of genes, 19,605. Mean hydrophobicity score, 4.177.
MASP, mucin-associated surface proteins.
Immunological dominance of T. cruzi TR.
For immunological analyses of T. cruzi TR, we focused on the 203 (1.04%) genes with a score of ≥500. Because the nearly 20,000 T. cruzi genes analyzed in this study are from its diploid genome (a haploid T. cruzi genome is estimated to have ∼12,000 protein-coding genes) (20), many of the genes, TR genes included, are represented twice in the pool of analyzed genes. After consolidating TR genes with 70% or greater identity, 106 genes with different TR domains were identified (the top 20 sequences are shown in Table 3). Of the 106 genes, 10 encoded previously characterized antigenic repeat motifs: clone 36, CRA, TcD, B12, B13, SAPA, FRA, TcLo1.2, TcE, and antigen 38 (1, 14, 18, 28, 32, 34, 42). The remaining 96 genes are previously uncharacterized as encoding antigens.
TABLE 3.
Top 20 TR genes of T. cruzi
| Gene no. | Identity | No. of similar TR genes | Size (kDa) | PSa (bp) | CNb | TR score | % TRc | MSd expression | Export propertye | Seroreactivityf | Leishmania homologg | Nameh | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Tc00.1047053510217.10 | 11 | 163 | 195 | 21.7 | 7,161 | 100 | No | No | Unknown | No | Tc8 | |
| 2 | Tc00.1047053511557.50 | 3 | 284 | 126 | 36.4 | 6,969 | 61 | No | TM | Unknown | No | Tc1 | |
| 3 | Tc00.1047053504019.3 | 1 | 206 | 42 | 103.4 | 6,904 | 80 | T | No | Unknown | No | Tc9 | |
| 4 | Tc00.1047053511633.79 | 1 | 126 | 114 | 22 | 4,849 | 77 | AEMT | SP | Clone 36 | No | 34 | |
| 5 | Tc00.1047053510827.40 | 0 | 158 | 486 | 7.1 | 4,802 | 88 | No | No | Unknown | No | ||
| 6 | Tc00.1047053506495.40 | 3 | 106 | 45 | 65.1 | 4,613 | 93 | No | SP, TM | Unknown | No | Tc10 | |
| 7 | Tc00.1047053506303.80 | 1 | 141 | 126 | 24.6 | 4,569 | 77 | No | No | CRA | No | 42 | |
| 8 | Tc00.1047053509265.110 | 4 | 185 | 15 | 178 | 4,386 | 51 | No | TM | TcD | No | 14 | |
| 9 | Tc00.1047053506491.20 | 2 | 399 | 126 | 71.2 | 4,384 | 41 | No | No | Unknown | No | Tc12 | |
| 10 | Tc00.1047053506559.559 | 0 | NA | 72 | 30.9 | 4,017 | 29 | M | NAi | Unknown | No | Tc13 | |
| 11 | Tc00.1047053508831.150 | 1 | 171 | 60 | 59.1 | 3,858 | 71 | No | No | B12 | Yes | 28 | |
| 12 | Tc00.1047053511671.60 | 1 | 125 | 36 | 72.4 | 3,842 | 69 | No | No | B13 | No | 28 | |
| 13 | Tc00.1047053511821.179 | 1 | 144 | 105 | 21.2 | 3,753 | 60 | AEMT | No | Unknown | No | Tc3 | |
| 14 | Tc00.1047053509157.120 | 1 | 116 | 126 | 15.5 | 3,555 | 62 | No | No | Unknown | No | Tc4 | |
| 15 | Tc00.1047053503617.20 | 1 | 218 | 30 | 99.9 | 3,524 | 51 | No | No | Unknown | Yes | ||
| 16 | Tc00.1047053511805.20 | 1 | 83 | 318 | 5.5 | 3,477 | 85 | No | No | Unknown | No | ||
| 17 | Tc00.1047053509269.4 | 1 | 150 | 27 | 111.1 | 3,392 | 73 | No | No | Unknown | Yes | ||
| 18 | Tc00.1047053506777.110 | 1 | 157 | 117 | 15.3 | 3,267 | 44 | A | No | Unknown | No | ||
| 19 | Tc00.1047053508119.200 | 0 | 103 | 117 | 14.1 | 3,191 | 55 | No | No | Unknown | No | Tc6 | |
| 20 | Tc00.1047053507049.119 | 1 | 128 | 60 | 30.5 | 3,160 | 49 | No | No | Unknown | No | Tc15 |
PS, period size.
CN, copy number.
Percentage of TR domains in nucleotide sequence of entire gene.
MS, mass spectroscopy-based protein expression evidence. A, amastigote; E, epimastigote; M, metacyclic trypomastigote; T, trypomastigote.
Presence of predicted signal sequence (SP) or transmembrane domain(s) (TM).
Unknown: serological reactivity not reported before; other entries are the names of previously characterized antigens.
Presence of Leishmania proteins with homologous repeat motifs with 60% amino acid sequence identity as the cutoff.
Names of recombinant proteins designated for this study.
NA, no data available.
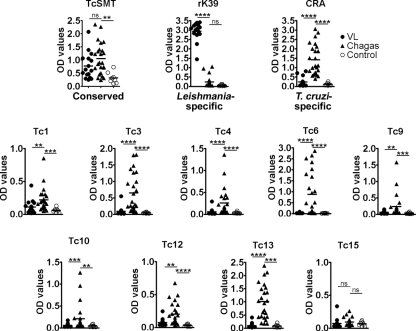
To examine whether previously uncharacterized T. cruzi TR proteins are also antigenic and potentially useful as diagnostics, we cloned and expressed recombinant forms of nine of the remaining proteins listed in Table 3. Two with homology to Leishmania proteins were excluded to avoid cross-reactivity with antibody from leishmaniasis patients. Eight of these detected antibodies in Chagas’ disease patient sera, and the responses were disease specific; the antibody recognition patterns by these antigens were similar to that of CRA and contrast with that of TcSMT or rK39 (Fig. 3). Therefore, at least 13 of the top 20 TR proteins, including the five previously characterized antigens, are antigenic.
FIG. 3.
Antigenic properties of T. cruzi TR proteins. Newly identified TR proteins and previously characterized Leishmania-specific antigen rK39, T. cruzi-specific antigen CRA, and conserved antigen TcSMT were evaluated by ELISA for reactivity with sera from VL patients (n = 16), Chagas’ disease patients (n = 24), and healthy controls (n = 8). OD, optical density; ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (by either unpaired t test or Mann-Whitney test).
DISCUSSION
TR proteins have been implicated in the ability to influence host immune responses to protozoan parasites and possibility to contribute to parasite survival. We previously described the antigenicity of TRs in Leishmania (26). In this study, we demonstrated that TR proteins from the related trypanosomatid parasite T. cruzi are also immunodominant, while there is little or no sequence similarity or apparent bias in cellular location. These features of TR proteins in the trypanosomatid parasites contrast with those in C. albicans, M. tuberculosis, and H. sapiens, which have biased cellular locations or belong to functional protein families. The immunological dominance of TR proteins in Leishmania and Trypanosoma parasites is supported by the fact that antigens of these parasites identified through serological screening of expression libraries are enriched for such proteins relative to the entire proteome. Forty-four percent of Leishmania antigens identified by serological screening of the expression library were TR proteins, whereas such proteins compose only 1% of the proteome (25, 26). There are 37 T. cruzi proteins cited as defined serological antigens in the review article by da Silveira et al. (19), and 9 (24%) of them are TR proteins. When the prevalence of TR proteins in the T. cruzi proteome is considered (<2%), the likelihood that such proteins are antigenic is significantly higher than that observed for non-TR proteins (P < 0.0001 by Fisher's exact test). The role of such proteins in parasite survival remains to be defined. A2, one of the Leishmania TR proteins, was shown to be a virulence factor of L. donovani, the causative agent of VL; it has more than 40 copies of a 10-amino-acid repeat, whereas A2 in L. major, which causes cutaneous leishmaniasis, has only 1 copy of the repeat, suggesting that multiple repeats in the A2 protein may play a role in the visceralization of the parasites (62, 63). We have found other pairs of TR genes in these Leishmania species; overall sequences are highly conserved in both non-TR and TR domains, with the major difference being in the copy number of the repeat (data not shown), suggesting that the chromosome bias was set before the divergence of these species or selective pressure on certain alleles caused the expansion or loss of TR regions in ancestor genes. Genetically distinct strains of T. cruzi are responsible for different clinical syndromes in humans (12, 46, 48). For example, the T. cruzi Z12 zymodeme tends to cause the acute form of Chagas’ disease, whereas the Z1 zymodeme preferentially causes chronic disease in humans (48). It is not known whether variability in TR genes is a primary factor giving different T. cruzi strains or zymodemes divergent characteristics relating to clinical outcomes, disease-induced immune responses, and preference for species of the insect vector reduviid bugs. Taken together, it is worthwhile analyzing characteristics of TR proteins such as the copy number and consensus sequence of the repeat(s) to determine whether they might explain certain biological properties of the parasite and possibly susceptibility to drugs.
Computational searches for such TR proteins may facilitate identifying novel antigens from parasite genomes. Identification of B-cell antigens from pathogens may facilitate the development of diagnostic tests or vaccines. Diagnostic methods for VL and Chagas’ disease often rely on the detection of parasite-specific antibodies (53, 56, 57). In Plasmodium, TR antigens including circumsporozoite protein and merozoite surface antigen 1 are promising malaria vaccine candidates (23, 55). Traditionally, such targets of B-cell responses have been identified from parasites through serological screening of an expression library or by immunoblotting of crude lysate separated by two-dimensional gel electrophoresis. In contrast, only a few attempts have been made to computationally predict serological antigens of pathogens from the proteome based on their sequences, such as predicting secreted or surface proteins (4, 11) and identifying proteins with α-helical coiled-coil domains (61). Although the prediction of secreted or surface proteins has shown some promise in identifying antigens from T. cruzi (11), it may not be powerful enough to reduce the number of candidates to a practical level when dealing with the whole genome. There are 3,141 T. cruzi genes containing sequences encoding predicted signal peptides, 5,169 with transmembrane domains, and 1,776 containing both. Because a signal sequence is either present or not, it is impossible to further prioritize these potentially secreted proteins. Therefore, new bioinformatic tools to screen sequences for antigen-encoding genes are needed and searching for TRs may serve as a powerful method of discovering novel antigens in combination with other computational methods.
There is a real need for improved diagnostics for Chagas’ disease. T. cruzi parasites are not readily detected during chronic infection. Therefore, indirect methods are required for the diagnosis of Chagas’ disease patients and screening for T. cruzi-contaminated samples. Serological tests for Chagas’ disease include the indirect fluorescent-antibody test and an ELISA with whole-cell or recombinant antigens. The Ortho T. cruzi ELISA Test System (www.orthoclinical.com/chagas/elisaTestSystem.aspx) is the first such test approved by the FDA and is used for blood screening. However, serological tests that use T. cruzi whole-cell lysate have cross-reactivity with leishmaniasis and other patient sera. This is likely because a number of proteins are conserved between the related kinetoplastid T. cruzi and Leishmania parasites. Leishmania is endemic in South America, often overlapping in distribution with T. cruzi. For this reason, we focused on T. cruzi TR proteins without homology to Leishmania proteins, and such antigenic proteins will be useful in the development of more accurate diagnostic tests for Chagas’ disease.
Acknowledgments
We thank Randy Howard for critical comments and Raodoh Mohamath and Alex Picone for technical assistance.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 14 July 2008.
REFERENCES
- 1.Affranchino, J. L., C. F. Ibanez, A. O. Luquetti, A. Rassi, M. B. Reyes, R. A. Macina, L. Aslund, U. Pettersson, and A. C. Frasch. 1989. Identification of a Trypanosoma cruzi antigen that is shed during the acute phase of Chagas’ disease. Mol. Biochem. Parasitol. 34221-228. [DOI] [PubMed] [Google Scholar]
- 2.Agüero, F., W. Zheng, D. B. Weatherly, P. Mendes, and J. C. Kissinger. 2006. TcruziDB: an integrated, post-genomics community resource for Trypanosoma cruzi. Nucleic Acids Res. 34D428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allred, D. R., T. C. McGuire, G. H. Palmer, S. R. Leib, T. M. Harkins, T. F. McElwain, and A. F. Barbet. 1990. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc. Natl. Acad. Sci. USA 873220-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aráoz, R., N. Honore, S. Cho, J. P. Kim, S. N. Cho, M. Monot, C. Demangel, P. J. Brennan, and S. T. Cole. 2006. Antigen discovery: a postgenomic approach to leprosy diagnosis. Infect. Immun. 74175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnaud, M. B., M. C. Costanzo, M. S. Skrzypek, G. Binkley, C. Lane, S. R. Miyasato, and G. Sherlock. 2005. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 33D358-D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atwood, J. A., III, D. B. Weatherly, T. A. Minning, B. Bundy, C. Cavola, F. R. Opperdoes, R. Orlando, and R. L. Tarleton. 2005. The Trypanosoma cruzi proteome. Science 309473-476. [DOI] [PubMed] [Google Scholar]
- 7.Beerli, R. R., and C. F. Barbas III. 2002. Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 20135-141. [DOI] [PubMed] [Google Scholar]
- 8.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berriman, M., E. Ghedin, C. Hertz-Fowler, G. Blandin, H. Renauld, D. C. Bartholomeu, N. J. Lennard, E. Caler, N. E. Hamlin, B. Haas, U. Bohme, L. Hannick, M. A. Aslett, J. Shallom, L. Marcello, L. Hou, B. Wickstead, U. C. Alsmark, C. Arrowsmith, R. J. Atkin, A. J. Barron, F. Bringaud, K. Brooks, M. Carrington, I. Cherevach, T. J. Chillingworth, C. Churcher, L. N. Clark, C. H. Corton, A. Cronin, R. M. Davies, J. Doggett, A. Djikeng, T. Feldblyum, M. C. Field, A. Fraser, I. Goodhead, Z. Hance, D. Harper, B. R. Harris, H. Hauser, J. Hostetler, A. Ivens, K. Jagels, D. Johnson, J. Johnson, K. Jones, A. X. Kerhornou, H. Koo, N. Larke, S. Landfear, C. Larkin, V. Leech, A. Line, A. Lord, A. Macleod, P. J. Mooney, S. Moule, D. M. Martin, G. W. Morgan, K. Mungall, H. Norbertczak, D. Ormond, G. Pai, C. S. Peacock, J. Peterson, M. A. Quail, E. Rabbinowitsch, M. A. Rajandream, C. Reitter, S. L. Salzberg, M. Sanders, S. Schobel, S. Sharp, M. Simmonds, A. J. Simpson, L. Tallon, C. M. Turner, A. Tait, A. R. Tivey, S. Van Aken, D. Walker, D. Wanless, S. Wang, B. White, O. White, S. Whitehead, J. Woodward, J. Wortman, M. D. Adams, T. M. Embley, K. Gull, E. Ullu, J. D. Barry, A. H. Fairlamb, F. Opperdoes, B. G. Barrell, J. E. Donelson, N. Hall, C. M. Fraser, S. E. Melville, and N. M. El-Sayed. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309416-422. [DOI] [PubMed] [Google Scholar]
- 10.Bhatia, A., N. S. Daifalla, S. Jen, R. Badaro, S. G. Reed, and Y. A. Skeiky. 1999. Cloning, characterization and serological evaluation of K9 and K26: two related hydrophilic antigens of Leishmania chagasi. Mol. Biochem. Parasitol. 102249-261. [DOI] [PubMed] [Google Scholar]
- 11.Bhatia, V., M. Sinha, B. Luxon, and N. Garg. 2004. Utility of the Trypanosoma cruzi sequence database for identification of potential vaccine candidates by in silico and in vitro screening. Infect. Immun. 726245-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenière, S. F., R. Carrasco, S. Revollo, G. Aparicio, P. Desjeux, and M. Tibayrenc. 1989. Chagas’ disease in Bolivia: clinical and epidemiological features and zymodeme variability of Trypanosoma cruzi strains isolated from patients. Am. J. Trop. Med. Hyg. 41521-529. [DOI] [PubMed] [Google Scholar]
- 13.Burns, J. M., Jr., W. G. Shreffler, D. R. Benson, H. W. Ghalib, R. Badaro, and S. G. Reed. 1993. Molecular characterization of a kinesin-related antigen of Leishmania chagasi that detects specific antibody in African and American visceral leishmaniasis. Proc. Natl. Acad. Sci. USA 90775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns, J. M., Jr., W. G. Shreffler, D. E. Rosman, P. R. Sleath, C. J. March, and S. G. Reed. 1992. Identification and synthesis of a major conserved antigenic epitope of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA 891239-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 16.Coppel, R. L., A. F. Cowman, R. F. Anders, A. E. Bianco, R. B. Saint, K. R. Lingelbach, D. J. Kemp, and G. V. Brown. 1984. Immune sera recognize on erythrocytes Plasmodium falciparum antigen composed of repeated amino acid sequences. Nature 310789-792. [DOI] [PubMed] [Google Scholar]
- 17.Dame, J. B., J. L. Williams, T. F. McCutchan, J. L. Weber, R. A. Wirtz, W. T. Hockmeyer, W. L. Maloy, J. D. Haynes, I. Schneider, D. Roberts, et al. 1984. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science 225593-599. [DOI] [PubMed] [Google Scholar]
- 18.DaRocha, W. D., D. C. Bartholomeu, C. D. Macedo, M. F. Horta, E. Cunha-Neto, J. E. Donelson, and S. M. Teixeira. 2002. Characterization of cDNA clones encoding ribonucleoprotein antigens expressed in Trypanosoma cruzi amastigotes. Parasitol. Res. 88292-300. [DOI] [PubMed] [Google Scholar]
- 19.da Silveira, J. F., E. S. Umezawa, and A. O. Luquetti. 2001. Chagas disease: recombinant Trypanosoma cruzi antigens for serological diagnosis. Trends Parasitol. 17286-291. [DOI] [PubMed] [Google Scholar]
- 20.El-Sayed, N. M., P. J. Myler, D. C. Bartholomeu, D. Nilsson, G. Aggarwal, A. N. Tran, E. Ghedin, E. A. Worthey, A. L. Delcher, G. Blandin, S. J. Westenberger, E. Caler, G. C. Cerqueira, C. Branche, B. Haas, A. Anupama, E. Arner, L. Aslund, P. Attipoe, E. Bontempi, F. Bringaud, P. Burton, E. Cadag, D. A. Campbell, M. Carrington, J. Crabtree, H. Darban, J. F. da Silveira, P. de Jong, K. Edwards, P. T. Englund, G. Fazelina, T. Feldblyum, M. Ferella, A. C. Frasch, K. Gull, D. Horn, L. Hou, Y. Huang, E. Kindlund, M. Klingbeil, S. Kluge, H. Koo, D. Lacerda, M. J. Levin, H. Lorenzi, T. Louie, C. R. Machado, R. McCulloch, A. McKenna, Y. Mizuno, J. C. Mottram, S. Nelson, S. Ochaya, K. Osoegawa, G. Pai, M. Parsons, M. Pentony, U. Pettersson, M. Pop, J. L. Ramirez, J. Rinta, L. Robertson, S. L. Salzberg, D. O. Sanchez, A. Seyler, R. Sharma, J. Shetty, A. J. Simpson, E. Sisk, M. T. Tammi, R. Tarleton, S. Teixeira, S. Van Aken, C. Vogt, P. N. Ward, B. Wickstead, J. Wortman, O. White, C. M. Fraser, K. D. Stuart, and B. Andersson. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309409-415. [DOI] [PubMed] [Google Scholar]
- 21.Gajria, B., A. Bahl, J. Brestelli, J. Dommer, S. Fischer, X. Gao, M. Heiges, J. Iodice, J. C. Kissinger, A. J. Mackey, D. F. Pinney, D. S. Roos, C. J. Stoeckert, Jr., H. Wang, and B. P. Brunk. 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 36(Database issue)D553-D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girard, M. P., Z. H. Reed, M. Friede, and M. P. Kieny. 2007. A review of human vaccine research and development: malaria. Vaccine 251567-1580. [DOI] [PubMed] [Google Scholar]
- 24.Goto, Y., L. Y. Bogatzki, S. Bertholet, R. N. Coler, and S. G. Reed. 2007. Protective immunization against visceral leishmaniasis using Leishmania sterol 24-c-methyltransferase formulated in adjuvant. Vaccine 257450-7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goto, Y., R. N. Coler, J. Guderian, R. Mohamath, and S. G. Reed. 2006. Cloning, characterization, and serodiagnostic evaluation of Leishmania infantum tandem repeat proteins. Infect. Immun. 743939-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto, Y., R. N. Coler, and S. G. Reed. 2007. Bioinformatic identification of tandem repeat antigens of the Leishmania donovani complex. Infect. Immun. 75846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravekamp, C., B. Rosner, and L. C. Madoff. 1998. Deletion of repeats in the alpha C protein enhances the pathogenicity of group B streptococci in immune mice. Infect. Immun. 664347-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruber, A., and B. Zingales. 1993. Trypanosoma cruzi: characterization of two recombinant antigens with potential application in the diagnosis of Chagas’ disease. Exp. Parasitol. 761-12. [DOI] [PubMed] [Google Scholar]
- 29.Hermans, P. W., D. van Soolingen, and J. D. van Embden. 1992. Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae. J. Bacteriol. 1744157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hertz-Fowler, C., C. S. Peacock, V. Wood, M. Aslett, A. Kerhornou, P. Mooney, A. Tivey, M. Berriman, N. Hall, K. Rutherford, J. Parkhill, A. C. Ivens, M. A. Rajandream, and B. Barrell. 2004. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 32D339-D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1986. Complete nucleotide sequence of type 6 M protein of the group A streptococcus. Repetitive structure and membrane anchor. J. Biol. Chem. 2611677-1686. [PubMed] [Google Scholar]
- 32.Houghton, R. L., D. R. Benson, L. D. Reynolds, P. D. McNeill, P. R. Sleath, M. J. Lodes, Y. A. Skeiky, D. A. Leiby, R. Badaro, and S. G. Reed. 1999. A multi-epitope synthetic peptide and recombinant protein for the detection of antibodies to Trypanosoma cruzi in radioimmunoprecipitation-confirmed and consensus-positive sera. J. Infect. Dis. 1791226-1234. [DOI] [PubMed] [Google Scholar]
- 33.Hoyer, L. L., C. B. Green, S. H. Oh, and X. Zhao. 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 461-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibañez, C. F., J. L. Affranchino, R. A. Macina, M. B. Reyes, S. Leguizamon, M. E. Camargo, L. Aslund, U. Pettersson, and A. C. Frasch. 1988. Multiple Trypanosoma cruzi antigens containing tandemly repeated amino acid sequence motifs. Mol. Biochem. Parasitol. 3027-33. [DOI] [PubMed] [Google Scholar]
- 35.Ivens, A. C., C. S. Peacock, E. A. Worthey, L. Murphy, G. Aggarwal, M. Berriman, E. Sisk, M. A. Rajandream, E. Adlem, R. Aert, A. Anupama, Z. Apostolou, P. Attipoe, N. Bason, C. Bauser, A. Beck, S. M. Beverley, G. Bianchettin, K. Borzym, G. Bothe, C. V. Bruschi, M. Collins, E. Cadag, L. Ciarloni, C. Clayton, R. M. Coulson, A. Cronin, A. K. Cruz, R. M. Davies, J. De Gaudenzi, D. E. Dobson, A. Duesterhoeft, G. Fazelina, N. Fosker, A. C. Frasch, A. Fraser, M. Fuchs, C. Gabel, A. Goble, A. Goffeau, D. Harris, C. Hertz-Fowler, H. Hilbert, D. Horn, Y. Huang, S. Klages, A. Knights, M. Kube, N. Larke, L. Litvin, A. Lord, T. Louie, M. Marra, D. Masuy, K. Matthews, S. Michaeli, J. C. Mottram, S. Muller-Auer, H. Munden, S. Nelson, H. Norbertczak, K. Oliver, S. O'Neil, M. Pentony, T. M. Pohl, C. Price, B. Purnelle, M. A. Quail, E. Rabbinowitsch, R. Reinhardt, M. Rieger, J. Rinta, J. Robben, L. Robertson, J. C. Ruiz, S. Rutter, D. Saunders, M. Schafer, J. Schein, D. C. Schwartz, K. Seeger, A. Seyler, S. Sharp, H. Shin, D. Sivam, R. Squares, S. Squares, V. Tosato, C. Vogt, G. Volckaert, R. Wambutt, T. Warren, H. Wedler, J. Woodward, S. Zhou, W. Zimmermann, D. F. Smith, J. M. Blackwell, K. D. Stuart, B. Barrell, and P. J. Myler. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 1017329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan, P., L. A. Snyder, and N. J. Saunders. 2003. Diversity in coding tandem repeats in related Neisseria spp. BMC Microbiol. 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katti, M. V., R. Sami-Subbu, P. K. Ranjekar, and V. S. Gupta. 2000. Amino acid repeat patterns in protein sequences: their diversity and structural-functional implications. Protein Sci. 91203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kemp, D. J., R. L. Coppel, and R. F. Anders. 1987. Repetitive proteins and genes of malaria. Annu. Rev. Microbiol. 41181-208. [DOI] [PubMed] [Google Scholar]
- 40.Koenen, M., A. Scherf, O. Mercereau, G. Langsley, L. Sibilli, P. Dubois, L. Pereira da Silva, and B. Muller-Hill. 1984. Human antisera detect a Plasmodium falciparum genomic clone encoding a nonapeptide repeat. Nature 311382-385. [DOI] [PubMed] [Google Scholar]
- 41.Kotera, Y., J. D. Fontenot, G. Pecher, R. S. Metzgar, and O. J. Finn. 1994. Humoral immunity against a tandem repeat epitope of human mucin MUC-1 in sera from breast, pancreatic, and colon cancer patients. Cancer Res. 542856-2860. [PubMed] [Google Scholar]
- 42.Lafaille, J. J., J. Linss, M. A. Krieger, T. Souto-Padron, W. de Souza, and S. Goldenberg. 1989. Structure and expression of two Trypanosoma cruzi genes encoding antigenic proteins bearing repetitive epitopes. Mol. Biochem. Parasitol. 35127-136. [DOI] [PubMed] [Google Scholar]
- 43.Levdansky, E., J. Romano, Y. Shadkchan, H. Sharon, K. J. Verstrepen, G. R. Fink, and N. Osherov. 2007. Coding tandem repeats generate diversity in Aspergillus fumigatus genes. Eukaryot. Cell 61380-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madoff, L. C., J. L. Michel, E. W. Gong, D. E. Kling, and D. L. Kasper. 1996. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc. Natl. Acad. Sci. USA 934131-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marri, P. R., J. P. Bannantine, and G. B. Golding. 2006. Comparative genomics of metabolic pathways in Mycobacterium species: gene duplication, gene decay and lateral gene transfer. FEMS Microbiol. Rev. 30906-925. [DOI] [PubMed] [Google Scholar]
- 46.Miles, M. A., R. A. Cedillos, M. M. Povoa, A. A. de Souza, A. Prata, and V. Macedo. 1981. Do radically dissimilar Trypanosoma cruzi strains (zymodemes) cause Venezuelan and Brazilian forms of Chagas’ disease? Lancet 11338-1340. [DOI] [PubMed] [Google Scholar]
- 47.Mollick, J. A., F. S. Hodi, R. J. Soiffer, L. M. Nadler, and G. Dranoff. 2003. MUC1-like tandem repeat proteins are broadly immunogenic in cancer patients. Cancer Immun. 33. [PubMed] [Google Scholar]
- 48.Montamat, E. E., G. M. De Luca D'Oro, R. H. Gallerano, R. Sosa, and A. Blanco. 1996. Characterization of Trypanosoma cruzi populations by zymodemes: correlation with clinical picture. Am. J. Trop. Med. Hyg. 55625-628. [DOI] [PubMed] [Google Scholar]
- 49.Pais, F. S., W. D. Darocha, R. M. Almeida, S. Y. Leclercq, M. L. Penido, S. P. Fragoso, D. C. Bartholomeu, R. T. Gazzinelli, and S. M. Teixeira. 2008. Molecular characterization of ribonucleoproteic antigens containing repeated amino acid sequences from Trypanosoma cruzi. Microbes Infect. 10716-725. [DOI] [PubMed] [Google Scholar]
- 50.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413848-852. [DOI] [PubMed] [Google Scholar]
- 51.Peacock, C. S., K. Seeger, D. Harris, L. Murphy, J. C. Ruiz, M. A. Quail, N. Peters, E. Adlem, A. Tivey, M. Aslett, A. Kerhornou, A. Ivens, A. Fraser, M. A. Rajandream, T. Carver, H. Norbertczak, T. Chillingworth, Z. Hance, K. Jagels, S. Moule, D. Ormond, S. Rutter, R. Squares, S. Whitehead, E. Rabbinowitsch, C. Arrowsmith, B. White, S. Thurston, F. Bringaud, S. L. Baldauf, A. Faulconbridge, D. Jeffares, D. P. Depledge, S. O. Oyola, J. D. Hilley, L. O. Brito, L. R. Tosi, B. Barrell, A. K. Cruz, J. C. Mottram, D. F. Smith, and M. Berriman. 2007. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 39839-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poulet, S., and S. T. Cole. 1995. Characterization of the highly abundant polymorphic GC-rich-repetitive sequence (PGRS) present in Mycobacterium tuberculosis. Arch. Microbiol. 16387-95. [DOI] [PubMed] [Google Scholar]
- 53.Reed, S. G. 1996. Diagnosis of leishmaniasis. Clin. Dermatol. 14471-478. [DOI] [PubMed] [Google Scholar]
- 54.Reeder, J. C., and G. V. Brown. 1996. Antigenic variation and immune evasion in Plasmodium falciparum malaria. Immunol. Cell Biol. 74546-554. [DOI] [PubMed] [Google Scholar]
- 55.Richie, T. L., and A. Saul. 2002. Progress and challenges for malaria vaccines. Nature 415694-701. [DOI] [PubMed] [Google Scholar]
- 56.Singh, S., and R. Sivakumar. 2003. Recent advances in the diagnosis of leishmaniasis. J. Postgrad. Med. 4955-60. [DOI] [PubMed] [Google Scholar]
- 57.Sundar, S., and M. Rai. 2002. Laboratory diagnosis of visceral leishmaniasis. Clin. Diagn. Lab. Immunol. 9951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van het Hoog, M., T. J. Rast, M. Martchenko, S. Grindle, D. Dignard, H. Hogues, C. Cuomo, M. Berriman, S. Scherer, B. B. Magee, M. Whiteway, H. Chibana, A. Nantel, and P. T. Magee. 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome. Biol. 8R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. Gabor Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. McKusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-Threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, A. Yao, J. Ye, M. Zhan, W. Zhang, H. Zhang, Q. Zhao, L. Zheng, F. Zhong, W. Zhong, S. Zhu, S. Zhao, D. Gilbert, S. Baumhueter, G. Spier, C. Carter, A. Cravchik, T. Woodage, F. Ali, H. An, A. Awe, D. Baldwin, H. Baden, M. Barnstead, I. Barrow, K. Beeson, D. Busam, A. Carver, A. Center, M. L. Cheng, L. Curry, S. Danaher, L. Davenport, R. Desilets, S. Dietz, K. Dodson, L. Doup, S. Ferriera, N. Garg, A. Gluecksmann, B. Hart, J. Haynes, C. Haynes, C. Heiner, S. Hladun, D. Hostin, J. Houck, T. Howland, C. Ibegwam, J. Johnson, F. Kalush, L. Kline, S. Koduru, A. Love, F. Mann, D. May, S. McCawley, T. McIntosh, I. McMullen, M. Moy, L. Moy, B. Murphy, K. Nelson, C. Pfannkoch, E. Pratts, V. Puri, H. Qureshi, M. Reardon, R. Rodriguez, Y. H. Rogers, D. Romblad, B. Ruhfel, R. Scott, C. Sitter, M. Smallwood, E. Stewart, R. Strong, E. Suh, R. Thomas, N. N. Tint, S. Tse, C. Vech, G. Wang, J. Wetter, S. Williams, M. Williams, S. Windsor, E. Winn-Deen, K. Wolfe, J. Zaveri, K. Zaveri, J. F. Abril, R. Guigo, M. J. Campbell, K. V. Sjolander, B. Karlak, A. Kejariwal, H. Mi, B. Lazareva, T. Hatton, A. Narechania, K. Diemer, A. Muruganujan, N. Guo, S. Sato, V. Bafna, S. Istrail, R. Lippert, R. Schwartz, B. Walenz, S. Yooseph, D. Allen, A. Basu, J. Baxendale, L. Blick, M. Caminha, J. Carnes-Stine, P. Caulk, Y. H. Chiang, M. Coyne, C. Dahlke, A. Mays, M. Dombroski, M. Donnelly, D. Ely, S. Esparham, C. Fosler, H. Gire, S. Glanowski, K. Glasser, A. Glodek, M. Gorokhov, K. Graham, B. Gropman, M. Harris, J. Heil, S. Henderson, J. Hoover, D. Jennings, C. Jordan, J. Jordan, J. Kasha, L. Kagan, C. Kraft, A. Levitsky, M. Lewis, X. Liu, J. Lopez, D. Ma, W. Majoros, J. McDaniel, S. Murphy, M. Newman, T. Nguyen, N. Nguyen, M. Nodell, S. Pan, J. Peck, M. Peterson, W. Rowe, R. Sanders, J. Scott, M. Simpson, T. Smith, A. Sprague, T. Stockwell, R. Turner, E. Venter, M. Wang, M. Wen, D. Wu, M. Wu, A. Xia, A. Zandieh, and X. Zhu. 2001. The sequence of the human genome. Science 2911304-1351. [DOI] [PubMed] [Google Scholar]
- 60.Verstrepen, K. J., A. Jansen, F. Lewitter, and G. R. Fink. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37986-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villard, V., G. W. Agak, G. Frank, A. Jafarshad, C. Servis, I. Nebie, S. B. Sirima, I. Felger, M. Arevalo-Herrera, S. Herrera, F. Heitz, V. Backer, P. Druilhe, A. V. Kajava, and G. Corradin. 2007. Rapid identification of malaria vaccine candidates based on alpha-helical coiled coil protein motif. PLoS ONE 2e645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, W. W., and G. Matlashewski. 1997. Loss of virulence in Leishmania donovani deficient in an amastigote-specific protein, A2. Proc. Natl. Acad. Sci. USA 948807-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, W. W., S. Mendez, A. Ghosh, P. Myler, A. Ivens, J. Clos, D. L. Sacks, and G. Matlashewski. 2003. Comparison of the A2 gene locus in Leishmania donovani and Leishmania major and its control over cutaneous infection. J. Biol. Chem. 27835508-35515. [DOI] [PubMed] [Google Scholar]