Abstract
Between 1 and 2% of the population in the developed world experiences a nonhealing or chronic wound characterized by an apparent arrest in a stage dominated by inflammatory processes. Lately, research groups have proposed that bacteria might be involved in and contribute to the lack of healing of these wounds. To investigate this, we collected and examined samples from chronic wounds obtained from 22 different patients, all selected because of suspicion of Pseudomonas aeruginosa colonization. These wound samples were investigated by standard culturing methods and peptide nucleic acid-based fluorescence in situ hybridization (PNA FISH) for direct identification of bacteria. By means of the culturing methods, Staphylococcus aureus was detected in the majority of the wounds, whereas P. aeruginosa was observed less frequently. In contrast, using PNA FISH, we found that a large fraction of the wounds contained P. aeruginosa. Furthermore, PNA FISH revealed the structural organization of bacteria in the samples. It appeared that P. aeruginosa aggregated as microcolonies imbedded in the matrix component alginate, which is a characteristic hallmark of the biofilm mode of growth. The present investigation suggests that bacteria present within these wounds tend to be aggregated in microcolonies imbedded in a self-produced matrix, characteristic of the biofilm mode of growth. Additionally, we must conclude that there exists no good correlation between bacteria detected by standard culturing methods and those detected by direct detection methods such as PNA FISH. This strongly supports the development of new diagnostic and treatment strategies for chronic wounds.
The number of patients developing chronic wounds is increasing with the worldwide increase in lifestyle diseases, such as obesity, diabetes, and cardiovascular diseases. In developed countries, it has been estimated that 1 to 2% of the population will experience a chronic wound during their lifetime (16). The socioeconomic consequences include patient suffering, loss of employment, and reduced life quality. In-depth knowledge of chronic wounds is urgently required and would significantly improve treatment and prognosis. Chronic wounds seem to be arrested in a stage dominated by inflammatory processes (13), among which the continuing influx of polymorphonuclear leukocytes (PMNs) is likely to play a significant role. Activated PMNs release cytotoxic enzymes, free oxygen radicals, and inflammatory mediators that cause extensive collateral damage to the host tissue (31). We recently suggested that within a chronic wound, the healing and destructive processes are out of balance and that, consequently, by manipulating and counterbalancing these processes the chronic wound might start to heal (4).
The presence of bacteria is most likely to influence this imbalance. A few studies of bacterial profiles in chronic wounds have been published; for example, chronic venous leg ulcers harbor Staphylococcus aureus (in 93.5% of the investigated ulcers), Enterococcus faecalis (71.7%), Pseudomonas aeruginosa (52.2%), coagulase-negative staphylococci (45.7%), Proteus species (41.3%), and anaerobic bacteria (39.1%) (15). A common dominator of previous studies describing the bacteriology of chronic wounds is that they agree on the almost universal presence of S. aureus (7, 15, 35). Although the deep dermal tissues of all chronic wounds harbor multiple species, more than half of the chronic wounds were colonized with P. aeruginosa (15). Furthermore, P. aeruginosa-infected wounds appeared significantly larger in terms of area than wounds that did not contain P. aeruginosa (15, 17, 27).
The presence of P. aeruginosa also seems to delay or even prevent the healing process (27). The presence of bacteria in wounds is a problem which is growing in pace with the increasing emergence of multidrug-resistant P. aeruginosa and S. aureus.
However, a significant contribution to the lack of successful antibiotic treatment might be the ability of colonizing bacteria to establish and proliferate in a biofilm in the wound. The clinical implications of bacterial biofilms are particularly pronounced in chronic infections (10). Microscopic investigations have revealed that biofilms develop commonly on inert surfaces of medical devices or on dead tissues, but they can also form on living tissues, as in the case of endocarditis. Common characteristics of bacterial biofilms are their resistance against the activities of the host immune system and tolerance of antibiotic intervention regimens. Unfortunately, a direct, simple method of identification of biofilms in clinical samples does not exist. What is possible is the identification of bacterial aggregates and matrix components combined with a persistent infection.
Here we report on the distribution and structural organization of bacteria located on the surface and within the chronic wound.
MATERIALS AND METHODS
Wound section preparation.
Material from chronic wounds (n = 22) was collected with acceptance of the patients and in accordance with the biomedical project protocol (KA-20051011), approved by the Danish scientific ethical board. The patients were not randomly or consecutively chosen but were selected because of clinical suspicion of P. aeruginosa colonization or recent P. aeruginosa-positive culture. The clinical suspicion of P. aeruginosa was made due to color, odor, or suspicion from an experienced surgeon. The wound biopsy material for microscopy was immediately fixed in phosphate-buffered saline with 4% paraformaldehyde and kept at 5°C before further preparation. The biopsy material was imbedded in paraffin, sectioned (4 μm), and mounted on glass slides. Before hybridization, the paraffin was removed from the sections by immersing the slides twice in xylene, twice in 99.9% ethanol, and twice in 96% ethanol and washing them three times with sterile water.
Tissue from chronically infected cystic fibrosis patients.
Lung tissues were collected with acceptance of the patients and in accordance with the biomedical project protocol (KF-01278432), approved by the Danish scientific ethical board.
PNA FISH.
The deparaffinized sections were systematically analyzed by fluorescence in situ hybridization (FISH) using peptide nucleic acid (PNA) probes (33). A mixture of a Texas Red-labeled P. aeruginosa-specific PNA probe and a fluorescein-labeled universal bacterium PNA probe or a mixture of a fluorescein-labeled S. aureus-specific PNA probe and a Texas Red-labeled universal bacterium PNA probe, both in hybridization solution (AdvanDx, Inc., Woburn, MA), was added to each section and hybridized in a PNA FISH workstation covered by a lid at 55°C for 90 min. The slides were washed for 30 min at 55°C in wash solution (AdvanDx), mounting medium and 4′,6′-diamidino-2-phenylindole (DAPI) were applied, and the slides were covered with a coverslip. The entire PNA FISH procedure required approximately 2.5 h. Slides were read using a fluorescence microscope equipped with fluorescein isothiocyanate, Texas Red, dual fluorescein isothiocyanate-Texas Red, and DAPI filters.
Three-dimensional imaging of the bacteria in the wounds was done by confocal laser scanning microscopy (CLSM) using a Zeiss LSM 510 system (Carl Zeiss GmbH, Jena, Germany) equipped with an argon laser and a helium-neon laser for excitation of the fluorophores. Simulated fluorescence projections of the biofilms were generated by using the IMARIS software package (Bitplane AG). Images were further processed for display by using Photoshop software (Adobe).
Standard culturing.
Routine swabs were taken intraoperatively for 19 of 22 patients; for the remaining 3 patients, swabs were not available and bacteriology was based on biopsies. The swabs and mechanically crushed tissue were cultured on aerobic and anaerobic standard agar plates (Statens Serum Institute [SSI], Copenhagen, Denmark). The aerobic cultures were grown on selective gram-negative plates (blue plates; SSI) as well as 5% horse blood agar (SSI). All plates were incubated at 37°C. Plates were inspected on days 2 and 4, and differentiation and identification of bacteria were carried out using standard methods.
Immunofluorescence microscopy for alginate.
Anti-alginate sera were obtained from pooled purified anti-alginate immunoglobulins G, obtained by regular subcutaneous immunization and bleeding of rabbits, using purified alginate from the mucoid cystic fibrosis P. aeruginosa strain NH57388A (23) as described previously (29). Since only alginate from one P. aeruginosa isolate was used, occasional non-cross-reactive alginates from other strains may not be demonstrated so easily with the immunoglobulin G preparation (29). The specificity of the anti-alginate sera was verified by comparing staining intensities of the mucoid strain PDO300 (28) to those of the nonmucoid strains PAO1 and PAO1 ΔalgD. Additionally, the staining with anti-alginate sera was compared to staining with nonimmunized rabbit immunoglobulin, which did not raise any signal. The titer of the anti-alginate sera was assessed by enzyme-linked immunosorbent assay, using the purified alginate from strain NH57388A as antigen and peroxidase-conjugated swine anti-rabbit antibody (Dako). The titer (highest reciprocal dilution that was still positive) was 1:320,0000.
The deparaffinized tissue sections were treated in Tris-EGTA buffer, pH 9 (microwave oven; 18 min at 600 W). The sections were washed in sterile water and Tris-buffered saline and then treated with 25% normal donkey serum to diminish background staining. Thereafter, the sections were incubated first with anti-alginate sera (1:1,000 dilution of the 24-g/liter stock solution) and then with donkey anti-rabbit Cy3-conjugated antibodies (1:200 dilution; Jackson ImmunoResearch). After being stained, the samples were mounted with “no fade” solution (25) (buffered paraphenylenediamine glycerol) and counterstained with Mayer's hematoxylin solution.
Statistical evaluation.
To compare the data gained from the different bacterial detection methods, chi-square tests were performed. P values of ≤0.05 were considered significant. For calculation of P values, the statistical program StatView (SAS Institute Inc., Cary, NC) was used.
RESULTS
Determination of distribution and organization of bacteria in wounds.
To investigate if bacteria were present, and if so, how they were organized spatially and structurally, PNA FISH was performed on fixed sections of multiple wounds. The bacteria were present in aggregates under the wound surface (Fig. 1). The depth at which the aggregates were found was correlated to the depth of the wound bed. If the wound bed was deep, aggregates were also deeply imbedded. On the other hand, if the wound bed was thin, the aggregates were found right under the surface. Very few single or planktonic cells were detected in any of the wounds. The aggregates imbedded in the investigated wounds were predominately single-species aggregates of P. aeruginosa (Fig. 1, frames a and b) or S. aureus (Fig. 1, frame e). The spatial organization of the bacteria was visualized using CLSM (Fig. 1, frames c and d). Only one wound was found to harbor mixed aggregates (P. aeruginosa together with unidentified bacteria) (Fig. 1, frame h). Using the available probe set with two specific probes together with the universal probes, unspecific aggregates were detected in many of the wounds (Fig. 1, frame g). It might very well be of great importance to know the identity of the bacteria in these aggregates. Current investigations are under way to solve this issue.
FIG. 1.
(a and b) Microcolonies of P. aeruginosa (identified by a specific PNA FISH probe [red stain]) surrounded by host cells (DAPI [blue stain]). (c) CLSM three-dimensional image of frame b. (d) Enlargement of frame c. (e) Large microcolony of S. aureus. (f) Single cells of S. aureus (identified by a specific PNA FISH probe [green stain]). Host cells are present (interspaced) (DAPI [blue stain]). (g) Microcolony of coccus-shaped bacteria (identified by a universal bacterial probe [green stain; appears purple here due to being mixed with the blue DAPI stain]). (h) Biofilm with mixed bacterial species (red stained bacteria are P. aeruginosa, and green stained bacteria are unidentified bacteria). White arrows point to bacterial aggregates (single cells in frame f), and yellow arrows point to the wound surface.
Identification of P. aeruginosa extracellular biofilm matrix material in wounds.
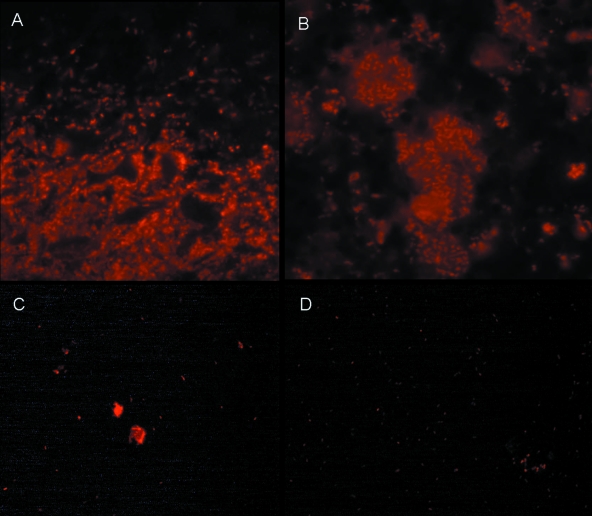
One of the hallmarks of bacterial biofilms is the presence of a self-produced exopolymeric substance surrounding the bacterial aggregates (8, 9, 26). We wanted to investigate whether the aggregates of P. aeruginosa could be considered biofilms in accordance with the commonly accepted biofilm definition. To do this, we employed antibodies against alginate, one of the major exopolymeric substances which P. aeruginosa is known to produce during chronic infection of the lungs of cystic fibrosis patients (30) (Fig. 2). In analyzing the wounds which were found to harbor large P. aeruginosa aggregates, alginate was detected surrounding most of these aggregates (Fig. 2).
FIG. 2.
Presence of exopolymeric substance alginate within and surrounding P. aeruginosa aggregates, as detected by alginate-specific immunostaining. (A) Wound; (B) cystic fibrosis lung; (C) in vitro smear of mucoid P. aeruginosa; (D) in vitro smear of alginate-negative P. aeruginosa.
Assessment of bacterial species distribution in wounds.
To investigate the bacterial prevalence in the wounds, we compared the results obtained using PNA FISH identification and standard culturing. We subdivided the wounds into several groups depending on the culture results and the presence of aggregates (Fig. 3). Due to the availability of specific PNA probes, we focused mainly on the presence of S. aureus and P. aeruginosa. Using standard culturing techniques, the sampled material indicated that 86% (19/22 samples) of the wounds were colonized by bacteria. More than 60% (12/19 samples) of wounds were colonized with S. aureus, and <30% (5/19 samples) of wounds were colonized with P. aeruginosa (P < 0.03). On the other hand, using PNA FISH, we detected large bacterial aggregates in close to 60% (13/22 samples) of the wounds; interestingly, only 15% (2/13 samples) were of S. aureus, whereas approximately 70% (9/13 samples) were of P. aeruginosa. In comparing the two detection methods, P. aeruginosa was detected more frequently with PNA FISH (P < 0.02). This was in contrast to the higher frequency of S. aureus detected by culture growth (P < 0.0001) from the wounds with evidence of S. aureus. Examination of the images obtained using the PNA FISH technique (Fig. 1) indicates that bacteria identified as S. aureus were present in connection with and on the surface of the wound, whereas P. aeruginosa was situated inside the wound bed, i.e., underneath the surface.
FIG. 3.
Number of wounds in each category. Among the wounds with evidence of P. aeruginosa colonization, P. aeruginosa was detected more frequently with PNA FISH (P < 0.02). This was in contrast to the higher frequency of S. aureus detected by culture growth (P < 0.0001) from the wounds with evidence of S. aureus. *, aggregates as described in the legend to Fig. 1.
DISCUSSION
Here we demonstrate the absence of a correlation between the bacterial species detected in wounds by culture and what is in fact present, as detected directly by PNA FISH. These observations may explain the overrepresentation of S. aureus found in most wounds by use of standard culturing swab techniques. If this is a general feature, it may have paramount importance for the interpretation of chronic wound swabs. However, further investigations of the bacterial species distribution in a large number of chronic wounds are under way in our laboratory to determine if this is a general characteristic. One thing which has to be taken into account is the fairly small area of each wound which we examined for aggregates/biofilms by means of PNA FISH. We previously proposed that bacteria are unevenly distributed in wounds (4), but investigating randomly picked samples from a large collection of wounds should statistically minimize any misinterpretation of the bacterial distribution and organization in the wounds.
The presence of large numbers of bacteria aggregated as microcolonies (Fig. 1) imbedded in a self-produced alginate matrix (Fig. 2) is indicative of the presence of P. aeruginosa biofilms in the deeper regions of chronic wounds. Alginate overproduction is known to protect bacteria against oxygen radicals produced and liberated by lysed PMNs. Even though the majority of P. aeruginosa strains isolated from chronic wounds show a nonmucoid phenotype upon culturing, they might still produce alginate in vivo (1, 6; T. Bjarnsholt, unpublished results).
It is generally accepted that when bacteria assume the biofilm phenotype, it offers increased protection against antibiotics and the activities of the host defense (2, 5, 9, 11, 12). Accordingly, our investigations suggest that the bacteria imbedded in the deeper regions of wounds reside in biofilms. These biofilms are the perfect setting for density-dependent gene regulation, such as quorum sensing (QS) (14). In vitro as well as in vivo investigations based on transcriptomics (19) and reporter gene expression (36) suggest that bacterial signaling or QS is enabled in biofilms. For many bacteria, the development of antibiotic and biocide tolerance or resistance phenotypes is regulated at least in part by QS, but the complete mechanism is not yet known (2, 3, 18, 20-22, 32). For P. aeruginosa, QS regulation is also the major regulatory mechanism for virulence factor production (24). As for protection against the host defense, we recently demonstrated that P. aeruginosa produces a strong leukocidal toxin, rhamnolipid B (24). Single cells probably stand little chance against the host defense, which may be why the bacteria form protective aggregates (Fig. 1, frames c and d). It is quite evident from all of the frames in Fig. 1 that the host cells do not penetrate these aggregates. Especially the aggregates of P. aeruginosa (Fig. 1, frames a and b) seem completely walled off from the host cells. Production of rhamnolipid is QS regulated and most likely functions as a shield against the cellular components of the host defense (24). Additionally, the presence of bacteria in aggregates could explain why application of antibiotics has little, if any, effect on the persistence of bacteria in chronic wounds.
It is well known that bacteria such as P. aeruginosa are almost impossible to eradicate from chronic wounds by the use of antibiotics. In addition to this, we recently tested different wound dressings containing silver salts as an antimicrobial agent, which can eradicate planktonic bacteria, but interestingly, none of these contained enough silver to eradicate biofilm bacteria in vitro (5).
In the evaluation of a wound swab or biopsy, it is of paramount importance to consider the discrepancy between an organism being culturable and being detectable by FISH. Our data suggest that bacterial profiling of chronic infected wounds by means of culture may underestimate the presence of important pathogens such as P. aeruginosa. Also, culturing from a biopsy does not reveal the presence of a bacterial biofilm. It is well known that bacteria imbedded in biofilms are difficult to recover and culture (34), a fact that may also contribute to “underdetection” per se. Although the present analysis has limitations, our data suggest that P. aeruginosa and possibly other opportunistic pathogens are of greater importance than S. aureus. We previously proposed that P. aeruginosa keeps infected wounds in a chronic state of inflammation similar to that seen in the lungs of cystic fibrosis patients (4). Due to this situation, we propose that the presence of bacteria plays a major role in the lack of healing for otherwise optimally treated chronic wounds. The realization that notorious opportunistic pathogens, such as P. aeruginosa, are growing as biofilms in chronic wounds urges a search for novel diagnostic and treatment strategies.
Acknowledgments
We thank Ulla Johansen, Lena Nørregaard Department for Clinical Microbiology, for assistance with bacteriology and sample preparation; Anne Jørgensen, Department for Pathology, Copenhagen University Hospital, for help during microscopy; and Bo Jørgensen, Copenhagen Wound Healing Center, for assisting in obtaining the wound samples.
This study received financial support from The Carlsberg Foundation and Lundbeck Foundation to T.B. and from the Strategic Danish Research Council to M.G.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Anastassiou, E. D., A. C. Mintzas, C. Kounavis, and G. Dimitracopoulos. 1987. Alginate production by clinical nonmucoid Pseudomonas aeruginosa strains. J. Clin. Microbiol. 25656-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjarnsholt, T., P. O. Jensen, M. Burmolle, M. Hentzer, J. A. Haagensen, H. P. Hougen, H. Calum, K. G. Madsen, C. Moser, S. Molin, N. Hoiby, and M. Givskov. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151373-383. [DOI] [PubMed] [Google Scholar]
- 3.Bjarnsholt, T., P. O. Jensen, T. B. Rasmussen, L. Christophersen, H. Calum, M. Hentzer, H. P. Hougen, J. Rygaard, C. Moser, L. Eberl, N. Hoiby, and M. Givskov. 2005. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 1513873-3880. [DOI] [PubMed] [Google Scholar]
- 4.Bjarnsholt, T., K. Kirketerp-Moller, P. O. Jensen, K. G. Madsen, R. Phipps, K. Krogfelt, N. Hoiby, and M. Givskov. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 162-10. [DOI] [PubMed] [Google Scholar]
- 5.Bjarnsholt, T., K. Kirketerp-Moller, S. Kristiansen, R. Phipps, A. K. Nielsen, P. O. Jensen, N. Hoiby, and M. Givskov. 2007. Silver against Pseudomonas aeruginosa biofilms. APMIS 115921-928. [DOI] [PubMed] [Google Scholar]
- 6.Bragonzi, A., D. Worlitzsch, G. B. Pier, P. Timpert, M. Ulrich, M. Hentzer, J. B. Andersen, M. Givskov, M. Conese, and G. Doring. 2005. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J. Infect. Dis. 192410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colsky, A. S., R. S. Kirsner, and F. A. Kerdel. 1998. Microbiologic evaluation of cutaneous wounds in hospitalized dermatology patients. Ostomy Wound Manage. 4440-46. [PubMed] [Google Scholar]
- 8.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41435-464. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 2841318-1322. [DOI] [PubMed] [Google Scholar]
- 10.Davies, D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2114-122. [DOI] [PubMed] [Google Scholar]
- 11.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drenkard, E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 51213-1219. [DOI] [PubMed] [Google Scholar]
- 13.Falanga, V. 2000. Classifications for wound bed preparation and stimulation of chronic wounds. Wound Repair Regen. 8347-352. [PubMed] [Google Scholar]
- 14.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50727-751. [DOI] [PubMed] [Google Scholar]
- 15.Gjodsbol, K., J. J. Christensen, T. Karlsmark, B. Jorgensen, B. M. Klein, and K. A. Krogfelt. 2006. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int. Wound J. 3225-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottrup, F. 2004. A specialized wound-healing center concept: importance of a multidisciplinary department structure and surgical treatment facilities in the treatment of chronic wounds. Am. J. Surg. 18738S-43S. [DOI] [PubMed] [Google Scholar]
- 17.Halbert, A. R., M. C. Stacey, J. B. Rohr, and A. Jopp-McKay. 1992. The effect of bacterial colonization on venous ulcer healing. Australas. J. Dermatol. 3375-80. [DOI] [PubMed] [Google Scholar]
- 18.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 341082-1093. [DOI] [PubMed] [Google Scholar]
- 19.Hentzer, M., L. Eberl, and M. Givskov. 2005. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation. Biofilms 237-61. [Google Scholar]
- 20.Hentzer, M., and M. Givskov. 2003. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Investig. 1121300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 14887-102. [DOI] [PubMed] [Google Scholar]
- 22.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 223803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann, N., T. B. Rasmussen, P. O. Jensen, C. Stub, M. Hentzer, S. Molin, O. Ciofu, M. Givskov, H. K. Johansen, and N. Hoiby. 2005. Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect. Immun. 732504-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen, P. O., T. Bjarnsholt, R. Phipps, T. B. Rasmussen, H. Calum, L. Christoffersen, C. Moser, P. Williams, T. Pressler, M. Givskov, and N. Hoiby. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 1531329-1338. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, G. D., and G. M. C. N. Araujo. 1981. A simple method of reducing the fading of immunofluorescence during microscopy. J. Immunol. Methods 43349-350. [DOI] [PubMed] [Google Scholar]
- 26.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madsen, S. M., H. Westh, L. Danielsen, and V. T. Rosdahl. 1996. Bacterial colonization and healing of venous leg ulcers. APMIS 104895-899. [DOI] [PubMed] [Google Scholar]
- 28.Mathee, K., O. Ciofu, C. Sternberg, P. W. Lindum, J. I. Campbell, P. Jensen, A. H. Johnsen, M. Givskov, D. E. Ohman, S. Molin, N. Hoiby, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 1451349-1357. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen, S. S., F. Espersen, N. Hoiby, and G. H. Shand. 1989. Purification, characterization, and immunological cross-reactivity of alginates produced by mucoid Pseudomonas aeruginosa from patients with cystic fibrosis. J. Clin. Microbiol. 27691-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen, S. S., N. Hoiby, F. Espersen, and C. Koch. 1992. Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax 476-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz, G. S., R. G. Sibbald, V. Falanga, E. A. Ayello, C. Dowsett, K. Harding, M. Romanelli, M. C. Stacey, L. Teot, and W. Vanscheidt. 2003. Wound bed preparation: a systematic approach to wound management. Wound Repair Regen. 11(Suppl. 1)S1-S28. [DOI] [PubMed] [Google Scholar]
- 32.Shih, P. C., and C. T. Huang. 2002. Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J. Antimicrob. Chemother. 49309-314. [DOI] [PubMed] [Google Scholar]
- 33.Stender, H. 2003. PNA FISH: an intelligent stain for rapid diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 3649-655. [DOI] [PubMed] [Google Scholar]
- 34.Trampuz, A., K. E. Piper, M. J. Jacobson, A. D. Hanssen, K. K. Unni, D. R. Osmon, J. N. Mandrekar, F. R. Cockerill, J. M. Steckelberg, J. F. Greenleaf, and R. Patel. 2007. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 357654-663. [DOI] [PubMed] [Google Scholar]
- 35.Valencia, I. C., R. S. Kirsner, and F. A. Kerdel. 2004. Microbiologic evaluation of skin wounds: alarming trend toward antibiotic resistance in an inpatient dermatology service during a 10-year period. J. Am. Acad. Dermatol. 50845-849. [DOI] [PubMed] [Google Scholar]
- 36.Wu, H., Z. Song, M. Hentzer, J. B. Andersen, A. Heydorn, K. Mathee, C. Moser, L. Eberl, S. Molin, N. Hoiby, and M. Givskov. 2000. Detection of N-acylhomoserine lactones in lung tissues of mice infected with Pseudomonas aeruginosa. Microbiology 1462481-2493. [DOI] [PubMed] [Google Scholar]